Molecular Genetic Markers in Acute Myeloid Leukemia
Abstract
:1. Introduction
2. Genetics and AML Classification
3. Cytogenetic Abnormalities and Prognosis
| AML with Recurrent Genetic Abnormalities | AML with Myelodysplasia Related Changes |
|---|---|
| RUNX1-RUNX1T1 t(8;21)(q22;q22) | Complex karyotype (≥3 unrelated abnormalities) |
| CBFB-MYH11 inv(16)(p12.1q22) or t(16;16)(p13.1;q22) | −7/del(7q), −5/del(5q) |
| PML-RARA t(15;17)(q22;q12) | −13/del(13q), del(11q), del(12p)/t(12p), del(9q) |
| MLLT3-MLL/KMT2A t(9;11)(q22;q23) | i(17q)/t(17p), idic(X)(q13) |
| DEK-NUP214 t(6;9)(p23;q34) | t(5;12)(q33;p12), t(5;7)(q33;q11.2) t(5;17)(q33;p13), t(5;10)(q33;q21) |
| RPN-EVI1 inv(3)(q21q26.2) or t(3;3)(q21;q26.2) | t(1;3)(p36.3;q21.2), t(3;5)(q25;q34) |
| RBM15-MKL1 t(1;22)(p13;q13) | t(11;16)(q23;p13.3) *, t(3;21)(q26.2;q22.1) * |
| NPM1 gene mutation (provisional entity) | t(2;11)(p21;q23) * |
| Mutated CEBPA (provisional entity) |
| Risk | Cytogenetics | Molecular |
|---|---|---|
| Favorable | inv(16) or t(16;16) | Normal cytogenetics with: |
| t(8;21) | Isolated biallelic CEBPA mutation | |
| t(15;17) | NPM1 mutation without FLT3 ITD | |
| Intermediate | Normal cytogenetics | KIT mutation in core binding factor leukemia: inv(16) or t(16;16) t(8;21) |
| Isolated +8 | ||
| t(9;11) | ||
| Other non-good and non-poor changes | ||
| Poor | Complex (≥3 clonal abnormalities) | Normal cytogenetics with: FLT3 ITD |
| Monosomal karyotype * | ||
| −5/−5q or −7/−7q | ||
| 11q23 rearrangements other than t(9;11) | ||
| inv(3) or t(3;3) | ||
| t(6;9) | ||
| t(9;22) |
4. Established Gene Mutations Associated with Prognosis
4.1. FLT3 (Fms-like Tyrosine Kinase 3)
4.2. NPM1 (Nucleophosmin 1)
4.3. CEBPA (CCAAT Enhancer Binding Protein)
4.4. KIT (v-KIT Hardy-Zuckerman 4 Feline Sa12rcoma Viral Oncogene Homolog)
5. Other Gene Mutations in AML
| Gene | Frequency | Effect |
|---|---|---|
| ASXL1 | 3%–5% | Associated with MDS, AML-MRC. Worse prognosis [19,29,42,43,44]. |
| BCOR | 4% CN-AML | Possible worse prognosis [45]. |
| DNMT3A | 20% | Possible worse prognosis. May respond to high dose anthracyclines [18,29]. |
| IDH1 | 6%–9% adult | Possible worse prognosis [29,46,47,48,49]. |
| 1% pediatric | ||
