Comparison of Clinical Manifestations, Antimicrobial Susceptibility Patterns, and Mutations of Fluoroquinolone Target Genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis Isolated in Taiwan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Setting and Design
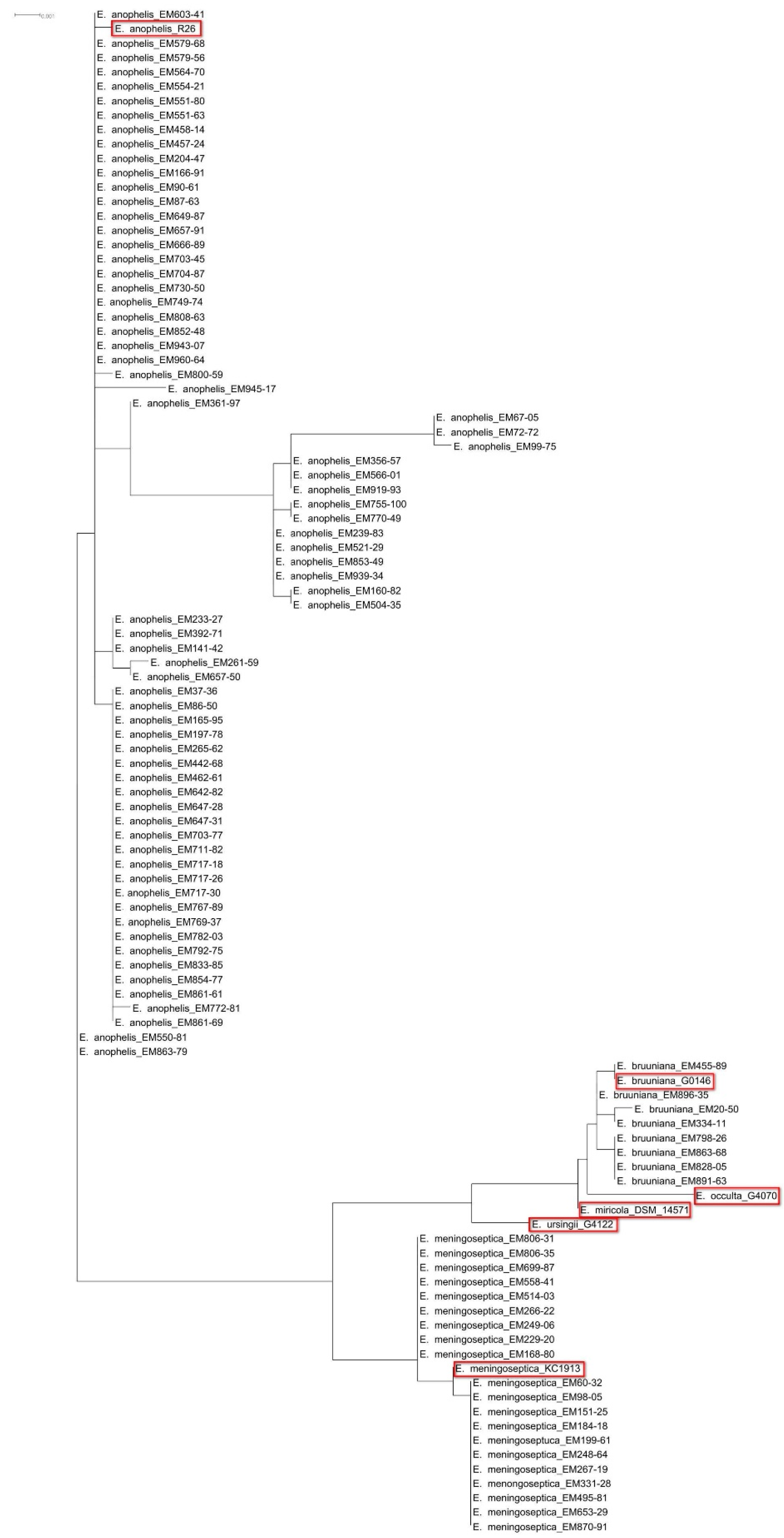
2.2. Species Identification Using 16S rRNA Gene Sequencing
2.3. Antimicrobial Susceptibility Testing
2.4. Identification of Mutations in the QRDRs
2.5. Data Analysis
3. Results
3.1. Species Identification
3.2. Site of Isolation
3.3. Clinical Characteristics of Elizabethkingia Infections
3.4. Factors Associated with Mortality
3.5. Antimicrobial Susceptibility Testing
3.6. Mutations in the QRDRs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Henriques, I.S.; Araújo, S.; Azevedo, J.S.; Alves, M.S.; Chouchani, C.; Pereira, A.; Correia, A. Prevalence and diversity of carbapenem-resistant bacteria in untreated drinking water in Portugal. Microb. Drug Resist. 2012, 18, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Lee, W.S.; Chen, F.L.; Ou, T.Y.; Hsueh, P.R. Elizabethkingia meningoseptica: An important emerging pathogen causing healthcare-associated infections. J. Hosp. Infect. 2014, 86, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.C.; Gulvik, C.A.; Whitney, A.M.; Humrighouse, B.W.; Graziano, J.; Emery, B.; Bell, M.; Loparev, V.; Juieng, P.; Gartin, J.; et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Anton. Van Leeuwenhoek 2017, 111, 55–72. [Google Scholar] [CrossRef] [PubMed]
- King, E.O. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am. J. Clin. Pathol. 1959, 31, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.; Tan, S.Y.-Y.; Tay, M.; Ding, Y.; Kjelleberg, S.; Givskov, M.; Lin, R.T.; Yang, L. First case of E anophelis outbreak in an intensive-care unit. Lancet 2013, 382, 855–856. [Google Scholar] [CrossRef]
- Lau, S.K.; Chow, W.N.; Foo, C.H.; Curreem, S.O.; Lo, G.C.; Teng, J.L.; Chen, J.H.; Ng, R.H.; Wu, A.K.; Cheung, I.Y.; et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 2016, 6, 26045. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.N.; Lai, C.H.; Yang, C.H.; Huang, Y.H.; Lin, H.H. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J. Antimicrob. Chemother. 2018, 73, 2497–2502. [Google Scholar] [CrossRef]
- Perrin, A.; Larsonneur, E.; Nicholson, A.C.; Edwards, D.J.; Gundlach, K.M.; Whitney, A.M.; Gulvik, C.A.; Bell, M.E.; Rendueles, O.; Cury, J.; et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 2017, 8, 15483. [Google Scholar] [CrossRef]
