Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis
Abstract
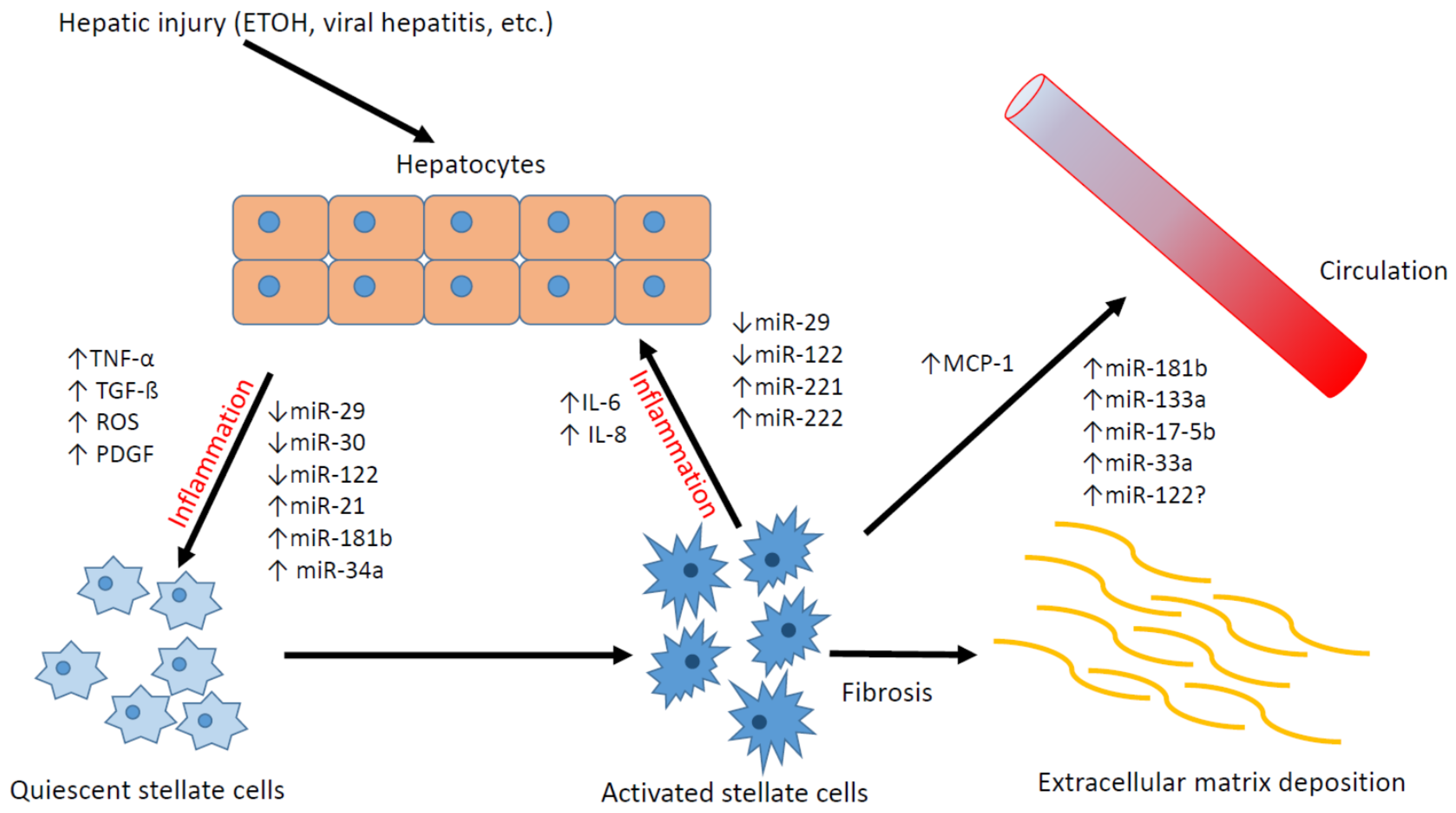
:1. Introduction
2. Biogenesis of microRNA
3. The Role of Hepatic Stellate Cells in the Development of Liver Fibrosis
4. Differentially Expressed MicroRNAs in Activated Hepatic Stellate Cells
5. The Role of MicroRNAs in Liver Fibrosis
5.1. microRNAs Regulate HSC Activation
5.2. microRNAs Regulate HSC Apoptosis and Proliferation
5.3. microRNAs Regulate HSC Migration
5.4. microRNAs Regulate ECM Deposition and Maturation
5.5. miRNAs Involved in Nonalcoholic Steatohepatitis
5.6. miRNAs Studied in Cholestatic Mouse Model
5.7. miRNAs in in vivo Portal Hypertension Model
5.8. Other Regulatory Mechanisms
6. microRNA as a Potential Therapeutic Target in Liver Fibrosis
7. microRNA as a Potential Biomarker of Liver Fibrosis
8. Conclusions
Author Contributions
Conflicts of Interest
References
- Friedman, S.L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 2000, 275, 2247–2250. [Google Scholar] [CrossRef] [PubMed]
- Bataller, R.; Brenner, D.A. Hepatic stellate cells as a target for the treatment of liver fibrosis. Semin. Liver Dis. 2001, 21, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008, 88, 125–172. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Engels, B.M.; Hutvagner, G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene 2006, 25, 6163–6169. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.L.; Wu, S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011, 300, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Homo Sapiens miRNAs. Available online: http://www.mirbase.org/cgi-bin/mirna_summary.pl?org=hsa (accessed on 17 March 2015).
