A Comprehensive Feature Analysis of the Fetal Heart Rate Signal for the Intelligent Assessment of Fetal State
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Description
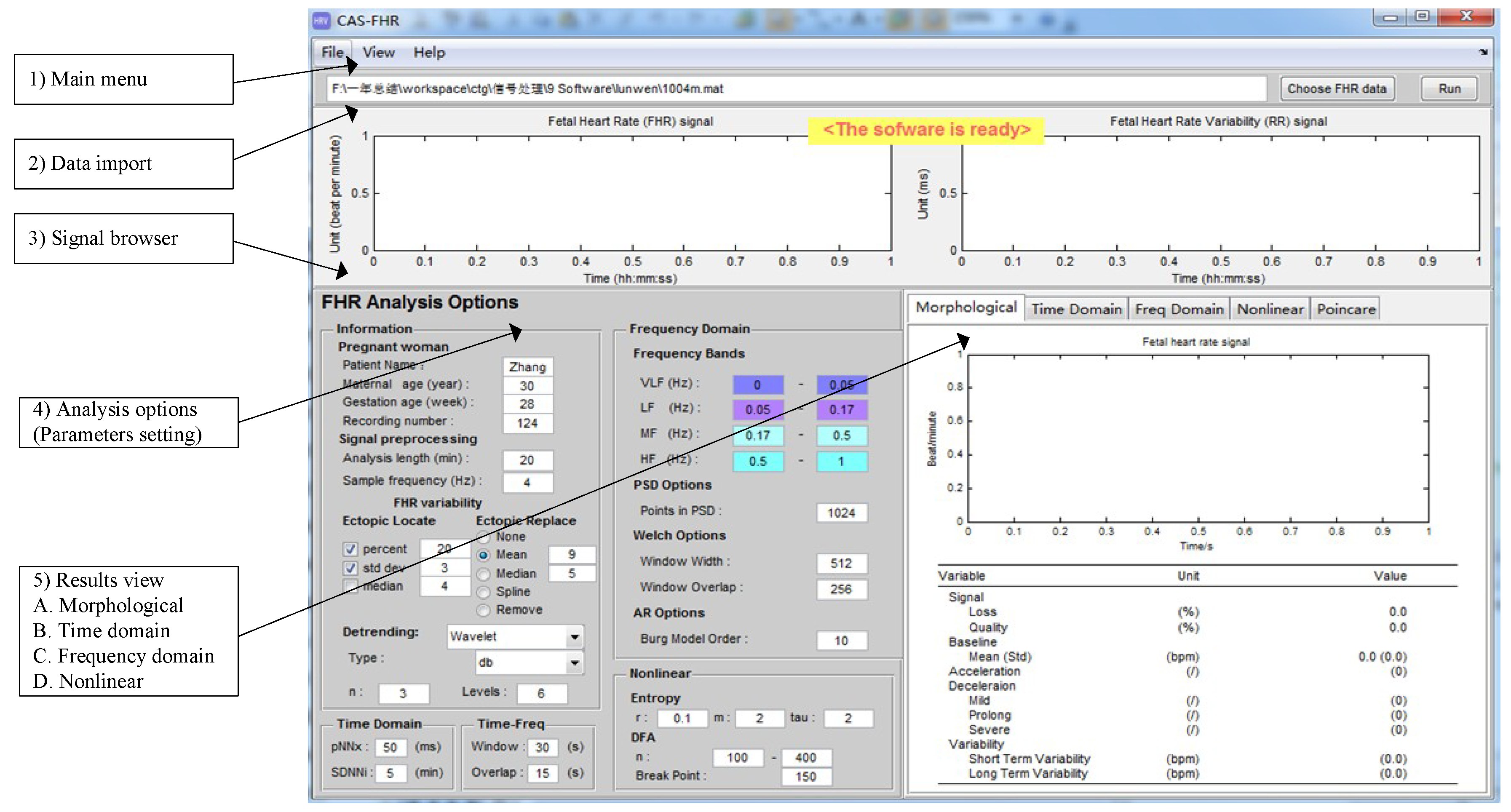
2.2. Software Interface
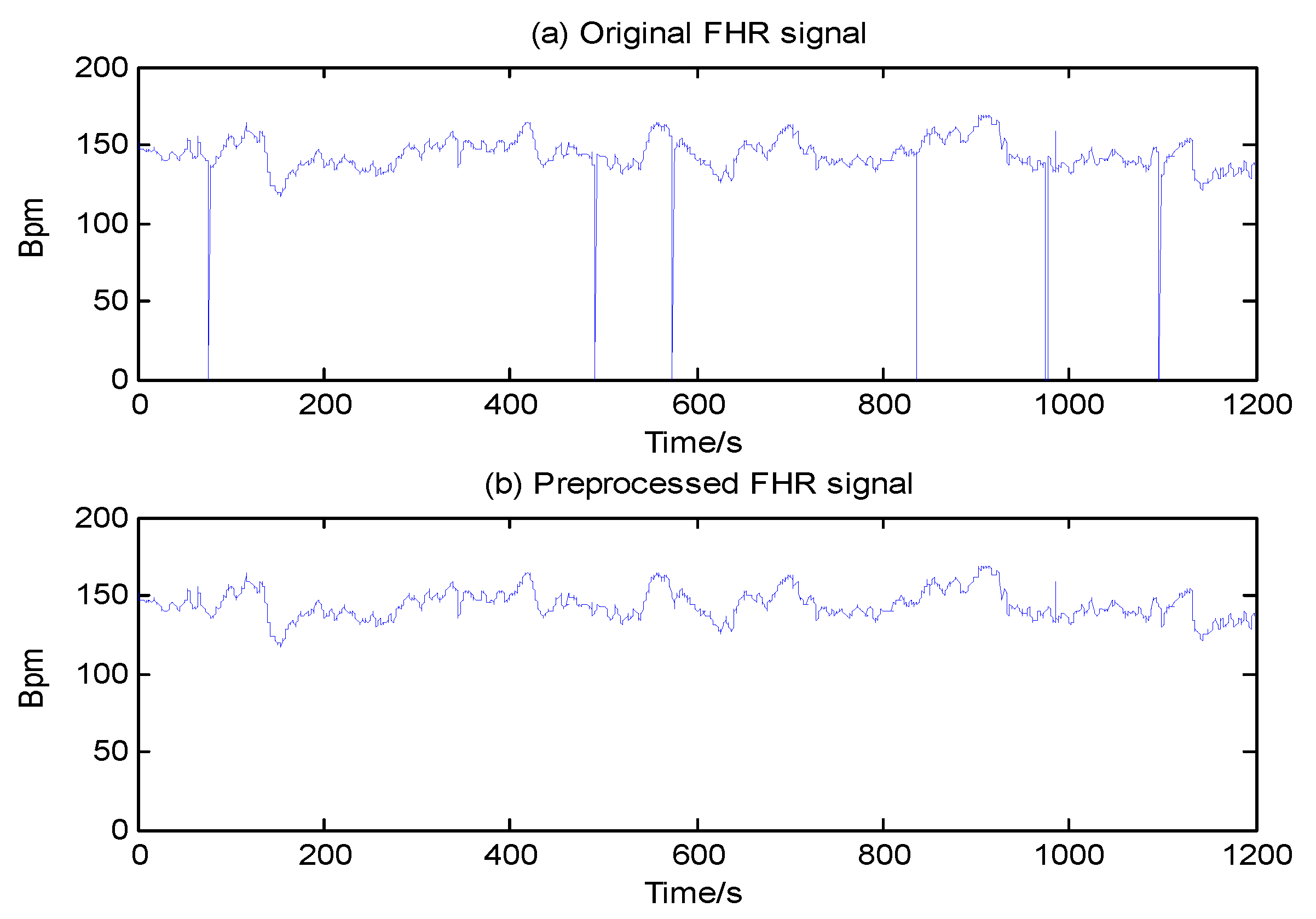
2.3. Signal Preprocessing
- A stable segment is chosen as the starting point; in such a segment, five adjacent samples do not differ by more than 10 bpm, and missing data are excluded when the length of x(i) = 0 is equal to or more than 10 s.