- Navon, L.; Clegg, W.J.; Morgan, J.; Austin, C.; McQuiston, J.R.; Blaney, D.D.; Walters, M.S.; Moulton-Meissner, H.; Nicholson, A. Notes from the field: Investigation of Elizabethkingia anophelis cluster—Illinois, 2014–2016. MMWR Morb. Mortal Wkly Rep. 2016, 65, 1380–1381. [Google Scholar] [CrossRef]
- Lin, J.N.; Lai, C.H.; Yang, C.H.; Huang, Y.H.; Lin, H.F.; Lin, H.H. Comparison of four automated microbiology systems with 16S rRNA gene sequencing for identification of Chryseobacterium and Elizabethkingia species. Sci. Rep. 2017, 7, 13824. [Google Scholar] [CrossRef]
- Holmes, B.; Steigerwalt, A.G.; Nicholson, A.C. DNA-DNA hybridization study of strains of Chryseobacterium, Elizabethkingia and Empedobacter and of other usually indole-producing non-fermenters of CDC groups IIc, IIe, IIh and IIi, mostly from human clinical sources, and proposals of Chryseobacterium bernardetii sp. nov., Chryseobacterium carnis sp. nov., Chryseobacterium lactis sp. nov., Chryseobacterium nakagawai sp. nov. and Chryseobacterium taklimakanense comb. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 4639–4662. [Google Scholar] [CrossRef] [PubMed]
- Felske, A.; Rheims, H.; Wolterink, A.; Stackebrandt, E.; Akkermans, A.D. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 1997, 143, 2983–2989. [Google Scholar] [CrossRef] [PubMed]
- Hantsis-Zacharov, E.; Shakéd, T.; Senderovich, Y.; Halpern, M. Chryseobacterium oranimense sp. nov., a psychrotolerant, proteolytic and lipolytic bacterium isolated from raw cow’s milk. Int. J. Syst. Evol. Microbiol. 2008, 58, 2635–2639. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Huson, D.H.; Richter, D.C.; Rausch, C.; Dezulian, T.; Franz, M.; Rupp, R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinform. 2007, 8, 460. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, Twenty-Sixth Informational Supplement, 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Kelesidis, T.; Karageorgopoulos, D.E.; Kelesidis, I.; Falagas, M.E. Tigecycline for the treatment of multidrug-resistant Enterobacteriaceae: A systematic review of the evidence from microbiological and clinical studies. J. Antimicrob. Chemother. 2008, 62, 895–904. [Google Scholar] [CrossRef]
- Hung, P.P.; Lin, Y.H.; Lin, C.F.; Liu, M.F.; Shi, Z.Y. Chryseobacterium meningosepticum infection: Antibiotic susceptibility and risk factors for mortality. J. Microbiol. Immunol. Infect. 2008, 41, 137–144. [Google Scholar]
- Lin, P.Y.; Chu, C.; Su, L.H.; Huang, C.T.; Chang, W.Y.; Chiu, C.H. Clinical and microbiological analysis of bloodstream infections caused by Chryseobacterium meningosepticum in nonneonatal patients. J. Clin. Microbiol. 2004, 42, 3353–3355. [Google Scholar] [CrossRef]