- Jovanovic, M.; Hengartner, M.O. MiRNAs and apoptosis: RNAs to die for. Oncogene 2006, 25, 6176–6187. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Wilfred, B.R.; Wang, W.X.; Nelson, P.T. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007, 91, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Kong, S.W.; Lu, J.; Bisping, E.; Zhang, H.; Allen, P.D.; Golub, T.R.; Pieske, B.; Pu, W.T. Altered microRNA expression in human heart disease. Physiol. Genomics 2007, 31, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Kasinski, A.L.; Slack, F.J. Epigenetics and genetics. microRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 2011, 11, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; De Strooper, B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009, 32, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Schoof, C.R.; Botelho, E.L.; Izzotti, A.; Vasques Ldos, R. MicroRNAs in cancer treatment and prognosis. Am. J. Cancer Res. 2012, 2, 414–433. [Google Scholar] [PubMed]
- Jeffrey, S.S. Cancer biomarker profiling with microRNAs. Nat. Biotechnol. 2008, 26, 400–401. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Chendrimada, T.P.; Shiekhattar, R. MicroRNA biogenesis: Isolation and characterization of the microprocessor complex. Methods Mol. Biol. 2006, 342, 33–47. [Google Scholar] [PubMed]
- Morishita, A.; Masaki, T. MiRNA in hepatocellular carcinoma. Hepatol. Res. 2015, 45, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, V.N. MicroRNA factory: RISC assembly from precursor microRNAs. Mol. Cell 2012, 46, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Geerts, A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin. Liver Dis. 2001, 21, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Evason, K.J.; Asahina, K.; Stainier, D.Y. Hepatic stellate cells in liver development, regeneration, and cancer. J. Clin. Investig. 2013, 123, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Krizhanovsky, V.; Yon, M.; Dickins, R.A.; Hearn, S.; Simon, J.; Miething, C.; Yee, H.; Zender, L.; Lowe, S.W. Senescence of activated stellate cells limits liver fibrosis. Cell 2008, 134, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva, T.; Brenner, D.A. Hepatic stellate cells and the reversal of fibrosis. J. Gastroenterol. Hepatol. 2006, 21 (Suppl. 3), S84–S87. [Google Scholar] [CrossRef] [PubMed]
- Senoo, H. Structure and function of hepatic stellate cells. Med. Electron. Microsc. 2004, 37, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Liao, Z.X.; Ping, J.; Xu, D.; Wang, H. Molecular mechanism of hepatic stellate cell activation and antifibrotic therapeutic strategies. J. Gastroenterol. 2008, 43, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Sicklick, J.K.; Choi, S.S.; Bustamante, M.; McCall, S.J.; Pérez, E.H.; Huang, J.; Li, Y.X.; Rojkind, M.; Diehl, A.M. Evidence for epithelial-mesenchymal transitions in adult liver cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291, G575–G583. [Google Scholar] [CrossRef] [PubMed]
- Miyata, E.; Masuya, M.; Yoshida, S.; Nakamura, S.; Kato, K.; Sugimoto, Y.; Shibasaki, T.; Yamamura, K.; Ohishi, K.; Nishii, K.; et al. Hematopoietic origin of hepatic stellate cells in the adult liver. Blood 2008, 111, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.; Fujii, H.; Hirose, T.; Yasuchika, K.; Azuma, H.; Hoppo, T.; Naito, M.; Machimoto, T.; Ikai, I. Commitment of bone marrow cells to hepatic stellate cells in mouse. J. Hepatol. 2004, 40, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Knittel, T.; Kobold, D.; Saile, B.; Grundmann, A.; Neubauer, K.; Piscaglia, F.; Ramadori, G. Rat liver myofibroblasts and hepatic stellate cells: Different cell populations of the fibroblast lineage with fibrogenic potential. Gastroenterology 1999, 117, 1205–1221. [Google Scholar] [CrossRef]
