Colorectal Cancer Prognosis Is Not Associated with BRAF and KRAS Mutations-A STROBE Compliant Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Selection
2.2. Definition of BRAF and KRAS Mutation Status
2.3. Kaplan–Meier Survival Analysis of BRAF and KRAS Mutations
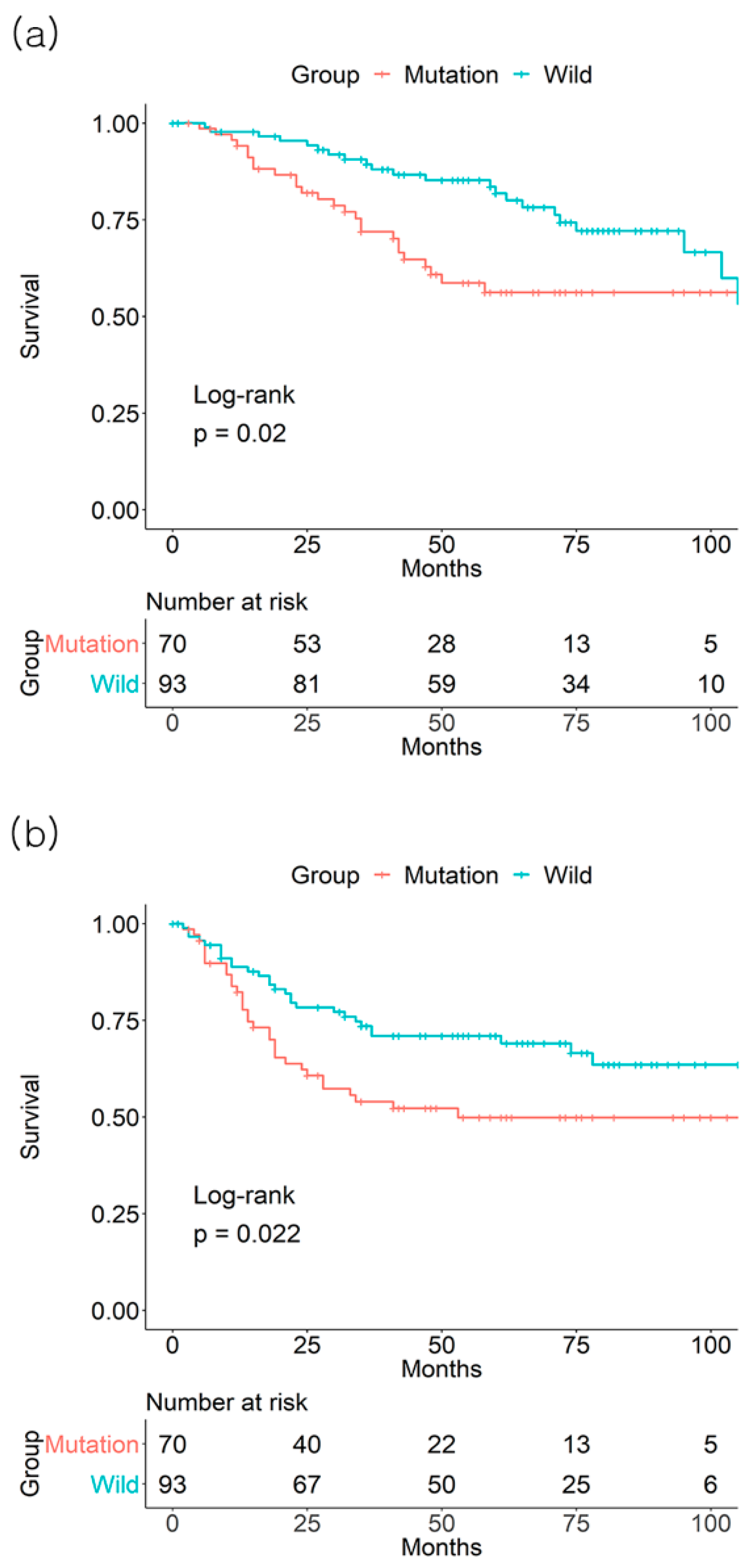
3. Results
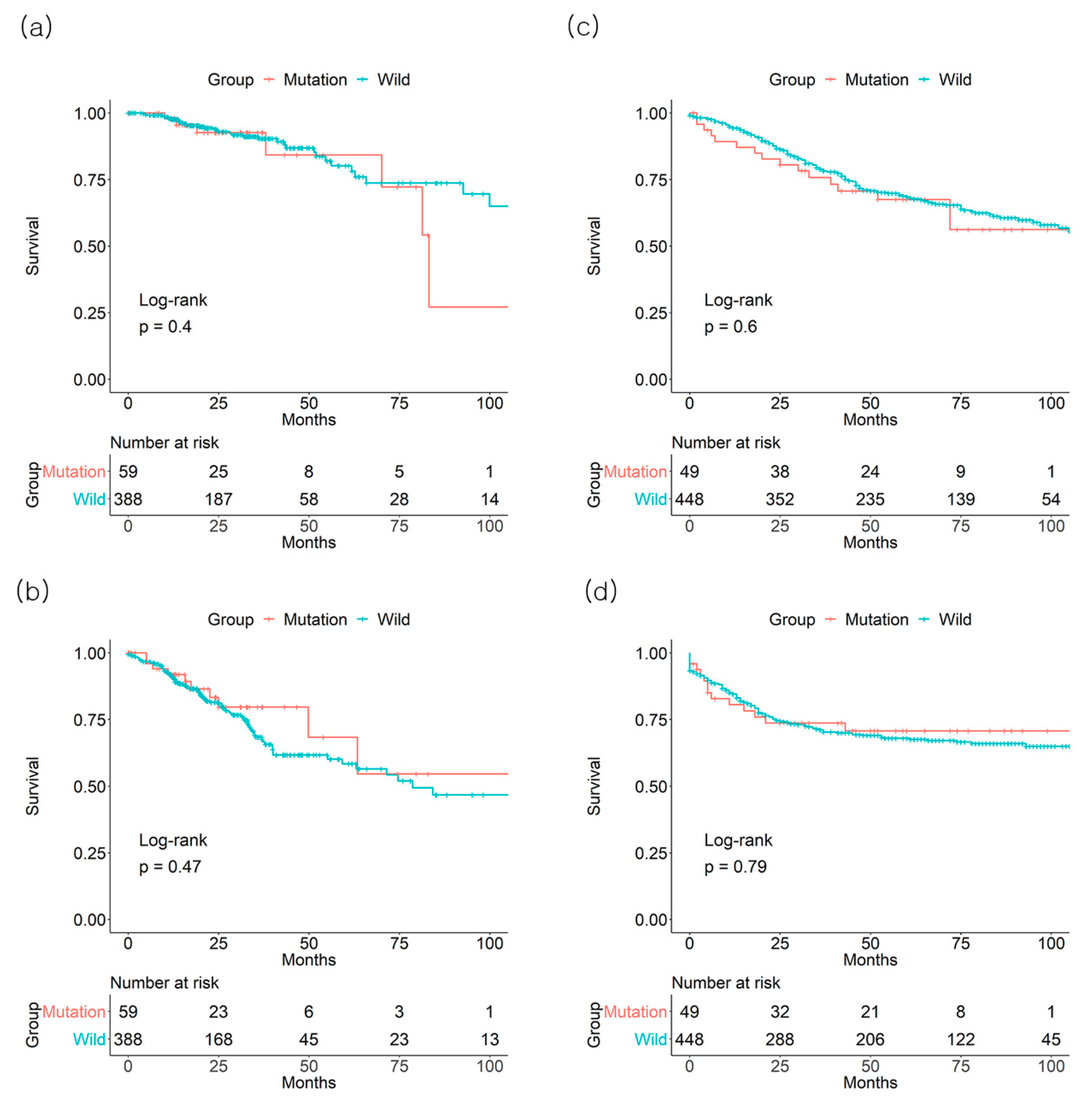
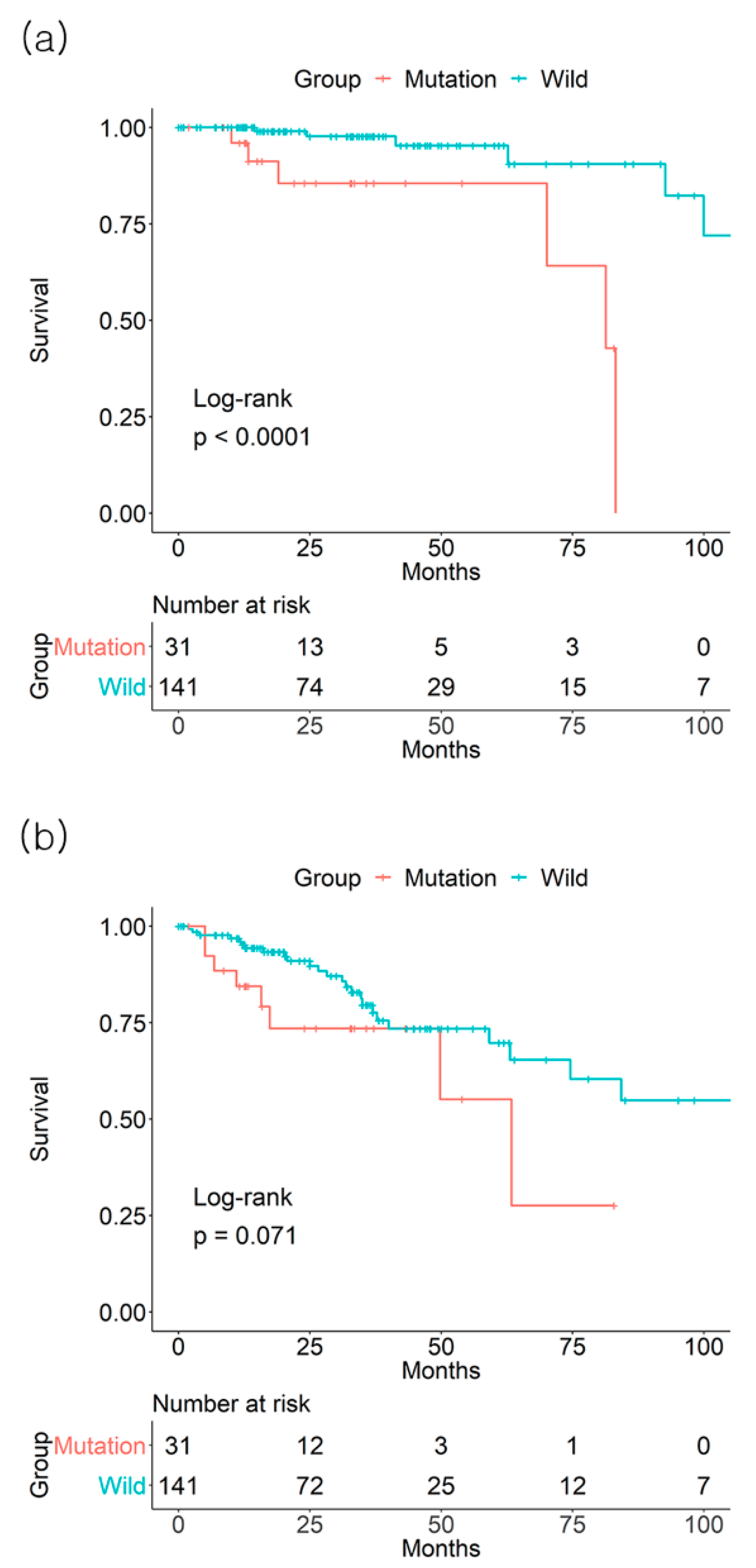
3.1. Individual Effect of BRAF Mutation on Survival Outcome
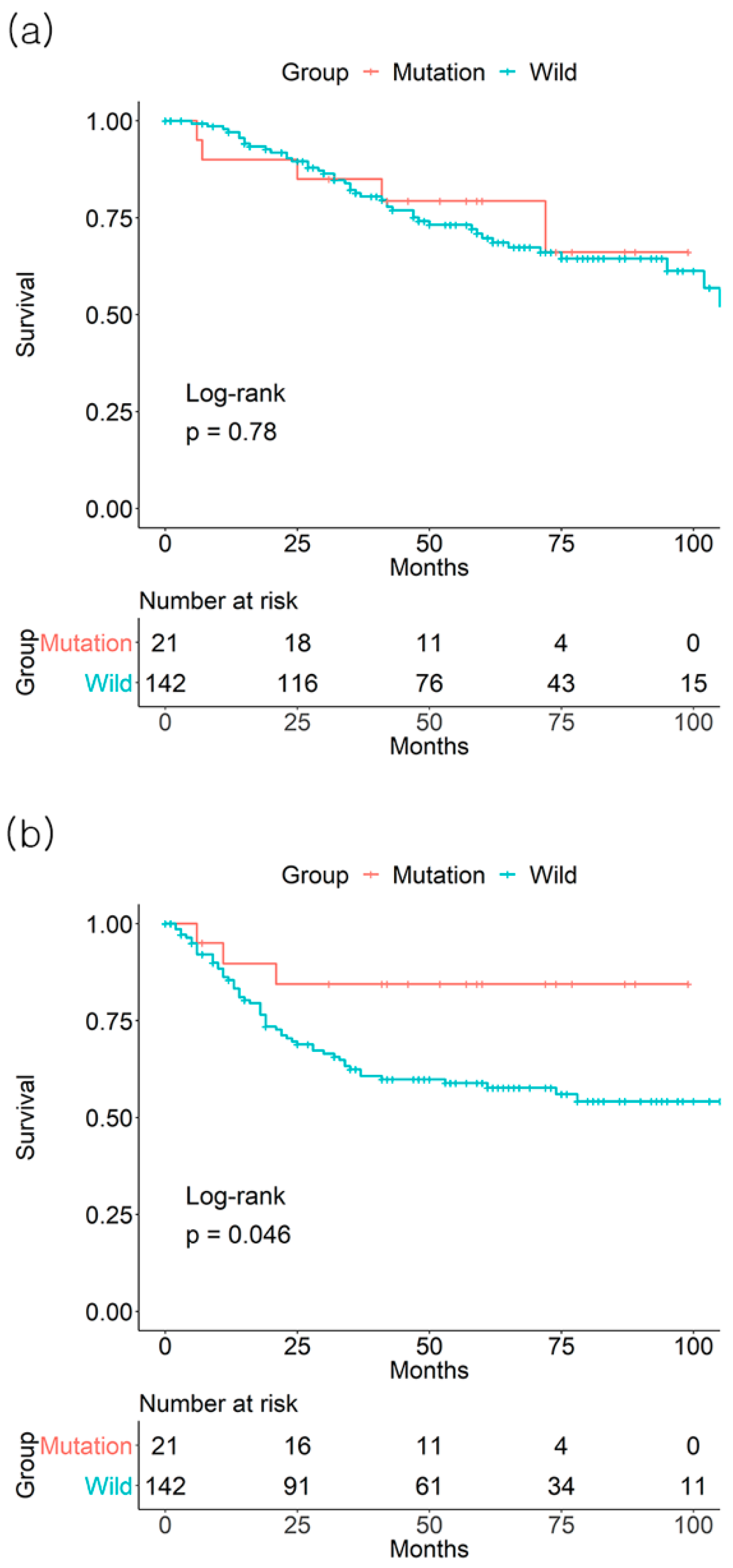
3.2. Individual Effect of KRAS Mutation on Survival Outcome
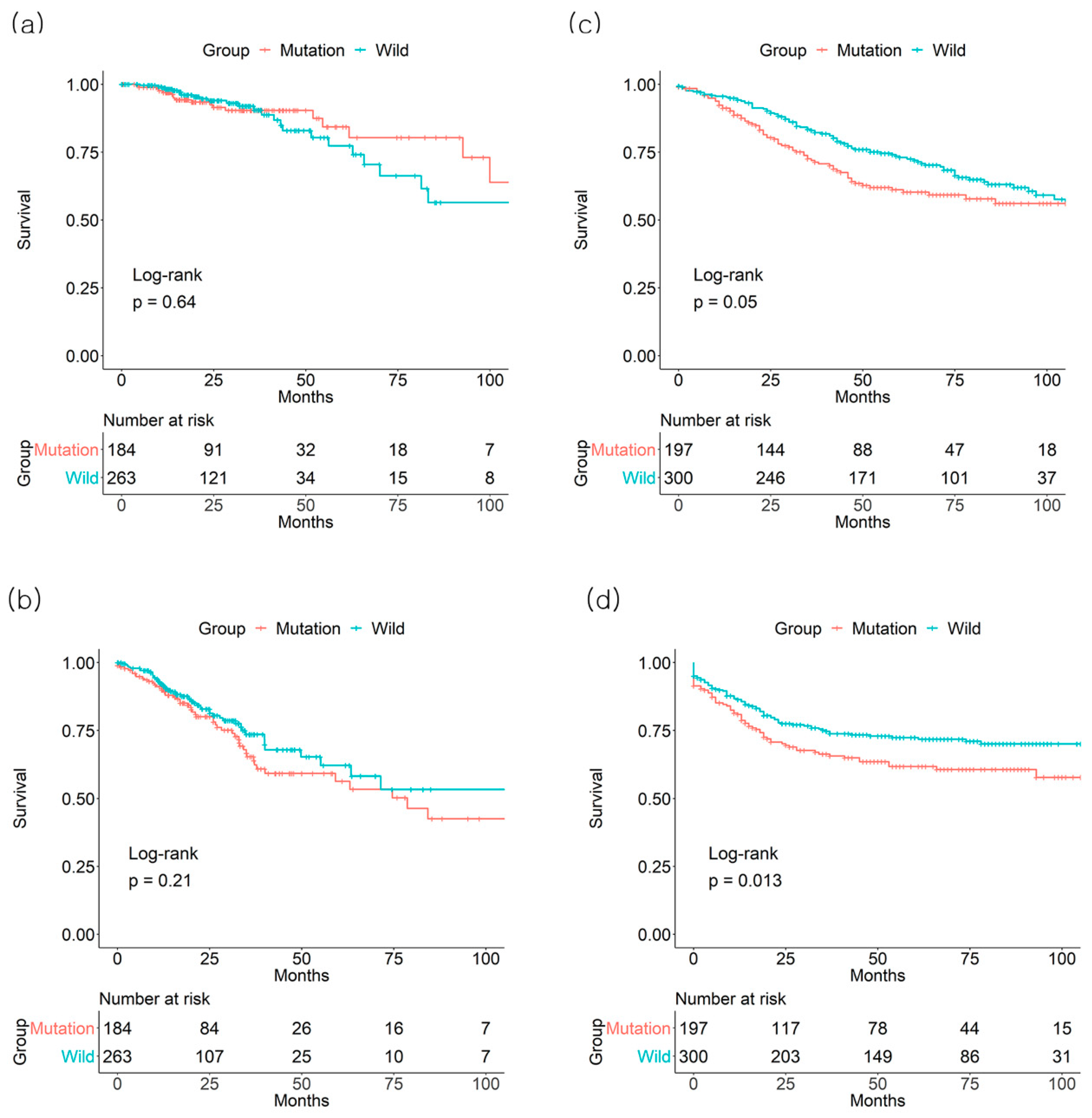
3.3. Combined Effect of BRAF and KRAS on Survival Outcome