- Lin, Y.T.; Chiu, C.H.; Chan, Y.J.; Lin, M.L.; Yu, K.W.; Wang, F.D.; Liu, C.Y. Clinical and microbiological analysis of Elizabethkingia meningoseptica bacteremia in adult patients in Taiwan. Scand. J. Infect. Dis. 2009, 41, 628–634. [Google Scholar] [CrossRef]
- Huang, Y.C.; Lin, Y.T.; Wang, F.D. Comparison of the therapeutic efficacy of fluoroquinolone and non-fluoroquinolone treatment in patients with Elizabethkingia meningoseptica bacteraemia. Int. J. Antimicrob. Agents 2018, 51, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Huang, Y.W.; Lin, Y.T.; Wang, F.D.; Chan, Y.J.; Yang, T.C. Risk factors and outcome of levofloxacin-resistant Elizabethkingia meningoseptica bacteraemia in adult patients in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Matthews, H.; Glaeser, S.P.; Martin, K.; Lodders, N.; Faye, I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol. 2011, 61, 2670–2675. [Google Scholar] [CrossRef]
- Frank, T.; Gody, J.C.; Nguyen, L.B.L.; Berthet, N.; Le Fleche-Mateos, A.; Bata, P.; Rafaï, C.; Kazanji, M.; Breurec, S. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet 2013, 381, 1876. [Google Scholar] [CrossRef]
- Han, M.S.; Kim, H.; Lee, Y.; Kim, M.; Ku, N.S.; Choi, J.Y.; Yong, D.; Jeong, S.H.; Lee, K.; Chong, Y. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J. Clin. Microbiol. 2017, 55, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005, 41 (Suppl. 2), 120–126. [Google Scholar] [CrossRef]
| Site of Isolation | All (n = 92) | Number of Episodes (%) | OR (95% CI) | p-Value | |
|---|---|---|---|---|---|
| E. meningoseptica (n = 20) | E. anophelis (n = 72) | ||||
| Cerebrospinal fluid | 2 (2.2) | 2 (10) | 0 | 0.045 | |
| Pleural effusion | 1 (1.1) | 0 | 1 (1.4) | 0.999 | |
| Respiratory tract | 11 (12) | 1 (5) | 10 (13.9) | 0.33 (0.04–2.72) | 0.445 |
| Ascites | 2 (2.2) | 0 | 2 (2.8) | 0.999 | |
| Bile | 6 (6.5) | 1 (5) | 5 (6.9) | 0.71 (0.08–6.41) | 0.999 |
| Blood | 56 (60.9) | 14 (70) | 42 (58.3) | 1.67 (0.58–4.84) | 0.441 |
| Tip of central venous catheter | 8 (8.7) | 1 (5) | 7 (9.7) | 0.49 (0.06–4.22) | 0.681 |
| Urine | 4 (4.3) | 1 (5) | 3 (4.2) | 1.21 (0.12–12.3) | 0.999 |
| Wound/Abscess | 2 (2.2) | 0 | 2 (2.8) | 0.999 | |
| Characteristics | All (n = 92) | Number of Episodes (%) | OR (95% CI) | p-Value | |
|---|---|---|---|---|---|
| E. meningoseptica (n = 20) | E. anophelis (n = 72) | ||||
| Sex | |||||
| Male | 64 (69.6) | 15 (75) | 49 (68.1) | 1.41 (0.46–4.35) | 0.55 |
| Female | 28 (30.4) | 5 (25) | 23 (31.9) | 0.71 (0.23–2.19) | 0.55 |
| Age | |||||
| Range (year) | 3–89 | 18–80 | 3–89 | ||