| IDH2 | 8%–12% adult | Controversial. IDH2 R140 mutation with NPM1 associated with a favorable prognosis in one study [29,46,47,48,49]. |
| 1%–2% pediatric | ||
| MLL/KMT2A | 4%–14% | MLL PTD shows worse prognosis in CN-AML [18,19,29,30,31]. |
| NRAS | 8%–13% adult and pediatric | No clear impact on prognosis [50,51]. |
| KRAS | 2% adult | No clear impact on prognosis [52]. |
| 9% pediatric | ||
| PHF6 | 2%–3% | Associated with adverse outcome [29]. |
| RUNX1 | 5%–18% | Possibly poorer prognosis. May do better with allogeneic transplant [19,29,53]. |
| TET2 | 7%–10% adult | Unclear, some studies show adverse outcome especially in intermediate risk AML with isolated CEBPA or NPM1 [18,29,54,55]. |
| 1.5%–4% pediatric | ||
| TP53 | 2%–9% adult | Unfavorable prognosis [18,19]. Mutations may be germline (Li-Fraumeni syndrome) and this possibility should be considered when testing especially in younger individuals. |
| 1% pediatric | ||
| WT1 | 4%–11% | Poorer outcome, especially in CN-AML [56,57]. |
5.1. ASXL1 (Additional Sex Combs like Transcriptional Regulator 1)
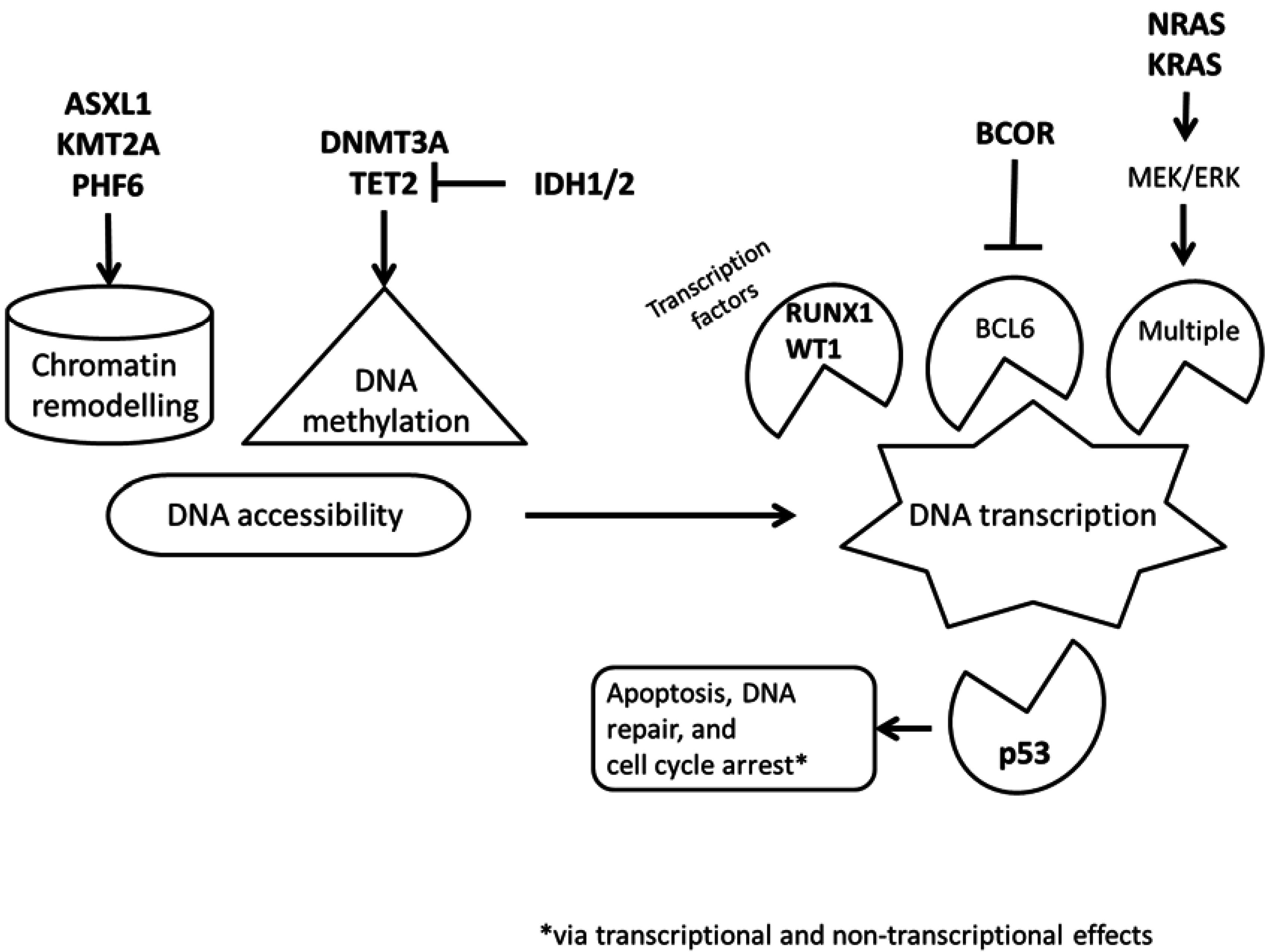
5.2. BCOR (BCL6 Corepressor)
5.3. DNMT3A (DNA Methyltransferase 3A)
5.4. IDH1 and IDH2 (Isocitrate Dehydrogenase 1 and 2)
5.5. MLL/KMT2A (Mixed Lineage Leukemia/Lysine (K)-Specific Methyltransferase 2A)
5.6. NRAS and KRAS (Neuroblastoma RAS Viral (v-ras) Oncogene Homolog and Kirsten Rat Sarcoma Viral Oncogene Homolog)
5.7. PHF6 (Plant Homeodomain Finger 6)
5.8. RUNX1 (Runt Related Transcription Factor 1)
5.9. TET2 (Tet Methylcytosine Dioxygenase 2)
5.10. TP53 (Tumor Protein p53)
5.11. WT1 (Wilms Tumor 1)
6. Conclusions
Author Contributions
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J.; Vardiman, J.W. Who Classification of Tumours of Hematopoietic and Lymphoid Tissues, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2008. [Google Scholar]
- O’Donnel, M.R.; Tallman, M.S.; Abboud, C.N.; Altman, J.K.; Appelbaum, F.R.; Arber, D.A.; Attar, E.; Borate, U.; Damon, L.E.; Gregory, K.; et al. National comprehensive cancer network: NCCN categories of evidence and consensus. Available online: http://www.nccn.org/professionals/physician_gls/categories_of_consensus.asp (accessed on 13 January 2015).