- Canbay, A.; Friedman, S.; Gores, G.J. Apoptosis: The nexus of liver injury and fibrosis. Hepatology 2004, 39, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Iredale, J.P.; Benyon, R.C.; Pickering, J.; McCullen, M.; Northrop, M.; Pawley, S.; Hovell, C.; Arthur, M.J. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J. Clin. Investig. 1998, 102, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Issa, R.; Williams, E.; Trim, N.; Kendall, T.; Arthur, M.J.; Reichen, J.; Benyon, R.C.; Iredale, J.P. Apoptosis of hepatic stellate cells: Involvement in resolution of biliary fibrosis and regulation by soluble growth factors. Gut 2001, 48, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Wallace, M.C.; Friedman, S.L. Hepatic fibrosis and the microenvironment: Fertile soil for hepatocellular carcinoma development. Gene Expr. 2014, 16, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Niu, X.; Wang, Y.; Kong, L.; Wang, R.; Zhang, Y.; Zhao, S.; Nan, Y. MiR-146a-5p suppresses activation and proliferation of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis through directly targeting Wnt1 and Wnt5a. Sci. Rep. 2015, 5, 16163. [Google Scholar] [CrossRef] [PubMed]
- Leti, F.; Malenica, I.; Doshi, M.; Courtright, A.; Van Keuren-Jensen, K.; Legendre, C.; Still, C.D.; Gerhard, G.S.; DiStefano, J.K. High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis. Transl. Res. 2015, 166, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Pan, Q.; Cheng, T.; Jiang, B.; Chen, G.Y.; Li, D.G. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009, 276, 5163–5176. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Pan, Q.; Li, D.G.; Sun, H.; Liu, B.W. MiR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J. Hepatol. 2009, 50, 766–778. [Google Scholar] [CrossRef] [PubMed]
- Lakner, A.M.; Steuerwald, N.M.; Walling, T.L.; Ghosh, S.; Li, T.; McKillop, I.H.; Russo, M.W.; Bonkovsky, H.L.; Schrum, L.W. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology 2012, 56, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Maubach, G.; Lim, M.C.; Chen, J.; Yang, H.; Zhuo, L. miRNA studies in in vitro and in vivo activated hepatic stellate cells. World J. Gastroenterol. 2011, 17, 2748–2773. [Google Scholar] [PubMed]
- Murakami, Y.; Toyoda, H.; Tanaka, M.; Kuroda, M.; Harada, Y.; Matsuda, F.; Tajima, A.; Kosaka, N.; Ochiya, T.; Shimotohno, K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE 2011, 6, e16081. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Cheung, O.; Puri, P.; Eicken, C.; Contos, M.J.; Mirshahi, F.; Maher, J.W.; Kellum, J.M.; Min, H.; Luketic, V.A.; Sanyal, A.J. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology 2008, 48, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- De Minicis, S.; Seki, E.; Uchinami, H.; Kluwe, J.; Zhang, Y.; Brenner, D.A.; Schwabe, R.F. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology 2007, 132, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Hu, J.F.; Liu, B.H.; Wu, C.Q.; Yu, H.Y.; Yao, D.K.; Zhu, L. MiR-9a-5p regulates proliferation and migration of hepatic stellate cells under pressure through inhibition of Sirt1. World J. Gastroenterol. 2015, 21, 9900–9915. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Guo, Y.; Chen, B.; Dong, P.; Zheng, J. MicroRNA-17–5p activates hepatic stellate cells through targeting of Smad7. Lab Investig. 2015, 95, 781–789. [Google Scholar] [PubMed]
- Wei, J.; Feng, L.; Li, Z.; Xu, G.; Fan, X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed. Pharmacother. 2013, 67, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zha, Y.; Hu, W.; Huang, Z.; Gao, Z.; Zang, Y.; Chen, J.; Dong, L.; Zhang, J. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (miR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J. Biol. Chem. 2013, 288, 37082–37093. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tang, N.; Wu, K.; Dai, W.; Ye, C.; Shi, J.; Zhang, J.; Ning, B.; Zeng, X.; Lin, Y. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLoS ONE 2014, 9, e108005. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, C.; Liu, Q.; Liu, B.; Song, C.; Zhu, S.; Wu, C.; Liu, S.; Yu, H.; Yao, D.; et al. The role of miR-31/FIH1 pathway in TGFbeta-induced liver fibrosis. Clin. Sci. (Lond.) 2015, 129, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Ou-Yang, P.H.; Han, X.P. Profibrotic effect of miR-33a with Akt activation in hepatic stellate cells. Cell. Signal. 2014, 26, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Teratani, T.; Suzuki, T.; Shimizu, M.; Sato, H.; Narimatsu, K.; Okada, Y.; Kurihara, C.; Irie, R.; Yokoyama, H.; et al. Free cholesterol accumulation in hepatic stellate cells: Mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 2014, 59, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Wu, S.; He, J.; Lou, L.; Ye, W.; Wang, J. MicroRNA-34a and microRNA-34c promote the activation of human hepatic stellate cells by targeting peroxisome proliferator-activated receptor gamma. Mol. Med. Rep. 2015, 11, 1017–1024. [Google Scholar] [PubMed]