| Median (year) | 61 | 61 | 62.5 | ||
| Mean ± standard deviation (year) | 61.1 ± 17 | 56.6 ± 15.6 | 62.4 ± 17.3 | 0.179 | |
| Comorbidity | |||||
| Diabetes mellitus | 24 (26.1) | 6 (30) | 18 (25) | 1.29 (0.43–3.84) | 0.652 |
| Hypertension | 26 (28.3) | 4 (20) | 22 (30.6) | 0.57 (0.17–1.9) | 0.354 |
| End-stage renal disease | 5 (5.4) | 1 (5) | 4 (5.6) | 0.9 (0.09–8.49) | 0.999 |
| Malignancy | 40 (43.5) | 8 (40) | 32 (44.4) | 0.83 (0.3-2.28) | 0.723 |
| Liver cirrhosis | 8 (8.7) | 3 (15) | 5 (6.9) | 2.37 (0.51–10.89) | 0.365 |
| Chronic obstructive pulmonary disease | 9 (9.8) | 0 | 9 (12.5) | 0.197 | |
| Type of infection acquisition | |||||
| Community-acquired infection | 9 (9.8) | 0 | 9 (12.5) | 0.197 | |
| Healthcare-associated infection | 83 (90.2) | 20 | 63 (87.5) | 0.197 | |
| Laboratory data | |||||
| White blood cell count (cells/mm3) | 13,281 ± 8740 | 13,353 ± 6687 | 13,261 ± 9271 | 0.967 | |
| Hemoglobin (g/dL) | 10.1 ± 2.1 | 9.8 ± 2.4 | 10.1 ± 2.1 | 0.585 | |
| Platelet count (×1000 cells/mm3) | 228,570 ± 131,056 | 216,550 ± 157,332 | 231,900 ± 123,846 | 0.69 | |
| Serum creatinine (mg/dL) | 1.8 ± 1.7 | 1.6 ± 1.3 | 1.9 ± 1.8 | 0.584 | |
| Empirical antimicrobial therapy | |||||
| β-lactams | 41 (44.6) | 11 (55) | 30 (41.7) | 1.71 (0.63–4.64) | 0.289 |
| β-lactam/lactamase inhibitors | 20 (21.7) | 4 (20) | 16 (22.2) | 0.88 (0.26–2.99) | 0.999 |
| Ciprofloxacin | 10 (10.9) | 1 (5) | 9 (12.5) | 0.37 (0.04–3.1) | 0.685 |
| Levofloxacin | 24 (26.1) | 1 (5) | 23 (31.9) | 0.11 (0.01–0.89) | 0.015 |
| Carbapenems | 17 (18.5) | 4 (20) | 13 (18.1) | 1.14 (0.33–3.96) | 0.999 |
| Aminoglycosides | 9 (9.8) | 3 (15) | 6 (8.3) | 1.94 (0.44–8.57) | 0.402 |
| Tigecycline | 8 (8.7) | 2 (10) | 6 (8.3) | 1.22 (0.23–6.58) | 0.999 |
| Colistin | 6 (6.5) | 1 (5) | 5 (6.9) | 0.71 (0.08–6.41) | 0.999 |
| Inappropriate empirical antimicrobial therapy | 74 (80.4) | 20 (100) | 54 (75) | 0.01 | |
| Shock | 42 (45.7) | 9 (45) | 33 (45.8) | 0.97 (0.36–2.62) | 0.999 |
| Admission to intensive care unit | 44 (47.8) | 9 (45) | 35 (48.6) | 0.87 (0.32–2.34) | 0.775 |
| Case fatality | 25 (27.2) | 6 (30) | 19 (26.4) | 1.2 (0.4–3.56) | 0.748 |
| Factor | Outcome | Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|---|---|
| Died | Survived | OR (95% CI) | p-Value | OR (95% CI) | p-Value | |
| All Isolates (n = 92) | ||||||
| Species | ||||||
| E. meningoseptica | 6 (24) | 14 (20.9) | 1.2 (0.4–3.56) | 0.748 | 1.76 (0.5–6.16) | 0.38 |
| E. anophelis | 19 (76) | 53 (79.1) | 0.748 | 0.57 (0.16–2) | 0.38 | |
| Age ≥65 years | 12 (48) | 29 (43.3) | 1.21 (0.48–3.04) | 0.686 | 2.89 (0.86–9.74) | 0.087 |