- Values of x(i) ≤ 50 or x(i) ≥ 200 are considered data spikes and are removed using linear interpolation.
- We interpolate x(i) using spline interpolation again when the difference of x(i) and x(i − 1) exceeds 25 bpm, a value used to define an unstable segment.
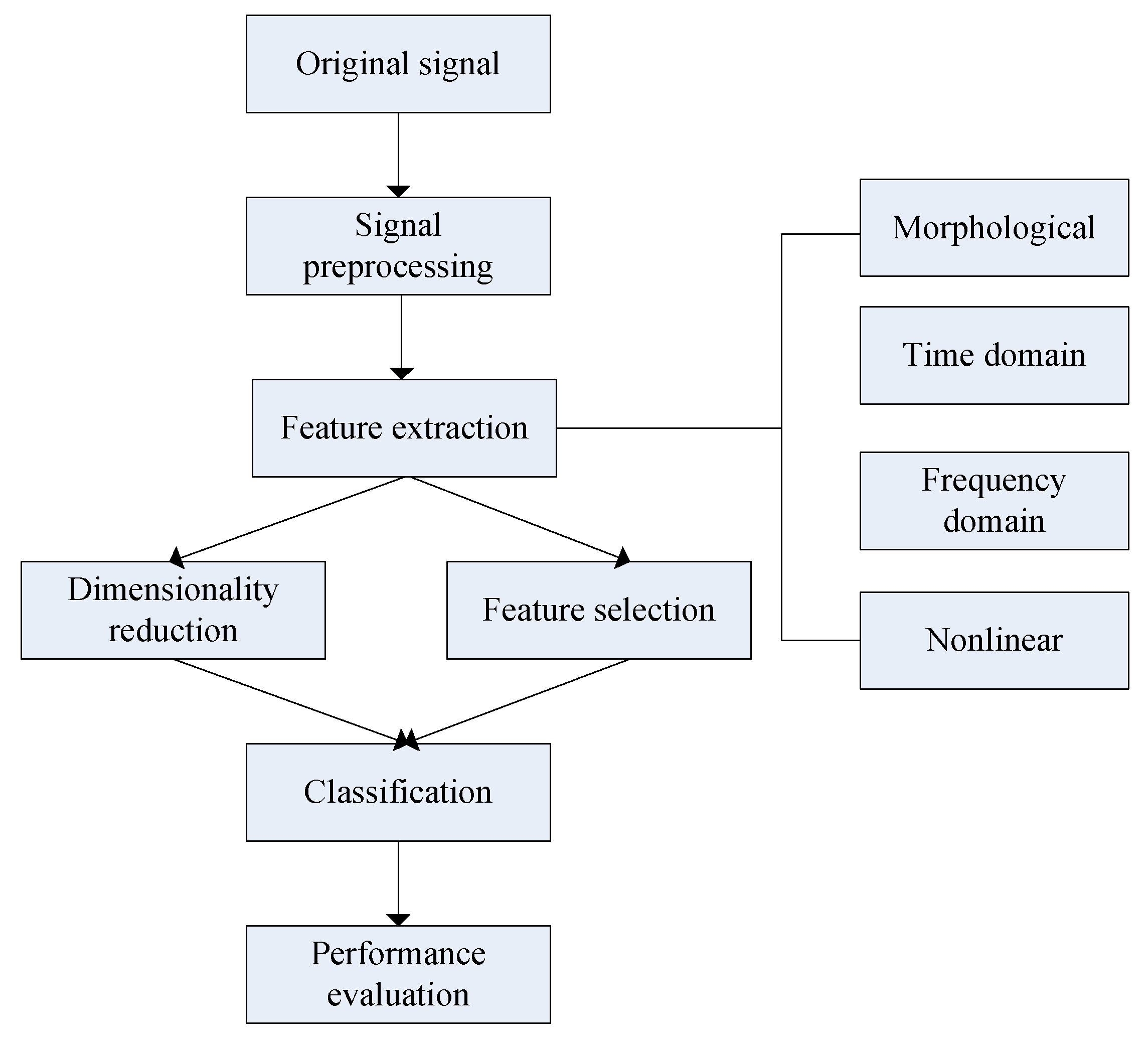
2.4. Feature Extraction
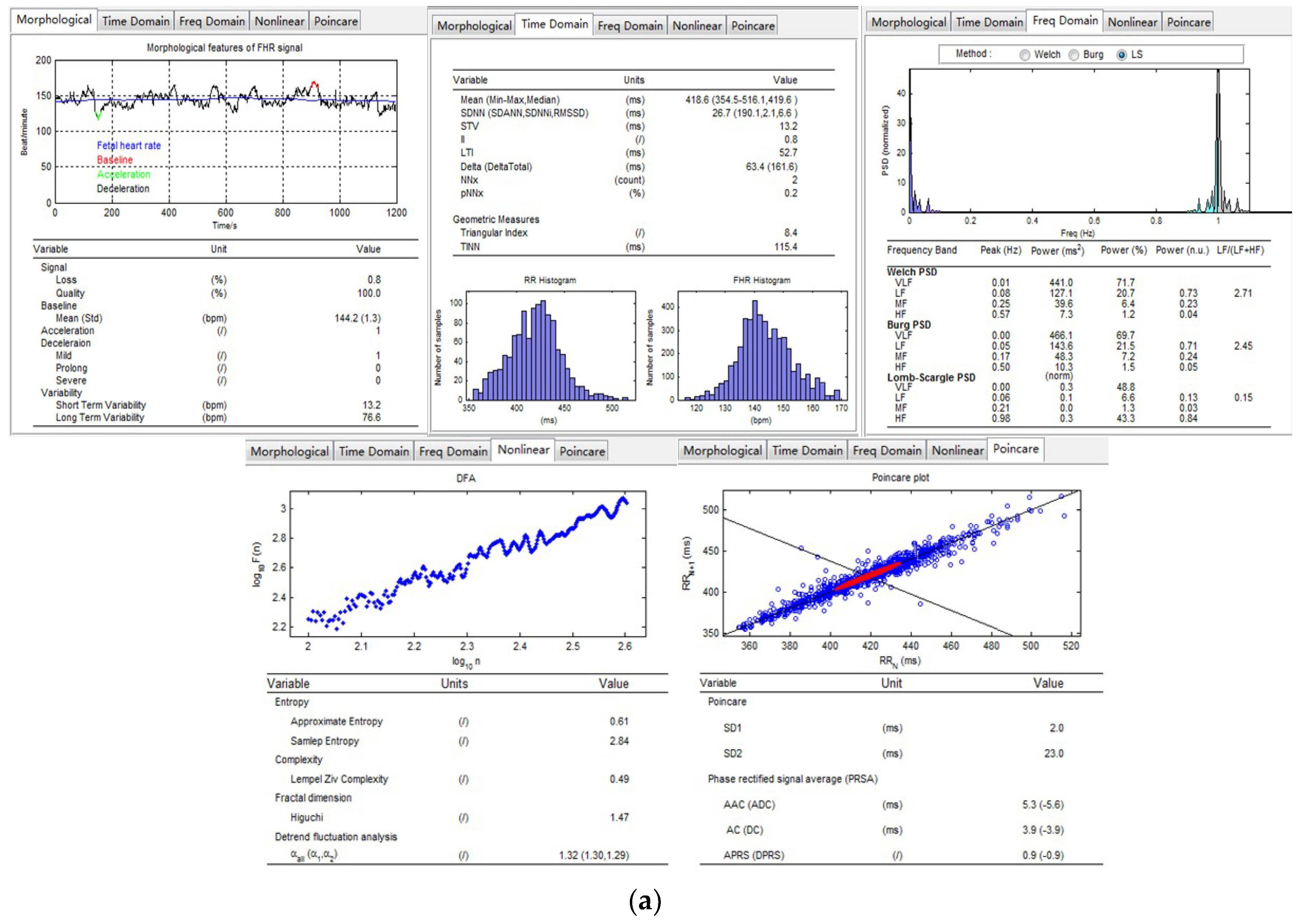
2.4.1. Morphological
- Set_1: {meanBL, sdBL, minBL, maxBL, ACC, DEC_mild, DEC_prolong, DEC_severe}.