- Yan, G.; Li, B.; Xin, X.; Xu, M.; Ji, G.; Yu, H. MicroRNA-34a Promotes Hepatic Stellate Cell Activation via Targeting ACSL1. Med. Sci. Monit. 2015, 21, 3008–3015. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Tan, W.; Cheng, S.; Wang, H.; Ye, S.; Yu, C.; He, Y.; Zeng, J.; Cen, J.; Hu, J. Upregulation of microRNA-126 in hepatic stellate cells may affect pathogenesis of liver fibrosis through the NF-kappaB pathway. DNA Cell Biol. 2015, 34, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wang, J.; Lu, H.; Zhang, G.; Liu, Y.; Zhang, Y.; Shang, H.; Ji, H.; Chen, X.; Duan, Y.; et al. MicroRNA-130a and -130b enhance activation of hepatic stellate cells by suppressing PPARgamma expression: A rat fibrosis model study. Biochem. Biophys. Res. Commun. 2015, 465, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Taketomi, A.; Shirabe, K.; Umeda, K.; Kayashima, H.; Ninomiya, M.; Uchiyama, H.; Soejima, Y.; Maehara, Y. Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 2011, 31, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.; Kemper, S.; Charrier, A.; Brigstock, D.R. Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G491–G499. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, M.; Ogawa, T.; Enomoto, M.; Motoyama, H.; Yoshizato, K.; Ikeda, K.; Kawada, N. Induction of microRNA-214-5p in human and rodent liver fibrosis. Fibrogenesis Tissue Repair 2012, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Enomoto, M.; Fujii, H.; Sekiya, Y.; Yoshizato, K.; Ikeda, K.; Kawada, N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut 2012, 61, 1600–1609. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Pan, Q.; Jiang, B.; Chen, G.Y.; Li, D.G. Effects of upregulated expression of microRNA-16 on biological properties of culture-activated hepatic stellate cells. Apoptosis 2009, 14, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Xie, J.; Liu, F.; He, J. MicroRNA-19b reduces hepatic stellate cell proliferation by targeting GRB2 in hepatic fibrosis models in vivo and in vitro as part of the inhibitory effect of estradiol. J. Cell. Biochem. 2015, 116, 2455–2464. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Friedman, R.C.; Marquez, R.T.; Keck, K.; Kong, B.; Icardi, M.S.; Brown, K.E.; Burge, C.B.; Schmidt, W.N.; Wang, Y.; et al. Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: MiR-29 overexpression reduces hepatitis C viral abundance in culture. J. Infect. Dis. 2011, 203, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Kwiecinski, M.; Noetel, A.; Elfimova, N.; Trebicka, J.; Schievenbusch, S.; Strack, I.; Molnar, L.; von Brandenstein, M.; Töx, U.; Nischt, R.; et al. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction. PLoS ONE 2011, 6, e24568. [Google Scholar] [CrossRef] [PubMed]
- Kwiecinski, M.; Elfimova, N.; Noetel, A.; Töx, U.; Steffen, H.M.; Hacker, U.; Nischt, R.; Dienes, H.P.; Odenthal, M. Expression of platelet-derived growth factor-C and insulin-like growth factor I in hepatic stellate cells is inhibited by miR-29. Lab Investig. 2012, 92, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Iizuka, M.; Sekiya, Y.; Yoshizato, K.; Ikeda, K.; Kawada, N. Suppression of type I collagen production by microRNA-29b in cultured human stellate cells. Biochem. Biophys. Res. Commun. 2010, 391, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Tiao, M.M.; Huang, L.T.; Chuang, J.H.; Kuo, K.C.; Yang, Y.L.; Wang, F.S. Activation of miR-29a in activated hepatic stellate cells modulates its profibrogenic phenotype through inhibition of histone deacetylases 4. PLoS ONE 2015, 10, e0136453. [Google Scholar] [CrossRef] [PubMed]