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017; pp. XVII, 1032. [Google Scholar]
- Huang, Z.; Duan, H.; Li, H. Identification of Gene Expression Pattern Related to Breast Cancer Survival Using Integrated TCGA Datasets and Genomic Tools. Biomed. Res. Int. 2015, 2015, 878546. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Devarakonda, S.; Morgensztern, D.; Govindan, R. Clinical applications of The Cancer Genome Atlas project (TCGA) for squamous cell lung carcinoma. Oncology 2013, 27, 899–906. [Google Scholar] [PubMed]
- Lee, H.; Flaherty, P.; Ji, H.P. Systematic genomic identification of colorectal cancer genes delineating advanced from early clinical stage and metastasis. BMC Med. Genomics 2013, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef]
- Ogino, S.; Goel, A. Molecular classification and correlates in colorectal cancer. J. Mol. Diagn. 2008, 10, 13–27. [Google Scholar] [CrossRef]
- Passiglia, F.; Bronte, G.; Bazan, V.; Galvano, A.; Vincenzi, B.; Russo, A. Can KRAS and BRAF mutations limit the benefit of liver resection in metastatic colorectal cancer patients? A systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2016, 99, 150–157. [Google Scholar] [CrossRef]
- Sanz-Garcia, E.; Argiles, G.; Elez, E.; Tabernero, J. BRAF mutant colorectal cancer: Prognosis, treatment, and new perspectives. Ann. Oncol. 2017, 28, 2648–2657. [Google Scholar] [CrossRef]