2.4.2. Time Domain
- Set_2: {meanRR, minRR, maxRR, medianRR, SDNN, SDANN, SDNNi, RMSSD, NNx, pNNx, STV, II, LTI, delta, delta_total, Tri, TINN}.
2.4.3. Frequency Domain
- Set_3: {Power_VLF, Power_LF, Power_MF, Power_HF, Power_Total, Percent_VLF, Percent_LF, Percent_MF, Percent_HF, Ratio_Band}.
2.4.4. Nonlinear
- Set_4: {FD_Hig, ApEn, SampEn, LZC, Hurst, alpha, AAC, ADC, APRS, DPRS, SD1, SD2}.
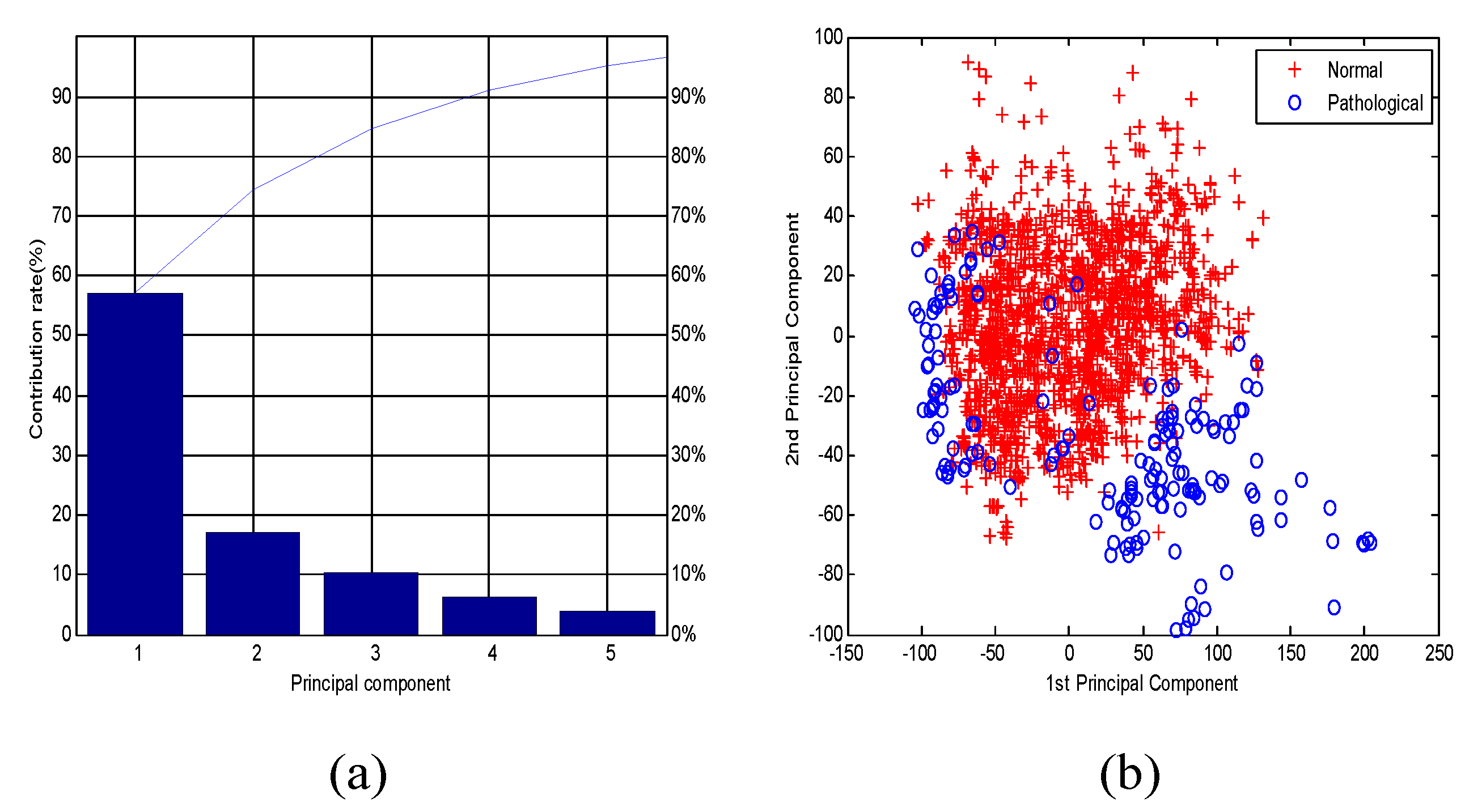
2.5. Feature Selection/Dimensionality Reduction and Classification
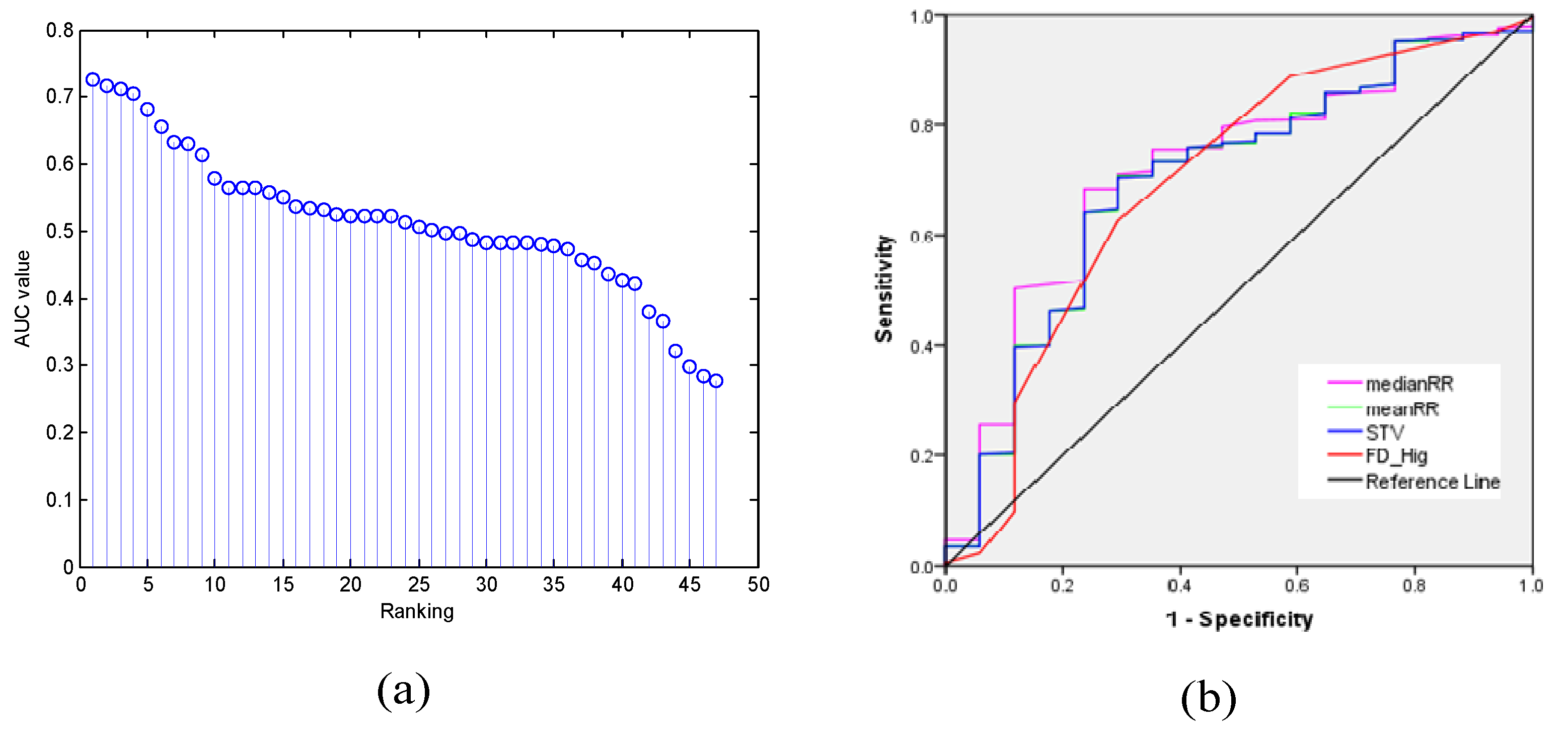
2.5.1. Feature Selection/Dimensionality Reduction