- Tiao, M.M.; Wang, F.S.; Huang, L.T.; Chuang, J.H.; Kuo, H.C.; Yang, Y.L.; Huang, Y.H. MicroRNA-29a protects against acute liver injury in a mouse model of obstructive jaundice via inhibition of the extrinsic apoptosis pathway. Apoptosis 2014, 19, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ghazwani, M.; Li, J.; Sun, M.; Stolz, D.B.; He, F.; Fan, J.; Xie, W.; Li, S. MiR-29b inhibits collagen maturation in hepatic stellate cells through down-regulating the expression of HSP47 and lysyl oxidase. Biochem. Biophys. Res. Commun. 2014, 446, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, C.; Lin, Z.; Guo, Y.; Shi, L.; Dong, P.; Lu, Z.; Gao, S.; Liao, Y.; Chen, B.; et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation—A novel mechanism suppressing liver fibrosis. FEBS J. 2014, 281, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.; Zheng, X.; Li, H.; Cao, Z.; Chang, H.; Luan, S.; Zhu, J.; Chen, J.; Zang, Y.; Zhang, J. MicroRNA-30 protect against CCl4-induced liver fibrosis by attenuating TGF-beta signaling in hepatic stellate cells. Toxicol. Sci. 2015, 146, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.; Zhang, H.; Zhang, J.; Zhao, S.; Zheng, X.; Zhang, Z.; Zhu, J.; Chen, J.; Dong, L.; Zang, Y. MicroRNA-101 suppresses liver fibrosis by targeting the TGF-beta signalling pathway. J. Pathol. 2014, 234, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ghazwani, M.; Zhang, Y.; Lu, J.; Fan, J.; Gandhi, C.R.; Li, S. MiR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J. Hepatol. 2013, 58, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Wang, Y.L.; Xie, C.; Sang, Y.; Li, T.J.; Zhang, M.; Wang, R.; Zhang, Q.; Zheng, L.; Zhuang, S.M. Identification of a novel TGF-beta-miR-122-fibronectin 1/serum response factor signaling cascade and its implication in hepatic fibrogenesis. Oncotarget 2015, 6, 12224–12233. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Pan, Q.; Xiong, H.; Qiao, Y.Q.; Bian, Z.L.; Zhong, W.; Sheng, L.; Li, H.; Shen, L.; Hua, J.; et al. Dynamic expression of miR-126* and its effects on proliferation and contraction of hepatic stellate cells. FEBS Lett. 2013, 587, 3792–3801. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Luedde, M.; Vargas, C.D.; Vucur, M.; Mollnow, T.; Zimmermann, H.W.; Koch, A.; Hellerbrand, C.; Weiskirchen, R.; Frey, N.; et al. MiR-133a mediates TGF-beta-dependent derepression of collagen synthesis in hepatic stellate cells during liver fibrosis. J. Hepatol. 2013, 58, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yi, J.; Ye, R.; Liu, J.; Duan, Q.; Xiao, J.; Liu, F. miR-144 regulates transforming growth factor-beta1 iduced hepatic stellate cell activation in human fibrotic liver. Int. J. Clin. Exp. Pathol. 2015, 8, 3994–4000. [Google Scholar]
- He, Y.; Huang, C.; Sun, X.; Long, X.R.; Lv, X.W.; Li, J. MicroRNA-146a modulates TGF-beta1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell. Signal. 2012, 24, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, S.K.; Jiang, J.; Kim, T.H.; Li, Y.; Wang, S.S.; Torok, N.J.; Wu, J.; Zern, M.A. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G101–G106. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Lin, Z.; Dong, P.; Lu, Z.; Gao, S.; Chen, X.; Wu, C.; Yu, F. Activation of hepatic stellate cells is suppressed by microRNA-150. Int. J. Mol. Med. 2013, 32, 17–24. [Google Scholar] [PubMed]
- Dai, W.; Zhao, J.; Tang, N.; Zeng, X.; Wu, K.; Ye, C.; Shi, J.; Lu, C.; Ning, B.; Zhang, J.; et al. MicroRNA-155 attenuates activation of hepatic stellate cell by simultaneously preventing EMT process and ERK1 signalling pathway. Liver Int. 2015, 35, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, Y.; Ogawa, T.; Iizuka, M.; Yoshizato, K.; Ikeda, K.; Kawada, N. Down-regulation of cyclin E1 expression by microRNA-195 accounts for interferon-beta-induced inhibition of hepatic stellate cell proliferation. J. Cell. Physiol. 2011, 226, 2535–2542. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Tao, H.; Hu, W.; Liu, L.P.; Shi, K.H.; Deng, Z.Y.; Li, J. MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis. Cell. Signal. 2014, 26, 2381–2389. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; He, Y.; Ma, T.T.; Huang, C.; Zhang, L.; Li, J. Participation of miR-200a in TGF-beta1-mediated hepatic stellate cell activation. Mol. Cell. Biochem. 