- Karagkounis, G.; Torbenson, M.S.; Daniel, H.D.; Azad, N.S.; Diaz, L.A., Jr.; Donehower, R.C.; Hirose, K.; Ahuja, N.; Pawlik, T.M.; Choti, M.A. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 2013, 119, 4137–4144. [Google Scholar] [CrossRef] [PubMed]
- Schirripa, M.; Bergamo, F.; Cremolini, C.; Casagrande, M.; Lonardi, S.; Aprile, G.; Yang, D.; Marmorino, F.; Pasquini, G.; Sensi, E.; et al. BRAF and RAS mutations as prognostic factors in metastatic colorectal cancer patients undergoing liver resection. Br. J. Cancer 2015, 112, 1921–1928. [Google Scholar] [CrossRef] [PubMed]
- Alwers, E.; Jia, M.; Kloor, M.; Blaker, H.; Brenner, H.; Hoffmeister, M. Associations Between Molecular Classifications of Colorectal Cancer and Patient Survival: A Systematic Review. Clin. Gastroenterol. Hepatol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Marisa, L.; de Reynies, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef] [PubMed]
- Von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gotzsche, P.C.; Vandenbroucke, J.P.; Initiative, S. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. J. Clin. Epidemiol. 2008, 61, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Therneau, T.M. A Package for Survival Analysis in S. Available online: https://CRAN.R-project.org/package=survival (accessed on 26 November 2018).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 25 October 2018).