2.5.2. Classification and Performance Evaluation
- F-measure (FM, Harmonic mean):
- Balanced error rate (BER):
- Quality index (QI, Geometric mean):
- Matthews correlation coefficient (MCC):
3. Results
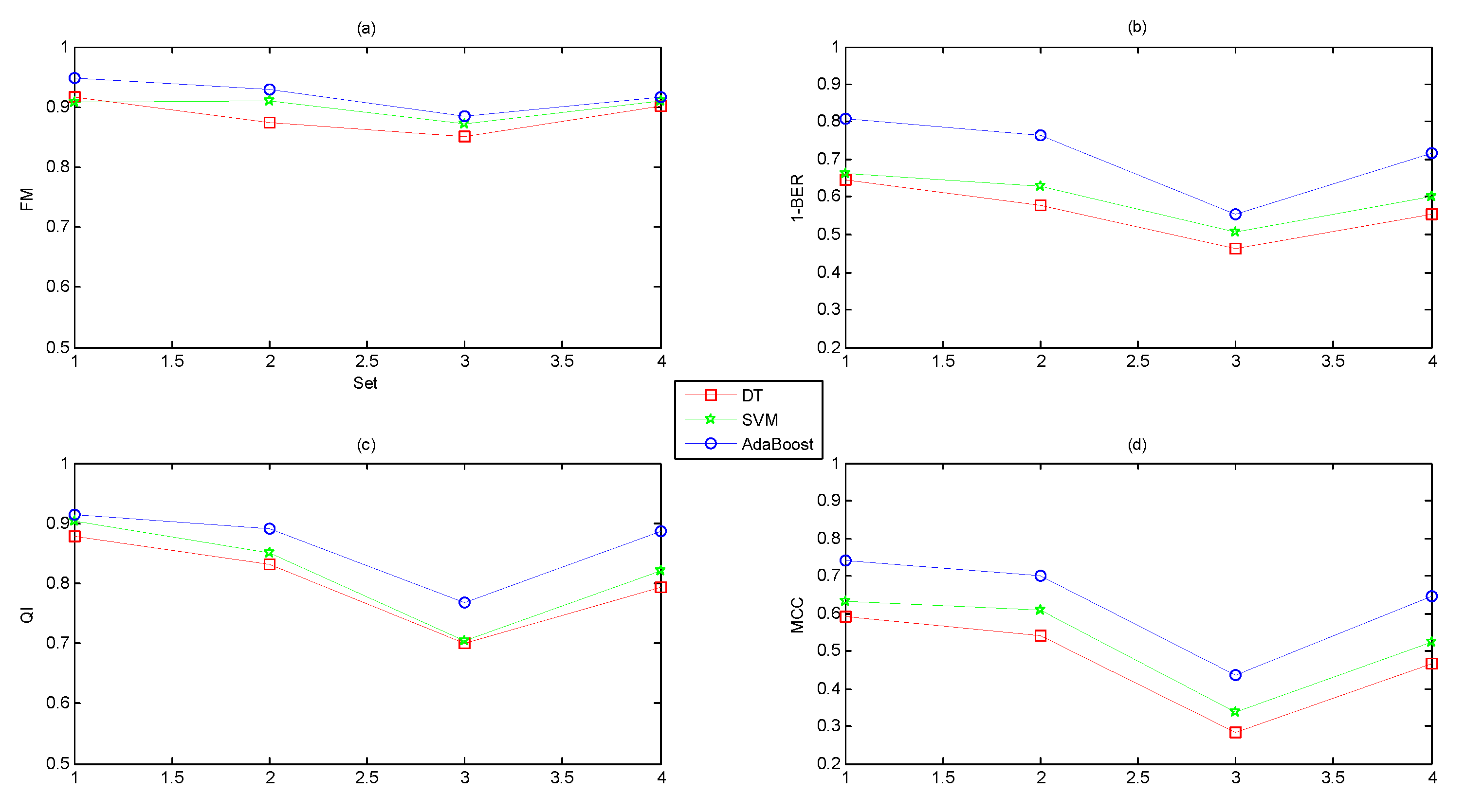
- Different feature domains contained different amounts of information regarding the fetal state.
- A combination of several feature domains could improve the performance, and the original feature set (Set_Complete) achieved the best performance.
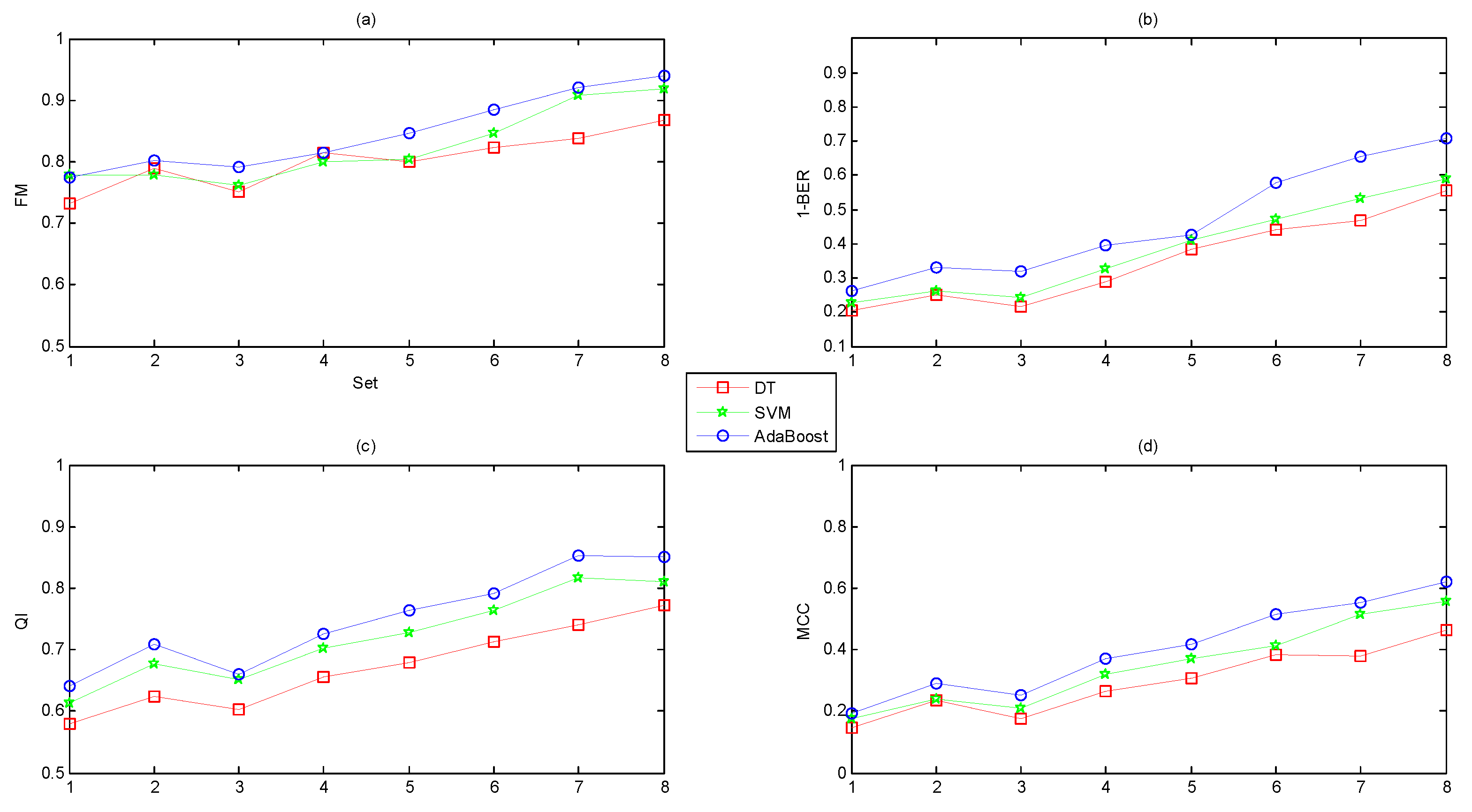
- The classification capacity of AdaBoost was better than DT and SVM.