2014, 388, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Charrier, A.; Zhou, Y.; Chen, R.; Yu, B.; Agarwal, K.; Tsukamoto, H.; Lee, L.J.; Paulaitis, M.E.; Brigstock, D.R. Epigenetic regulation of connective tissue growth factor by microRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology 2014, 59, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, C.Q.; Zhang, Z.Q.; Yao, D.K.; Zhu, L. Loss of expression of miR-335 is implicated in hepatic stellate cell migration and activation. Exp. Cell Res. 2011, 317, 1714–1725. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Hou, Q.R.; Deng, L.F.; Fei, C.; Xu, W.P.; Zhang, Q.; Wu, K.M.; Ning, B.F.; Xie, W.F.; Zhang, X. MicroRNA-370 Attenuates Hepatic Fibrogenesis by Targeting Smoothened. Dig. Dis. Sci. 2015, 60, 2038–2048. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; He, X.; Duan, Y.; Chen, J.; Wang, J.; Sun, X.; Qian, H.; Feng, J.; Sun, W.; Xu, F.; et al. Expression of microRNA-454 in TGF-beta1-stimulated hepatic stellate cells and in mouse livers infected with Schistosoma japonicum. Parasit. Vectors 2014, 7, 148. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ma, N.; Zhao, R.; Wu, G.; Zhang, Y.; Qiao, Y.; Han, D.; Xu, Y.; Xiang, Y.; Yan, B.; et al. Overexpression of miR-483–5p/3p cooperate to inhibit mouse liver fibrosis by suppressing the TGF-beta stimulated HSCs in transgenic mice. J. Cell. Mol. Med. 2014, 18, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, E.S.; Chen, H.Y.; Man, K.; Go, M.Y.; Huang, X.R.; Lan, H.Y.; Sung, J.J.; Yu, J. MicroRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 2015, 6, 7325–7338. [Google Scholar] [CrossRef] [PubMed]
- Mannaerts, I.; Eysackers, N.; Onyema, O.O.; Van Beneden, K.; Valente, S.; Mai, A.; Odenthal, M.; van Grunsven, L.A. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS ONE 2013, 8, e55786. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, Y.; Ogawa, T.; Yoshizato, K.; Ikeda, K.; Kawada, N. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem. Biophys. Res. Commun. 2011, 412, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, C.; Xu, Z.; Xia, P.; Dong, P.; Chen, B.; Yu, F. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol. Cell. Biochem. 2015, 398, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.M.; Chang, Y.; Jia, A. Effects of interferon-alpha on expression of hepatic stellate cell and transforming growth factor-beta1 and alpha-smooth muscle actin in rats with hepatic fibrosis. World J. Gastroenterol. 2005, 11, 2634–2636. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Kawada, N.; Ikeda, K. Effect of natural interferon alpha on proliferation and apoptosis of hepatic stellate cells. Hepatol. Int. 2009, 3, 497–503. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanabe, J.; Izawa, A.; Takemi, N.; Miyauchi, Y.; Torii, Y.; Tsuchiyama, H.; Suzuki, T.; Sone, S.; Ando, K. Interferon-beta reduces the mouse liver fibrosis induced by repeated administration of concanavalin A via the direct and indirect effects. Immunology 2007, 122, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zeisberg, M.; Mosterman, B.; Sudhakar, A.; Yerramalla, U.; Holthaus, K.; Xu, L.; Eng, F.; Afdhal, N.; Kalluri, R. Liver fibrosis: Insights into migration of hepatic stellate cells in response to extracellular matrix and growth factors. Gastroenterology 2003, 124, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lin, Z.; Zheng, J.; Gao, S.; Lu, Z.; Dong, P. Suppression of collagen synthesis by Dicer gene silencing in hepatic stellate cells. Mol. Med. Rep. 2014, 9, 707–714. [Google Scholar] [PubMed]
- Yu, F.; Lu, Z.; Chen, B.; Wu, X.; Dong, P.; Zheng, J. Salvianolic acid B-induced microRNA-152 inhibits liver fibrosis by attenuating DNMT1-mediated Patched1 methylation. J. Cell. Mol. Med. 2015, 19, 2617–2632. [Google Scholar] [CrossRef] [PubMed]
- Knabel, M.K.; Ramachandran, K.; Karhadkar, S.; Hwang, H.W.; Creamer, T.J.; Chivukula, R.R.; Sheikh, F.; Clark, K.R.; Torbenson, M.; Montgomery, R.A.; et al. Systemic Delivery of scAAV8-Encoded MiR-29a Ameliorates Hepatic Fibrosis in Carbon Tetrachloride-Treated Mice. PLoS ONE 2015, 10, e0124411. [Google Scholar]