- R Core Team. R: A Language and Environment for Statistical Computing; 3.4.1; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Dienstmann, R.; Mason, M.J.; Sinicrope, F.A.; Phipps, A.I.; Tejpar, S.; Nesbakken, A.; Danielsen, S.A.; Sveen, A.; Buchanan, D.D.; Clendenning, M.; et al. Prediction of overall survival in stage II and III colon cancer beyond TNM system: A retrospective, pooled biomarker study. Ann. Oncol. 2017, 28, 1023–1031. [Google Scholar] [CrossRef]
- Jass, J.R. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007, 50, 113–130. [Google Scholar] [CrossRef]
- De Sousa, E.M.F.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; de Rooij, L.P.; de Jong, J.H.; de Boer, O.J.; van Leersum, R.; Bijlsma, M.F.; et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef]
- Sadanandam, A.; Lyssiotis, C.A.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.; Lannon, W.A.; Grotzinger, C.; Del Rio, M.; et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef]
- Won, D.D.; Lee, J.I.; Lee, I.K.; Oh, S.T.; Jung, E.S.; Lee, S.H. The prognostic significance of KRAS and BRAF mutation status in Korean colorectal cancer patients. BMC Cancer 2017, 17, 403. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Petrelli, F.; Coinu, A.; Di Bartolomeo, M.; Borgonovo, K.; Maggi, C.; Cabiddu, M.; Iacovelli, R.; Bossi, I.; Lonati, V.; et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. Eur. J. Cancer 2015, 51, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.; Dias, M.M.; Wiese, M.D.; Kichenadasse, G.; McKinnon, R.A.; Karapetis, C.S.; Sorich, M.J. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br. J. Cancer 2015, 112, 1888–1894. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Huang, J.F.; Liu, K.; Zhang, L.Q.; Yang, Z.; Chuai, Z.R.; Wang, Y.X.; Shi, D.C.; Huang, Q.; Fu, W.L. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: A systematic review and meta-analysis. PLoS ONE 2014, 9, e90607. [Google Scholar] [CrossRef] [PubMed]
- Seligmann, J.F.; Fisher, D.; Smith, C.G.; Richman, S.D.; Elliott, F.; Brown, S.; Adams, R.; Maughan, T.; Quirke, P.; Cheadle, J.; et al. Investigating the poor outcomes of BRAF-mutant advanced colorectal cancer: Analysis from 2530 patients in randomised clinical trials. Ann. Oncol. 2017, 28, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Poulogiannis, G.; Luo, F.; Arends, M.J. RAS signalling in the colorectum in health and disease. Cell Commun. Adhes. 2012, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Arrington, A.K.; Heinrich, E.L.; Lee, W.; Duldulao, M.; Patel, S.; Sanchez, J.; Garcia-Aguilar, J.; Kim, J. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int. J. Mol. Sci. 2012, 13, 12153–12168. [Google Scholar] [CrossRef]
- Tran, N.H.; Cavalcante, L.L.; Lubner, S.J.; Mulkerin, D.L.; LoConte, N.K.; Clipson, L.; Matkowskyj, K.A.; Deming, D.A. Precision medicine in colorectal cancer: The molecular profile alters treatment strategies. Ther. Adv. Med. Oncol. 2015, 7, 252–262. [Google Scholar] [CrossRef]
- Shan, L.; Li, M.; Ma, J.; Zhang, H. PCR-based assays versus direct sequencing for evaluating the effect of KRAS status on anti-EGFR treatment response in colorectal cancer patients: A systematic review and meta-analysis. PLoS ONE 2014, 9, e107926. [Google Scholar] [CrossRef]