- The feature selection algorithms of ST and AUC improved the performance while the dimensionality reduction method of PCA reduced the performance.
- The classification abilities of the three classifiers ranked in the following order: AdaBoost > SVM > DT.
4. Discussion
- Each feature domain reflected physiological information to different degrees: Nonlinear > time domain > frequency domain > morphological.
- The more features we used, the better the performance. A combination of 47 features yielded better performance than other combinations of individual feature subsets.
- The suitable feature selection algorithms (ST and AUC) improved the performance, but the dimensionality reduction approach (PCA) reduced the performance.
- The classification capacity of the ensemble learning algorithm (AdaBoost) was more powerful than common base classifiers (DT and SVM).
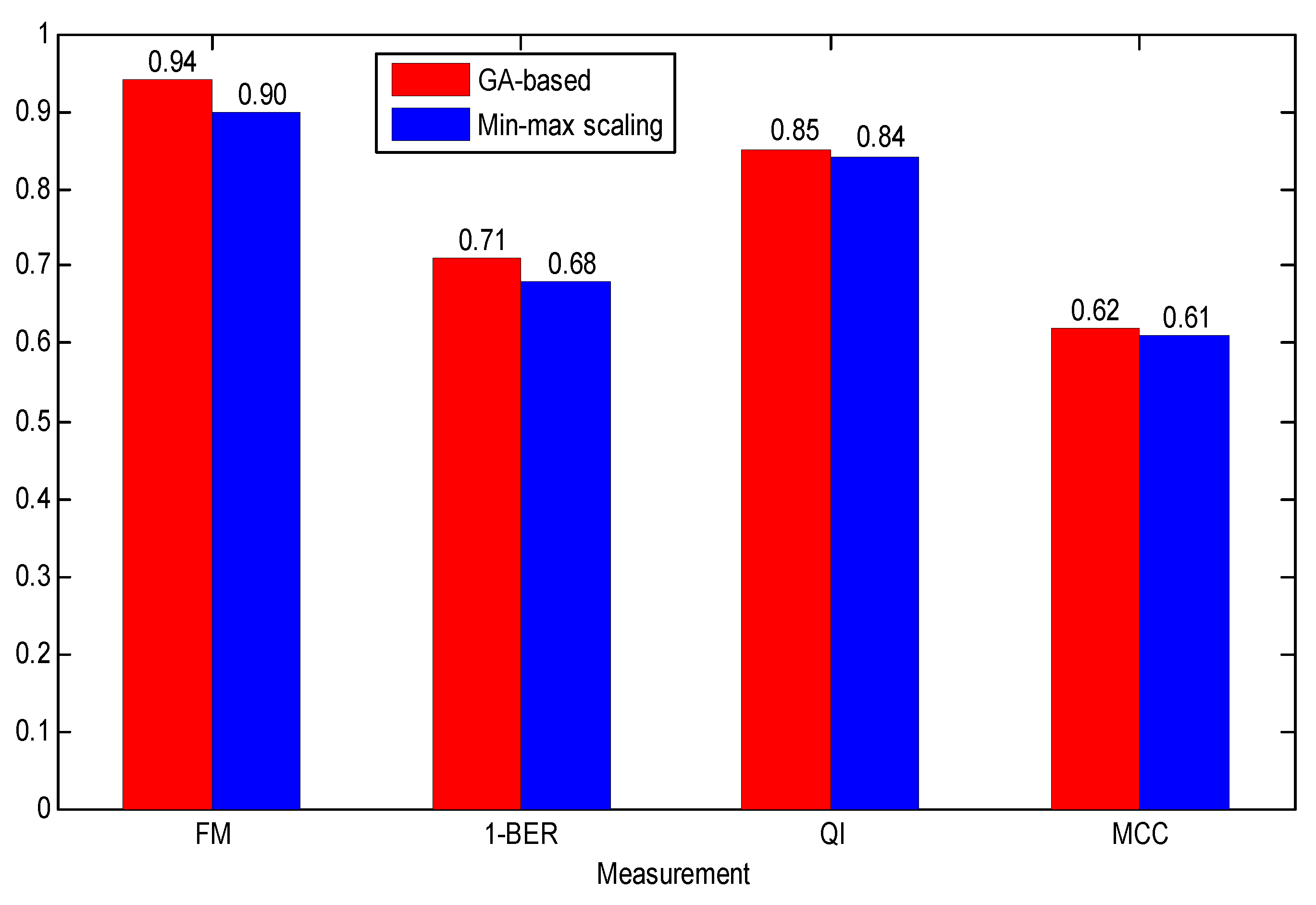
- Due to the relationship between the FHR parameters and GA, the GA-based data normalization method achieved better performance than common min-max scaling method.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nageotte, M.P. Fetal heart rate monitoring. Semin. Fetal Neonatal Med. 2015, 20, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Steer, P.J. Has electronic fetal heart rate monitoring made a difference. Semin. Fetal Neonatal Med. 2008, 13, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.C.; Pereira, A.C.; Bernardes, J. Agreement studies in obstetrics and gynaecology: Inappropriateness, controversies and consequences. BJOG Int. J. Obstet. Gynaecol. 2005, 112, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Ayres-de-Campos, D.; Spong, C.Y.; Chandraharan, E. FIGO consensus guidelines on intrapartum fetal monitoring: Cardiotocography. In J. Gynecol. Obstet. 2015, 131, 13–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parer, J.T.; Ikeda, T.; King, T.L. The 2008 national institute of child health and human development report on fetal heart rate monitoring. Obstet. Gynecol. 2008, 114, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Hruban, L.; Spilka, J.; Ek, V.C.; Jank, P.; Huptych, M. Agreement on intrapartum cardiotocogram recordings between expert obstetricians. J. Eval. Clin. Pract. 2015, 21, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Krupa, N.; Ali, M.; Zahedi, E.; Ahmed, S.; Hassan, F.M. Antepartum fetal heart rate feature extraction and classification using empirical mode decomposition and support vector machine. Biomed. Eng. Online 2011, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Spilka, J.; Chudacek, V.; Koucky, M.; Lhotska, L.; Huptych, M.; Janku, P.; Georgoulas, G.; Stylios, C. Using nonlinear features for fetal heart rate classification. Biomed. Signal Process. 2012, 7, 350–357. [Google Scholar] [CrossRef]
- Doret, M.; Spilka, J.; Chudáček, V.; Gonçalves, P.; Abry, P. Fractal analysis and hurst parameter for intrapartum fetal heart rate variability analysis: A versatile alternative to frequency bands and LF/HF ratio. PLoS ONE 2015, 10, e0136661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comert, Z.; Kocamaz, A.F.; Subha, V. Prognostic model based on image-based time-frequency features and genetic algorithm for fetal hypoxia assessment. Comput. Biol. Med. 2018, 99, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Low, J.A.; Mcgrath, M.J.; Marshall, S.J.; Fischer-Fay, A.; Karchmar, E.J. The relationship between antepartum fetal heart rate, intrapartum fetal heart rate, and fetal acid-base status. Am. J. Obstet. Gynecol. 1986, 154, 769–775. [Google Scholar] [CrossRef]
- Chudáček, V.; Spilka, J.; Bursa, M.; Janku, P.; Hruban, L.; Huptych, L.; Lhotska, L. Open access intrapartum CTG database. BMC Pregnancy Childbirth 2014, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Sampson, M.B.; Mudaliar, N.A.; Lele, A.S. Fetal heart rate variability. Postgrad. Med. 1980, 67, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Jovic, A.; Bogunovic, N. Electrocardiogram analysis using a combination of statistical, geometric, and nonlinear heart rate variability features. Artif. Intell. Med. 2011, 51, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, E215–E220. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, R.L.; Huddleston, J.F.; Nelson, K.G. Apgar scores and umbilical arterial pH in preterm newborn infants. Am. J. Obstet. Gynecol. 1984, 149, 651–654. [Google Scholar] [CrossRef]