- Li, J.; Zhang, Y.; Kuruba, R.; Gao, X.; Gandhi, C.R.; Xie, W.; Li, S. Roles of microRNA-29a in the antifibrotic effect of farnesoid X receptor in hepatic stellate cells. Mol. Pharmacol. 2011, 80, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Jiang, H.J.; Sun, X.M.; Cai, X.K.; He, X.X.; Li, P.Y.; Tang, W.X.; Song, Y.H.; Lin, J.S. Hepatic stellate cell-specific gene silencing induced by an artificial microRNA for antifibrosis in vitro. Dig. Dis. Sci. 2010, 55, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gao, Z.; Hu, W.; Yin, S.; Wang, C.; Zang, Y.; Chen, J.; Zhang, J.; Dong, L. 3,3′-Diindolylmethane ameliorates experimental hepatic fibrosis via inhibiting miR-21 expression. Br. J. Pharmacol. 2013, 170, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.C.; He, Y.J.; Lv, X.; Ye, G.R.; Wang, L.X. Curcumin prevents liver fibrosis by inducing apoptosis and suppressing activation of hepatic stellate cells. J. Nat. Med. 2009, 63, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. Newe Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Van der Ree, M.H.; van der Meer, A.J.; de Bruijne, J.; Maan, R.; van Vliet, A.; Welzel, T.M.; Zeuzem, S.; Lawitz, E.J.; Rodriguez-Torres, M.; Kupcova, V.; et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014, 111, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O'Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Xu, Y.; Hao, J.; Wang, S.; Li, C.; Meng, S. MiR-122 in hepatic function and liver diseases. Protein Cell 2012, 3, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Bihrer, V.; Friedrich-Rust, M.; Kronenberger, B.; Forestier, N.; Haupenthal, J.; Shi, Y.; Peveling-Oberhag, J.; Radeke, H.H.; Sarrazin, C.; Herrmann, E.; et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 2011, 106, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jia, Y.; Zheng, R.; Guo, Y.; Wang, Y.; Guo, H.; Fei, M.; Sun, S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 2010, 56, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wang, J.; Katayama, H.; Sen, S.; Liu, S.M. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma 2013, 60, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Trebicka, J.; Anadol, E.; Elfimova, N.; Strack, I.; Roggendorf, M.; Viazov, S.; Wedemeyer, I.; Drebber, U.; Rockstroh, J.; Sauerbruch, T.; et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 2013, 58, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, W.; Guo, K.; Xiao, Y.; Wang, Y.; Fan, J. miR-181b promotes hepatic stellate cells proliferation by targeting p27 and is elevated in the serum of cirrhosis patients. Biochem. Biophys. Res. Commun. 2012, 421, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Luedde, T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J. Hepatol. 2014, 61, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Sun, C.C.; Zhao, F.; Zhang, Y.D.; Li, D.J. MiR-33a levels in hepatic and serum after chronic HBV-induced fibrosis. J. Gastroenterol. 2015, 50, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Petrasek, J.; Mundkur, S.; Catalano, D.; Levin, I.; Ward, J.; Alao, H.; Kodys, K.; Szabo, G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012, 56, 1946–1957. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; Eischeid, H.; Goertzen, J.; Schierwagen, R.; Anadol, E.; Strassburg, C.P.; Sauerbruch, T.; Odenthal, M.; Trebicka, J. The role of miRNA-34a as a prognostic biomarker for cirrhotic patients with portal hypertension receiving TIPS. PLoS ONE 2014, 9, e103779. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; Reiberger, T.; Huang, J.; Eischeid, H.; Schierwagen, R.; Mandorfer, M.; Anadol, E.; Schwabl, P.; Schwarze-Zander, C.; Warnecke-Eberz, U.; et al. Circulating miRNA-122 levels are associated with hepatic necroinflammation and portal hypertension in HIV/HCV coinfection. PLoS ONE 2015, 10, e0116768. [Google Scholar] [CrossRef] [PubMed]
- Anadol, E.; Schierwagen, R.; Elfimova, N.; Tack, K.; Schwarze-Zander, C.; Eischeid, H.; Noetel, A.; Boesecke, C.; Jansen, C.; Dold, L.; et al. Circulating microRNAs as a marker for liver injury in human immunodeficiency virus patients. Hepatology 2015, 61, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Chronic Liver Disease and Cirrhosis. Available online: http://www.cdc.gov/nchs/fastats/liver-disease.htm (accessed on 17 June 2015).
- ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/results?term=microrna&Search=Search (accessed on 11 June 2015).
| Ref. | miRNA Detection Method | Species | Experimental Model | Number of Significantly Dysregulated miRNAs | Upregulated miRNAs | Downregulated miRNA | Examples |
|---|---|---|---|---|---|---|---|
| [47] | Microarray | Human | Biopsy samples from individuals with NASH | 46 | 23 | 23 | miR-34a, -122, -146b, -451 |
| [39] | Microarray | Mouse | NASH model using methionine-choline deficient diet | 37 | 19 | 18 | miR-146a-5p |
| [41,42] | Microarray | Rat | Culture activated | 21 | 12 | 9 | miR-15b, -16 |
| [43] | Microarray | Rat | Culture activated | 55 | NR | NR | miR-19a, -19b, -92a |
| [40] | HT sequencing | Human | Biopsy samples from individuals with NAFLD | 75 | 30 | 45 | miR-182 |
| [44] | Microarray | Rat | Culture activated | 42 | 16 | 26 | miR-26a, -29a, -214 |
| [45] | Microarray | Mouse | CCL4-induced fibrosis | NR | 11 | NR | miR-200a, -200b |
| [46] | Microarray | Mouse | CCL4-induced fibrosis | 31 | 10 | 21 | miR-29 |
| miRNA | Target Genes | Potential Roles | References |
|---|---|---|---|
| miR-9a-5p | Sirt1 | Proliferation and migration | [49] |
| miR-17-5p | Smad7 | HSC activation and proliferation | [50] |
| miR-21 | PTEN, AP-1, SPRY2, HNF4α, PDCD4 | Collagen synthesis and HSC activation | [51,52,53] |
| miR-31 | FIH1 | Activation, proliferation, and migration | [54] |
| miR-33a | PPARΥ | HSC activation | [55,56] |
| miR-34a and -34c | PPARΥ, ACSL1 | HSC activation | [57,58] |
| miR-126 | IκBα | HSC activation and TGF-β expression | [59] |
| miR-130a and -130b | PPARΥ | HSC activation and proliferation | [60] |
| miR-181b | p27, PTEN | Cell proliferation | [61] |
| miR-214 | CCN2 | HSC activation | [62] |
| miR-214-5p | MMP-2, MMP-9, α-SMA, TGF-β1 | HSC ctivation | [63] |
| miR-221 and -222 | CDKN1B | Collagen deposition | [64] |
| miRNA | Target Genes | Potential Roles | References |
|---|---|---|---|
| miR-16 | CD1 | Apoptosis | [65] |
| miR-19b | GRB2, TGF-β1 | HSC proliferation | [43,66] |
| miR-29 | IGF-I, PDGF-C, HSP47, Col1A1 | Collagen expression and cell proliferation | [67,68,69,70] |
| miR-29a | HDAC4, Various ECM producing genes | HSC activation and ECM production | [71,72] |
| miR-29b | HSP47, Col1A1, SP1, PTEN | Collagen production and maturation | [73,74] |
| miR-30 | KLF11 | TGF-β1 expression | [75] |
| miR-101 | TβRI, KLF6 | Fibrinogenesis, proliferation, and migration | [76] |
| miR-122 | P4HA1, FN1, SRF, | Collagen production, maturation and ECM production | [77,78] |
| miR-126* | VEGF | Proliferation and cell contraction | [79] |
| miR-133a | ECM | Collagen production | [80] |
| miR-144 | TGF-β1 | HSC activation | [81] |
| miR-146a | SMAD4, Wnt1, Wnt5a | Proliferation and proliferation | [39,82] |
| miR-150 | c-myb, SP1, Col4A4 | HSC activation and collagen production | [83,84] |
| miR-155 | TCF4 and AGTR1 | HSC activation | [85] |
| miR-195 | cyclin E1, p21 | Cell proliferation | [86] |
| miR-200a | α-SMA, β-catenin, TGF-β2, Keap1 | HSC activation and proliferation | [87,88] |
| miR-214 | CCN2 | Hepatic fibrosis through α-SMA | [89] |
| miR-335 | TNC | HSC migration, production of α-SMA, and collagen type I | [90] |
| miR-370 | SMO | Proliferation | [91] |
| miR-454 | Smad4 | HSC activation | [92] |
| miR-483 | PDGF-β, TIM2 | HSC activation | [93] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitano, M.; Bloomston, P.M. Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis. J. Clin. Med. 2016, 5, 38. https://doi.org/10.3390/jcm5030038
Kitano M, Bloomston PM. Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis. Journal of Clinical Medicine. 2016; 5(3):38. https://doi.org/10.3390/jcm5030038
Chicago/Turabian StyleKitano, Mio, and P. Mark Bloomston. 2016. "Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis" Journal of Clinical Medicine 5, no. 3: 38. https://doi.org/10.3390/jcm5030038
APA StyleKitano, M., & Bloomston, P. M. (2016). Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis. Journal of Clinical Medicine, 5(3), 38. https://doi.org/10.3390/jcm5030038