| Sex, male | 20 (80) | 44 (65.7) | 2.09 (0.7–6.3) | 0.184 | 2.99 (0.78–11.52) | 0.112 |
| Underlying disease | ||||||
| Diabetes mellitus | 8 (32) | 16 (23.9) | 1.5 (0.55–4.12) | 0.43 | 1.56 (0.44–5.49) | 0.491 |
| Hypertension | 4 (16) | 22 (32.8) | 0.39 (0.12–1.27) | 0.111 | 0.27 (0.07–1.11) | 0.069 |
| End-stage renal disease | 1 (4) | 4 (6) | 0.66 (0.07-6.17) | 0.999 | 0.45 (0.03–6.77) | 0.566 |
| Malignancy | 12 (48) | 28 (41.8) | 1.29 (0.51–3.24) | 0.593 | 1.11 (0.35–3.47) | 0.86 |
| Liver cirrhosis | 5 (20) | 3 (4.5) | 5.33 (1.17–24.31) | 0.032 | 4.67 (0.9–24.3) | 0.067 |
| Chronic obstructive pulmonary disease | 4 (16) | 5 (7.5) | 2.36 (0.58–9.62) | 0.248 | 1.98 (0.28–18.87) | 0.492 |
| Inappropriate empirical antimicrobial therapy | 24 (96) | 50 (74.6) | 8.16 (1.03–64.97) | 0.02 | 12.45 (1.33–116.77) | 0.027 |
| E. meningoseptica (n = 20) | ||||||
| Age ≥65 years | 3 (50) | 4 (28.6) | 2.5 (0.35–18.04) | 0.613 | 0.999 | |
| Sex, male | 5 (83.3) | 10 (71.4) | 2 (0.17–22.95) | 0.999 | 0.999 | |
| Underlying disease | ||||||
| Diabetes mellitus | 2 (33.3) | 4 (28.6) | 1.25 (0.16–9.77) | 0.999 | 4.5 (0.31–65.23) | 0.27 |
| Hypertension | 0 | 4 (28.6) | 0.267 | 0.999 | ||
| End-stage renal disease | 0 | 1 (7.1) | 0.999 | 0.999 | ||
| Malignancy | 2 (33.3) | 6 (42.9) | 0.67 (0.09–4.93) | 0.999 | 0.68 (0.03–17.96) | 0.814 |
| Liver cirrhosis | 2 (33.3) | 1 (7.1) | 6.5 (0.46–91.92) | 0.202 | 5.11 (0.3–87.96) | 0.261 |
| Chronic obstructive pulmonary disease | 0 | 0 | ||||
| Inappropriate empirical antimicrobial therapy | 6 (100) | 0 | ||||
| E. anophelis (n = 72) | ||||||
| Age ≥65 years | 9 (47.4) | 25 (47.2) | 1.01 (0.35–2.88) | 0.988 | 1.81 (0.36–8.97) | 0.47 |
| Sex, male | 15 (78.9) | 34 (64.2) | 2.1 (0.61–7.22) | 0.235 | 2.09 (0.53–8.19) | 0.291 |
| Underlying disease | ||||||
| Diabetes mellitus | 6 (31.6) | 12 (22.6) | 1.58 (0.49–5.04) | 0.539 | 1.44 (0.36–5.76) | 0.606 |
| Hypertension | 4 (21.1) | 18 (34) | 0.52 (0.15–1.79) | 0.295 | 0.36 (0.09–1.46) | 0.153 |
| End-stage renal disease | 1 (5.3) | 3 (5.7) | 0.93 (0.09–9.48) | 0.999 | 0.66 (0.04–10.92) | 0.773 |
| Malignancy | 10 (52.6) | 22 (41.5) | 1.57 (0.55–4.49) | 0.403 | 1.3 (0.34–4.98) | 0.706 |
| Liver cirrhosis | 3 (15.8) | 2 (3.8) | 4.78 (0.73–31.19) | 0.111 | 3.4 (0.51–22.5) | 0.204 |
| Chronic obstructive pulmonary disease | 4 (21.1) | 5 (9.4) | 2.56 (0.61–10.77) | 0.231 | 3.06 (0.59–15.9) | 0.183 |
| Inappropriate empirical antimicrobial therapy | 18 (94.7) | 36 (67.9) | 8.5 (1.05–69.04) | 0.021 | 8.5 (1.05–69.04) | 0.045 |