- Ross, M.G.; Gala, R. Use of umbilical artery base excess: Algorithm for the timing of hypoxic injury. Am. J. Obstet. Gynecol. 2002, 187, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chudácčk, V.; Huptych, M.; Koucký, M.; Spilka, J.; Bauer, L.; Lhotská, L. Fetal heart rate data pre-processing and annotation. In Proceedings of the International Conference on Information Technology and Applications in Biomedicine, Larnaca, Cyprus, 4–7 November 2009; pp. 1–4. [Google Scholar]
- Dawes, G.S.; Houghton, C.R.; Redman, C.W. Baseline in human fetal heart-rate records. BJOG Int. J. Obstet. Gynecol. 1982, 89, 270–275. [Google Scholar] [CrossRef]
- Andersson, S. Acceleration and Deceleration and Baseline Estimation. Master’s Thesis, Chalmers University of Technology, Gothenburg, Sweden, 2011. [Google Scholar]
- Ayres-de Campos, D.; Bernardes, J.; Garrido, A.; Marques-de-Sa, J.; Pereira-Leite, L. SisPorto 2.0: A program for automated analysis of cardiotocograms. J. Mater. Med. 2000, 9, 311–318. [Google Scholar]
- Dalton, K.J.; Dawes, G.S.; Patrick, J.E. The autonomic nervous system and fetal heart rate variability. Am. J. Obstet. Gynecol. 1983, 146, 456–462. [Google Scholar] [CrossRef]
- Goldberger, J.J.; Johnson, N.P.; Subacius, H.; Ng, J.; Greenland, P. Comparison of the physiologic and prognostic implications of the heart rate versus the RR interval. Heart. Rhythm. 2014, 11, 1925–1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czabanski, R.; Jzezwski, J.; Matonia, A.; Jezewski, M. Computerized analysis of fetal heart rate signals as the predictor of neonatal academia. Expert Syst. Appl. 2012, 39, 11846–11860. [Google Scholar] [CrossRef]
- Torres, B.C.; Lopez, C.L.; Orellans, J.N. Analysis of heart rate variability at rest and during aerobic exercise: A study in healthy people and cardiac patients. Br. J. Sports Med. 2008, 42, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Signorini, M.G.; Magenes, G.; Cerutti, S.; Arduini, D. Linear and nonlinear parameters for the analysis of fetal heart rate signal from cardiotocographic recordings. IEEE Trans. Biomed. Eng. 2003, 50, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Van Laar, J.O.; Porath, M.M.; Peters, C.H.; Oei, S.G. Spectral analysis of fetal heart rate variability for fetal surveillance: Review of the literature. AOGS 2008, 87, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Warmerdam, G.J.J.; Vullings, R.; Bergmans, J.W.M.; Oei, S.G. Reliability of spectral analysis of fetal heart rate variability. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 2817–2820. [Google Scholar]
- Goncalves, H.; Bernardes, J.; Rocha, A.P.; Ayres-De-Campos, D. Linear and nonlinear analysis of heart rate pattern associated with fetal behavioral states in the antepartum period. Early Hum. Dev. 2007, 83, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Germán-Salló, Z.; Germán-Salló, M. Non-linear Methods in HRV Analysis. Proced. Technol. 2016, 22, 645–651. [Google Scholar] [CrossRef]
- Theiler, J. Estimating fractal dimension. J. Opt. Soc. Am. A 1990, 7, 1055–1073. [Google Scholar] [CrossRef]
- Raghavendra, B.S.; Dutt, D.N. Multiscale fractal dimension technique for characterization and analysis of biomedical signals. In Proceedings of the 2011 Digital Signal Processing and Signal Processing Education Meeting (DSP/SPE), Sedona, AZ, USA, 4–7 January 2011; pp. 370–374. [Google Scholar]
- Higuchi, T. Approach to an irregular time series on the basis of the fractal theory. Phys. D Nonlinear Phenom. 1998, 31, 277–283. [Google Scholar] [CrossRef]
- Ferrario, M.; Signorini, M.G.; Magenes, G.; Cerutti, S. Comparison of Entropy-Based Regularity Estimators: Application to the Fetal Heart Rate Signal for the Identification of Fetal Distress. IEEE Trans. Biomed. Eng. 2006, 53, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Pincus, S. Approximate entropy (ApEn) as a complexity measure. Chaos 1995, 5, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Phys. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Lempel, A.; Ziv, J. On the Complexity of Finite Sequences. IEEE Trans. Inf. Theory 1976, 22, 75–81. [Google Scholar] [CrossRef]
- Ferrario, M.; Signorini, M.G.; Magenes, G. Complexity analysis of the fetal heart rate for the identification of pathology in fetuses. In Proceedings of the Computers in Cardiology, Lyon, France, 25–28 September 2005; pp. 989–992. [Google Scholar] [CrossRef]
- Kantelhardt, J.W.; Zschiegner, S.A.; Koscielny-Bunde, E.; Havlin, S.; Bunde, A.; Stanley, H.E. Multifractal detrended fluctuation analysis of nonstationary time series. Phys. A Stat. Med. Appl. 2002, 316, 87–114. [Google Scholar] [CrossRef] [Green Version]