- Morkel, M.; Riemer, P.; Blaker, H.; Sers, C. Similar but different: Distinct roles for KRAS and BRAF oncogenes in colorectal cancer development and therapy resistance. Oncotarget 2015, 6, 20785–20800. [Google Scholar] [CrossRef]
- Edge, S.; Byrd, D.; Compton, C.; Fritz, A.; Greene, F.; Trotti, A. AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
| TCGA | GSE39582 | ||
|---|---|---|---|
| Total number of patients studied | 447 | 497 | |
| Age at diagnosis (mean ± SD), years old | 65.0 ± 12.9 | 66.8 ± 13.1 | |
| Gender | |||
| Male | 213 (47.7%) | 271 (54.5%) | |
| Female | 234 (52.3%) | 226 (45.5%) | |
| Number of BRAF mutations | 59 (13.2%) | 49 (9.9%) | |
| Number of KRAS mutations | 384 (85.9%) | 197 (39.6%) | |
| AJCC TNM stage (number of BRAF/KRAS mutations) | |||
| I | 83 (10/36) | 32 (1/15) | |
| II | 172 (31/77) | 242 (21/83) | |
| III | 134 (14/50) | 163 (21/70) | |
| IV | 58 (4/21) | 60 (6/29) | |
| Survival event | |||
| Dead | 46 (10.3%) | 40 (8.0%) | |
| Alive | 404 (90.4%) | 457 (92.0%) | |
| Median follow-up time, months (range) | 23.9 (0–147.9) | 52 (0–201) | |
| Median time to survival event, months (range) | 24.4 (4.0–99.9) | 31 (0–183) |
| Author, Year | Country | Time Frame | Patients | Stage | Outcomes | Poor Prognosis |
|---|---|---|---|---|---|---|
| BRAF | ||||||
| Won et al. 2017 | South Korea | 2010–2013 | 1049 | I, II, III | OS *, DFS ** | Mutant type |
| Seligmann et al., 2017 | United Kingdom | 2000–2003 | 231 | IV | PFS *** | Mutant type |
| Roth et al. 2010 | Switzerland | (−) | 1404 | II, III | OS *, DFS ** | Mutant type |
| KRAS | ||||||
| De Roock et al. 2010 | Belgium | 2001–2008 | 229 | IV | PFS *** | No difference |
| Bokemeyer et al. 2011 | Germany | 2006–2007 | 315 | IV | PFS *** | Mutant type |
| Schwartzberg et al. 2014 | Spain | 2009–2011 | 278 | IV | PFS ***, OS * | Mutant type |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-H.; Ahn, J.; Park, W.S.; Choe, E.K.; Kim, E.; Shin, R.; Heo, S.C.; Jung, S.; Kim, K.; Chai, Y.J.; et al. Colorectal Cancer Prognosis Is Not Associated with BRAF and KRAS Mutations-A STROBE Compliant Study. J. Clin. Med. 2019, 8, 111. https://doi.org/10.3390/jcm8010111
Lee J-H, Ahn J, Park WS, Choe EK, Kim E, Shin R, Heo SC, Jung S, Kim K, Chai YJ, et al. Colorectal Cancer Prognosis Is Not Associated with BRAF and KRAS Mutations-A STROBE Compliant Study. Journal of Clinical Medicine. 2019; 8(1):111. https://doi.org/10.3390/jcm8010111
Chicago/Turabian StyleLee, Joon-Hyop, Jiyoung Ahn, Won Seo Park, Eun Kyung Choe, Eunyoung Kim, Rumi Shin, Seung Chul Heo, Sohee Jung, Kwangsoo Kim, Young Jun Chai, and et al. 2019. "Colorectal Cancer Prognosis Is Not Associated with BRAF and KRAS Mutations-A STROBE Compliant Study" Journal of Clinical Medicine 8, no. 1: 111. https://doi.org/10.3390/jcm8010111
APA StyleLee, J.-H., Ahn, J., Park, W. S., Choe, E. K., Kim, E., Shin, R., Heo, S. C., Jung, S., Kim, K., Chai, Y. J., & Chae, H. (2019). Colorectal Cancer Prognosis Is Not Associated with BRAF and KRAS Mutations-A STROBE Compliant Study. Journal of Clinical Medicine, 8(1), 111. https://doi.org/10.3390/jcm8010111