| Characteristics | All Elizabethkingia (n = 92) | E. meningoseptica (n = 20) | E. anophelis (n = 72) | OR (95% CI) d | p-Value d | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 a | MIC90 b | S, n (%) c | MIC50 a | MIC90 b | S, n (%) c | MIC50 a | MIC90 b | S, n (%) c | |||
| Piperacillin | 64 | >64 | 17 (18.5) | 64 | >64 | 3 (15) | 64 | >64 | 14 (19.4) | 0.73 (0.19–2.85) | 0.756 |
| Piperacillin tazobactam | 128/4 | >128/4 | 22 (23.9) | 128/4 | >128/4 | 1 (5) | 64/4 | >128/4 | 22 (30.6) | 0.12 (0.02–0.95) | 0.02 |
| Ticarcillin-clavulanic acid | >64/2 | >64/2 | 0 | >64/2 | >64/2 | 0 | >64/2 | >64/2 | 0 | ||
| Ceftazidime | >16 | >16 | 0 | >16 | >16 | 0 | >16 | >16 | 0 | ||
| Cefepime | >32 | >32 | 1 (2.2) | >32 | >32 | 0 | >32 | >32 | 2 (2.8) | 0.999 | |
| Ceftriaxone | >32 | >32 | 0 | >32 | >32 | 0 | >32 | >32 | 0 | ||
| Aztreonam | >16 | >16 | 0 | >16 | >16 | 0 | >16 | >16 | 0 | ||
| Imipenem | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Meropenem | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Gentamicin | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Tobramycin | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Amikacin | >32 | >32 | 4 (4.3) | >32 | >32 | 0 | >32 | >32 | 4 (5.6) | 0.573 | |
| Tetracycline | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Minocycline | <1 | 4 | 84 (91.3) | 2 | 4 | 12 (60) | <1 | 2 | 72 (100) | <0.001 | |
| Tigecycline | 4 | >8 | 22 (23.9) | 4 | >8 | 3 (15) | 4 | 8 | 19 (26.4) | 0.49 (0.13–1.87) | 0.382 |
| Ciprofloxacin | 2 | >2 | 9 (9.8) | >2 | >2 | 2 (10) | 2 | >2 | 7 (9.7) | 1.03 (0.2–5.4) | 0.999 |
| Levofloxacin | 2 | >8 | 48 (52.2) | 8 | >8 | 6 (30) | 2 | >8 | 42 (58.3) | 0.31 (0.11–0.89) | 0.025 |
| Trimethoprim-sulfamethoxazole | >4/76 | >4/76 | 11 (12) | >4/76 | >4/76 | 2 (10) | >4/76 | >4/76 | 9 (12.5) | 0.78 (0.15–3.93) | 0.999 |
| Number of Susceptible Antibiotics | All Isolates (n = 92) | Number of Episodes (%) | OR (95% CI) | p-Value | |
|---|---|---|---|---|---|
| E. meningoseptica (n = 20) | E. anopheles (n = 72) | ||||
| 0 | 6 (6.5) | 6 (30) | 0 | <0.001 | |
| 1 | 22 (23.9) | 5 (25) | 17 (23.6) | 0.93 (0.29–2.93) | 0.009 |
| 2 | 28 (30.4) | 5 (25) | 23 (31.9) | 1.41 (0.46–4.35) | 0.55 |
| 3 | 19 (20.7) | 3 (15) | 16 (22.2) | 1.62 (0.42–6.23) | 0.755 |
| 4 | 7 (7.6) | 0 | 7 (9.7) | 0.34 | |
| 5 | 5 (5.4) | 1 (5) | 4 (5.6) | 1.12 (0.12–10.6) | 0.999 |
| 6 | 3 (3.3) | 0 | 3 (4.2) | 0.999 | |
| 7 | 2 (2.2) | 0 | 2 (2.8) | 0.999 | |
| Amino Acid | Susceptibility of Levofloxacin | No. of Episode (%) | |||||
|---|---|---|---|---|---|---|---|
| Susceptible (n = 48) | Non-Susceptible (n = 44) | OR (95% CI) | p-Value | E. meningoseptica (n = 20) | E. anopheles (n = 72) | p-Value | |