- Bauer, A.; Kantelhardt, J.W.; Bunde, A.; Barthel, P.; Schneider, R.; Malik, M.; Schmidt, G. Phase-rectified signal averaging detects quasi-periodicities in non-stationary data. Phys. A Stat. Med. Appl. 2006, 364, 423–434. [Google Scholar] [CrossRef]
- Fanelli, A.; Magenes, G.; Campanile, M.; Signorini, M.G. Quantitative assessment of fetal well-being through CTG recordings: A new parameter based on Phase Rectified Signal Average. IEEE J. Biomed. Health 2013, 17, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, S.; Gaitán, M.J.; González, R.; Yánez, O. Correlation among Poincare plot indexes and time and frequency domain measures of heart rate variability. J. Med. Eng. Technol. 2001, 25, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, D.; Zebrowski, J.; Cysarz, D.; Goncalve, H.; Pytilks, A.; Amorim-Costa, C.; Bernardes, J.; Ayres-de-Campos, D.; Witte, O.W.; Ekkehard, S.; et al. Monitoring fetal maturation-objectives, techniques and indices of autonomic function. Physiol. Meas. 2017, 38, R61–R88. [Google Scholar] [CrossRef] [PubMed]
- Magenes, G.; Bellazzi, R.; Malovini, A.; Signorini, M.G. Comparison of data mining techniques applied to fetal heart rate parameters for the early identification of IUGR fetuses. In Proceedings of the International Conference of the Engineering in Medicine and Biology Society, Orlando, FL, USA, 16–20 August 2016; pp. 916–919. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An Introduction to Variable Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar] [CrossRef]
- Winter, D.; Joost, C.; Dodou, D. Five-Point Likert Items: t Test versus Mann-Whitney-Wilcoxon. Prac. Assess. Res. Eval. 2010, 15, 1–16. [Google Scholar]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal component analysis. J. Mark. Res. 2002, 87, 513. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutmann, P.; Witten, I.H. The weka data mining software: An update. ACM SIG Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Morgan, M.B., Leyba, C., Hammett, J., Eds.; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 1993; ISBN 1-55860-23. [Google Scholar]
- Ukil, A. Support Vector Machine. Comput. Sci. 2002, 1, 1–28. [Google Scholar] [CrossRef]
- Margineantu, D.D.; Dietterich, T.G. Pruning adaptive boosting. In Proceedings of the Fourteenth International Conference on Machine Learning, San Francisco, CA, USA, 8–12 July 1997; pp. 211–218. [Google Scholar]
- Mitchell, T.M. Machine Learning; McGraw-Hill Publishers Inc.: New York, NY, USA, 1998; ISBN 0070428077. [Google Scholar]
- Xu, L.; Redman, C.W.; Payne, S.J.; Georgieva, A. Feature selection using genetic algorithms for fetal heart rate analysis. Physiol. Meas. 2014, 35, 1357. [Google Scholar] [CrossRef] [PubMed]
- Spilka, J.; Georgoulas, G.; Karvelis, P.; Chudáček, V.; Stylios, C.D.; Lhotská, L. Discriminating normal from “abnormal” pregnancy cases using an automated fhr evaluation method. In Proceedings of the Artificial Intelligence: Methods and Applications, SETN 2014, Ioannina, Greece, 15–17 May 2014; LNAI 8445. pp. 521–531. [Google Scholar]
- Georgoulas, G.; Karvelis, P.; Spilka, J.; Chudáček, V.; Stylios, C.D.; Lhotská, L. Investigating pH based evaluation of fetal heart rate (FHR) recordings. Health Technol. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]

| Information | Mean | Min | Max |
|---|---|---|---|
| Maternal age (MA, year) | 29.6 | 18 | 46 |
| Gestational age (GA, week) | 40.0 | 37 | 43 |
| pH | 7.23 | 6.85 | 7.47 |
| BDecf (mmol/L) | 4.60 | −3.40 | 26.11 |
| pCO2 | 7.07 | 0.70 | 12.30 |
| BE | −6.38 | −26.80 | −0.20 |
| Apgar 1 min | 8.3 | 1 | 10 |
| Apgar 5 min | 9.1 | 4 | 10 |
| Gravidity | 1.4 | 1 | 11 |
| Parity | 0.4 | 0 | 7 |
| Diabetes | No = 515, Yes = 37 | ||
| Birth weight (BW, g) | 3401 | 1970 | 4750 |
| Infant sex | Male = 286, Female = 266 | ||
| Delivery type | Vaginal = 506, Cesarean section = 46 | ||
| Positive | Negative | Evaluation Focus | |
|---|---|---|---|
| Predicted as positive | TP | FP | / |
| Predicted as negative | FN | TN | / |
| Acc | The overall efficiency of a classifier | ||
| Se (Recall) | The efficiency of a classifier to categorize positively labeled data | ||
| Sp | The efficiency of a classifier to categorize negatively labeled data | ||
| Precision | The data with positive labels correctly classified by the classifier | ||
| Set | Parameter (Unit) | Normal (447) | Pathological (105) | p |
|---|---|---|---|---|
| Set_1 | meanBL (bpm) | 136.0 (14.4) | 142.0 (16.2) | 0.004 |
| sdBL (bpm) | 2.4 (1.3) | 3.2 (2.6) | 0.318 | |
| minBL (bpm) | 132.0 (15.2) | 135.9 (17.2) | 0.008 | |
| maxBL (bpm) | 140.7 (14.7) | 148.0 (16.6) | 0.002 | |
| ACC | 1.45 (2.24) | 2.24 (3.10) | 0.185 | |