| Position 83 of GyrA | |||||||
| Serine | 48 (100) | 28 (63.6) | <0.001 | 16 (80) | 60 (83.3) | 0.478 | |
| Isoleucine | 0 | 13 (29.5) | <0.001 | 4 (20) | 9 (12.5) | ||
| Arginine | 0 | 3 (6.8) | 0.105 | 0 | 3 (4.2) | ||
| Position 95 of GyrA | |||||||
| Serine | 5 (10.4) | 12 (27.3) | 0.31 (0.1–0.97) | 0.037 | 17 (85) | 72 (100) | 0.009 |
| Proline | 1 (2.1) | 2 (4.5) | 0.45 (0.04–5.11) | 0.605 | 3 (15) | 0 | |
| Position 102 of GyrA | |||||||
| Lysine | 3 (6.3) | 9 (20.5) | 0.26 (0.07–1.03) | 0.055 | 12 (60) | 72 (100) | <0.001 |
| Glutamine | 3 (6.3) | 5 (11.4) | 0.52 (0.12–2.32) | 0.473 | 8 (40) | 0 | |
| Position 425 of GyrB | |||||||
| Isoleucine | 2 (4.2) | 7 (15.9) | 0.23 (0.05–1.17) | 0.081 | 9 (45) | 72 (100) | <0.001 |
| Lysine | 4 (8.3) | 7 (15.9) | 0.48 (0.13–1.77) | 0.263 | 11(55) | 0 | |
| Position 452 of GyrB | |||||||
| Arginine | 6 (12.5) | 13 (29.5) | 0.341 (0.12–0.996) | 0.044 | 19 (95) | 72 (100) | 0.217 |
| Serine | 0 | 1 (2.3) | 0.478 | 1 (5) | 0 | ||
| Position 470 of GyrB | |||||||
| Glutamate | 5 (10.4) | 12 (27.3) | 0.31 (0.1–0.97) | 0.037 | 17 (85) | 72 (100) | 0.009 |
| Aspartate | 1 (2.1) | 2 (4.5) | 0.45 (0.04-5.11) | 0.605 | 3 (15) | 0 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.-N.; Lai, C.-H.; Yang, C.-H.; Huang, Y.-H. Comparison of Clinical Manifestations, Antimicrobial Susceptibility Patterns, and Mutations of Fluoroquinolone Target Genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis Isolated in Taiwan. J. Clin. Med. 2018, 7, 538. https://doi.org/10.3390/jcm7120538
Lin J-N, Lai C-H, Yang C-H, Huang Y-H. Comparison of Clinical Manifestations, Antimicrobial Susceptibility Patterns, and Mutations of Fluoroquinolone Target Genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis Isolated in Taiwan. Journal of Clinical Medicine. 2018; 7(12):538. https://doi.org/10.3390/jcm7120538
Chicago/Turabian StyleLin, Jiun-Nong, Chung-Hsu Lai, Chih-Hui Yang, and Yi-Han Huang. 2018. "Comparison of Clinical Manifestations, Antimicrobial Susceptibility Patterns, and Mutations of Fluoroquinolone Target Genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis Isolated in Taiwan" Journal of Clinical Medicine 7, no. 12: 538. https://doi.org/10.3390/jcm7120538
APA StyleLin, J.-N., Lai, C.-H., Yang, C.-H., & Huang, Y.-H. (2018). Comparison of Clinical Manifestations, Antimicrobial Susceptibility Patterns, and Mutations of Fluoroquinolone Target Genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis Isolated in Taiwan. Journal of Clinical Medicine, 7(12), 538. https://doi.org/10.3390/jcm7120538