| DEC_mild | 1.40 (1.84) | 2.0 (1.85) | 0.098 | |
| DEC_prolong | 0.07 (0.30) | 0.17 (0.47) | 0.657 | |
| DEC_severe | 0.03 (0.19) | 0.00 (0.00) | 1.000 | |
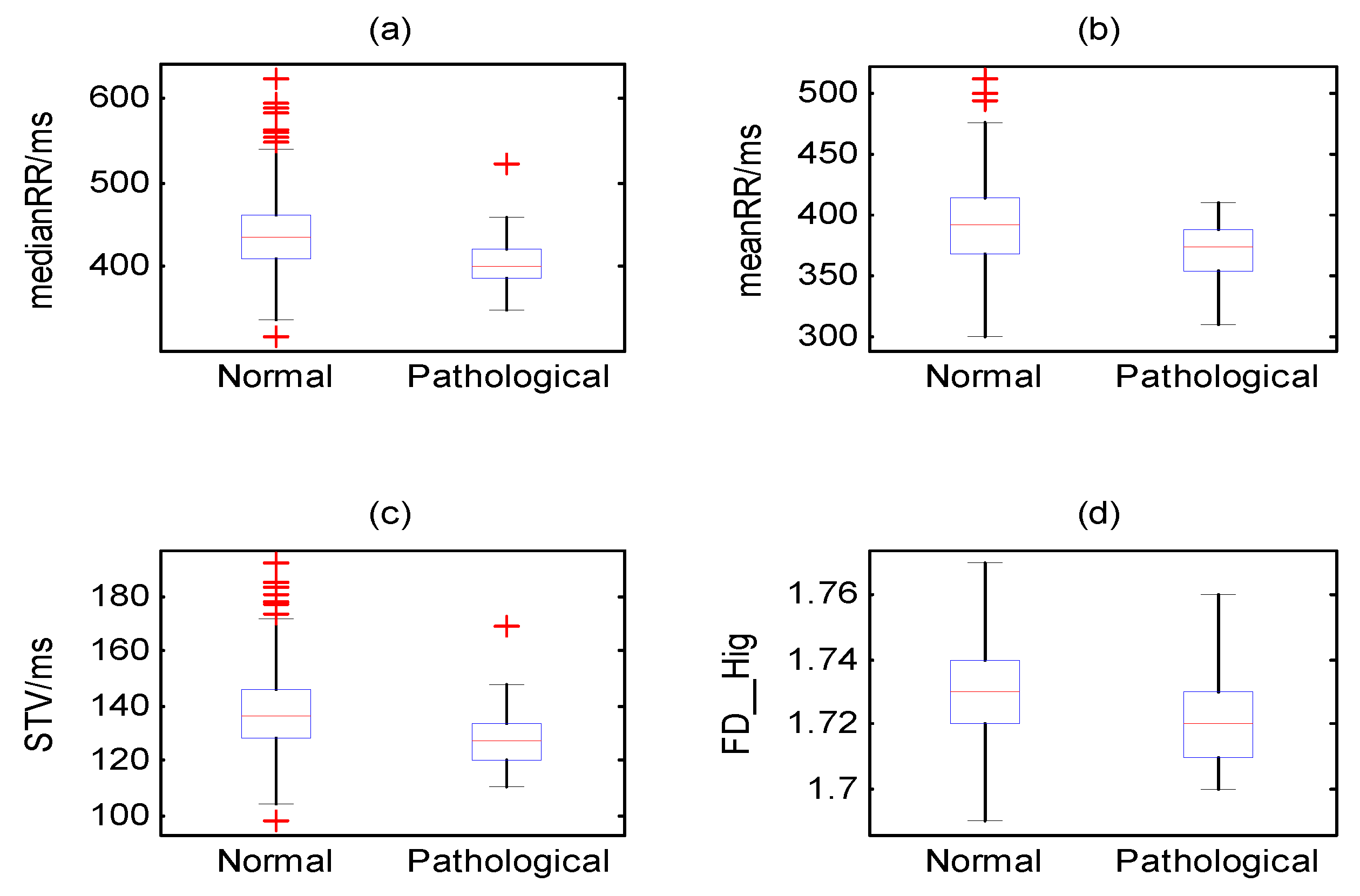
| Set_2 | meanRR (ms) | 447.4 (43.3) | 431.0 (52.2) | 0.005 |
| minRR (ms) | 370.4 (31.8) | 349.9 (27.2) | 0.019 | |
| maxRR (ms) | 640.5 (158.2) | 654.1 (165.8) | 0.341 | |
| medianRR (ms) | 439.3 (40.6) | 418.5 (50.2) | 0.002 | |
| SDNN (ms) | 42.6 (27.8) | 51.4 (32.7) | 0.896 | |
| SDANN (ms) | 29.7 (24.1) | 38.3 (27.4) | 0.983 | |
| SDNNi (ms) | 25.1 (14.5) | 28.9 (18.1) | 0.879 | |
| RMSSD (ms) | 10.7 (5.4) | 12.0 (7.3) | 0.912 | |
| NNx | 12.5 (16.0) | 17.5 (22.2) | 0.596 | |
| pNNx | 1.1 (1.3) | 1.5 (1.9) | 0.596 | |
| STV (ms) | 12.1 (8.7) | 14.1 (17.6) | 0.005 | |
| II | 0.9 (0.2) | 0.9 (0.2) | 0.079 | |
| LTI (ms) | 640.5 (158.2) | 654.1 (165.8) | 0.732 | |
| delta (ms) | 82.5 (42.3) | 92.9 (55.2) | 0.746 | |
| delta_total (ms) | 270.1 (154.6) | 304.2 (158.3) | 0.927 | |
| FHRVTi | 6.2 (2.6) | 6.6 (2.8) | 0.394 | |
| TINN | 81.7 (40.6) | 81.9 (37.7) | 0.978 |
| Set | Parameter (Unit) | Normal (447) | Pathological (105) | p |
|---|---|---|---|---|
| Set_3 | Power_VLF (ms2) | 1477 (2747) | 2708 (4734) | 0.757 |
| Power_LF (ms2) | 639 (998) | 927 (1464) | 0.359 | |
| Power_MF (ms2) | 180 (233) | 201 (300) | 0.527 | |
| Power_HF (ms2) | 105 (139) | 120 (173) | 0.536 | |
| Power_Total (ms2) | 2401 (3707) | 3956 (5989) | 0.498 | |
| Percent_VLF (%) | 86.3 (8.4) | 87.5 (8.3) | 0.513 | |
| Percent_LF (%) | 11.1 (7.0) | 10.1 (6.9) | 0.256 | |
| Percent_MF (%) | 1.8 (1.2) | 1.8 (1.2) | 0.935 | |
| Percent_HF (%) | 0.8 (0.7) | 0.7 (0.5) | 0.382 | |
| Ratio_Band | 4.6 (1.7) | 4.4 (1.5) | 0.340 | |
| Set_4 | FD_Hig | 1.54 (0.09) | 1.52 (0.11) | 0.005 |
| ApEn | 0.41 (0.00) | 0.41 (0.00) | 0.168 | |
| SampEn | 2.44 (0.38) | 2.37 (0.35) | 0.102 | |
| LZC | 1.13 (0.11) | 1.14 (0.12) | 0.046 | |
| Hurst | 0.93 (0.04) | 0.94 (0.03) | 0.099 | |
| alpha | 1.32 (0.12) | 1.32 (0.13) | 0.006 | |
| ACC (ms) | 8.95 (8.97) | 8.54 (8.74) | 0.603 | |
| ADC (ms) | −8.10 (8.88) | −8.35 (8.21) | 0.449 | |
| APRS | 2.98 (2.24) | 2.78 (2.56) | 0.774 | |
| DPRS | −2.77 (2.29) | −2.45 (2.73) | 0.856 | |
| SD1 (ms) | 8.45 (5.56) | 8.76 (5.21) | 0.579 | |
| SD2 (ms) | 54.57 (44.59) | 55.13 (46.32) | 0.548 |
| Reference (Year) | Database | Distribution (N/P) | Method | Performance (%) |
|---|---|---|---|---|
| [7] (2011) | Private | Imbalance (30/60) | FE: Empirical mode decomposition C: Support vector machine | Acc = 87 |
| [8] (2012) | Private | Imbalance (123/94) | FE: 12 (nonlinear) FS: Information gain C: Support vector machine, naïve Bayes, C4.5 | Se = 73 Sp = 76 FM = 71 |
| [54] (2014) | Private | Balance (255/255) | FE: 64 (morphological and linear) FS: genetic algorithm C: Support vector machine | Se = 83 Sp = 66 |
| [9] (2015) | Private | Imbalance (30/15) | FE: Ratio and Hurst C: Statistical test (p-value) | AUC = 81 |
| [55] (2014) | CTU-UHB | Imbalance (175/377) | FE: 33 (morpholical, linear and nonlinear) C: Latent class analysis+random forest | Se = 72 Sp = 78 |
| [56] (2017) | CTU-UHB | Imbalance (508/44) | FE: 42 (morpholical, linear and nonlinear) FE: Relevance in estimating features C: Least square support vector machine | Se = 72 Sp = 65 |
| [10] (2018) | CTU-UHB | Imbalance (439/113) | FE: Image-based time-frequency features FS: genetic algorithm C: Least square support vector machine | Se = 63 Sp = 66 |
| Current work | CTU-UHB | Imbalance (447/105) | FE: 47 (morpholical, linear and nonlinear) FS: Statistcal test, AUC C: Adaptive boosting | Acc = 92 Se = 92 Sp = 90 AUC = 91 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Zhang, Y.; Deng, Y. A Comprehensive Feature Analysis of the Fetal Heart Rate Signal for the Intelligent Assessment of Fetal State. J. Clin. Med. 2018, 7, 223. https://doi.org/10.3390/jcm7080223
Zhao Z, Zhang Y, Deng Y. A Comprehensive Feature Analysis of the Fetal Heart Rate Signal for the Intelligent Assessment of Fetal State. Journal of Clinical Medicine. 2018; 7(8):223. https://doi.org/10.3390/jcm7080223
Chicago/Turabian StyleZhao, Zhidong, Yang Zhang, and Yanjun Deng. 2018. "A Comprehensive Feature Analysis of the Fetal Heart Rate Signal for the Intelligent Assessment of Fetal State" Journal of Clinical Medicine 7, no. 8: 223. https://doi.org/10.3390/jcm7080223
APA StyleZhao, Z., Zhang, Y., & Deng, Y. (2018). A Comprehensive Feature Analysis of the Fetal Heart Rate Signal for the Intelligent Assessment of Fetal State. Journal of Clinical Medicine, 7(8), 223. https://doi.org/10.3390/jcm7080223