- Ofran, Y.; Rowe, J.M. Genetic profiling in acute myeloid leukaemia—Where are we and what is its role in patient management. Br. J. Haematol. 2013, 160, 303–320. [Google Scholar]
- Martelli, M.P.; Sportoletti, P.; Tiacci, E.; Martelli, M.F.; Falini, B. Mutational landscape of AML with normal cytogenetics: Biological and clinical implications. Blood Rev. 2013, 27, 13–22. [Google Scholar] [PubMed]
- Whitman, S.P.; Archer, K.J.; Feng, L.; Baldus, C.; Becknell, B.; Carlson, B.D.; Carroll, A.J.; Mrózek, K.; Vardiman, J.W.; George, S.L.; et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A cancer and leukemia group B study. Cancer Res. 2001, 61, 7233–7239. [Google Scholar] [PubMed]
- Gale, R.E.; Green, C.; Allen, C.; Mead, A.J.; Burnett, A.K.; Hills, R.K.; Linch, D.C.; Party, M.R.C.A.L.W. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood 2008, 111, 2776–2784. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Bacher, U.; Haferlach, C.; Alpermann, T.; Kern, W.; Haferlach, T. Diversity of the juxtamembrane and TKD1 mutations (exons 13–15) in the FLT3 gene with regards to mutant load, sequence, length, localization, and correlation with biological data. Genes Chromosomes Cancer 2012, 51, 910–924. [Google Scholar] [CrossRef] [PubMed]
- Schlenk, R.F.; Kayser, S.; Bullinger, L.; Kobbe, G.; Casper, J.; Ringhoffer, M.; Held, G.; Brossart, P.; Lübbert, M.; Salih, H.R.; et al. Differential impact of allelic ratio and insertion site in FLT3-ITD positive AML with respect to allogeneic transplantation. Blood 2014, 124, 3441–3449. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Schlenk, R.F.; Londono, M.C.; Breitenbuecher, F.; Wittke, K.; Du, J.; Groner, S.; Späth, D.; Krauter, J.; Ganser, A.; et al. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood 2009, 114, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Blau, O.; Berenstein, R.; Sindram, A.; Blau, I.W. Molecular analysis of different FLT3-ITD mutations in acute myeloid leukemia. Leuk. Lymphoma 2013, 54, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Meshinchi, S.; Stirewalt, D.L.; Alonzo, T.A.; Boggon, T.J.; Gerbing, R.B.; Rocnik, J.L.; Lange, B.J.; Gilliland, D.G.; Radich, J.P. Structural and numerical variation of FLT3-ITD in pediatric AML. Blood 2008, 111, 4930–4933. [Google Scholar] [CrossRef] [PubMed]
- Stirewalt, D.L.; Kopecky, K.J.; Meshinchi, S.; Engel, J.H.; Pogosova-Agadjanyan, E.L.; Linsley, J.; Slovak, M.L.; Willman, C.L.; Radich, J.P. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood 2006, 107, 3724–3726. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, V.; Gianfaldoni, G.; Mannelli, F.; Leoni, F.; Ciolli, S.; Guglielmelli, P.; Antonioli, E.; Longo, G.; Bosi, A.; Vannucchi, A.M. The size of duplication does not add to the prognostic significance of FLT3 internal tandem duplication in acute myeloid leukemia patients. Leukemia 2006, 20, 2074–2076. [Google Scholar] [CrossRef] [PubMed]
- Breitenbuecher, F.; Schnittger, S.; Grundler, R.; Markova, B.; Carius, B.; Brecht, A.; Duyster, J.; Haferlach, T.; Huber, C.; Fischer, T. Identification of a novel type of itd mutations located in nonjuxtamembrane domains of the FLT3 tyrosine kinase receptor. Blood 2009, 113, 4074–4077. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, Y.; Kantarjian, H.M.; Luthra, R.; Ravandi, F.; Borthakur, G.; Garcia-Manero, G.; Konopleva, M.; Estrov, Z.; Andreeff, M.; Cortes, J.E. Treatment with FLT3 inhibitor in patients with FLT3-mutated acute myeloid leukemia is associated with development of secondary FLT3-tyrosine kinase domain mutations. Cancer 2014, 120, 2142–2149. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Albiero, E.; Bolli, N.; De Marco, M.F.; Madeo, D.; Martelli, M.; Nicoletti, I.; Rodeghiero, F. Aberrant cytoplasmic expression of C-terminal-truncated NPM leukaemic mutant is dictated by tryptophans loss and a new NES motif. Leukemia 2007, 21, 2052–2054. [Google Scholar] [CrossRef] [PubMed]
- Schlenk, R.F.; Döhner, K.; Krauter, J.; Fröhling, S.; Corbacioglu, A.; Bullinger, L.; Habdank, M.; Späth, D.; Morgan, M.; Benner, A.; et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008, 358, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Kihara, R.; Nagata, Y.; Kiyoi, H.; Kato, T.; Yamamoto, E.; Suzuki, K.; Chen, F.; Asou, N.; Ohtake, S.; Miyawaki, S.; et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia 2014, 28, 1586–1595. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Schnittger, S.; Kohlmann, A.; Eder, C.; Roller, A.; Dicker, F.; Schmid, C.; Wendtner, C.M.; Staib, P.; Serve, H.; et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood 2012, 120, 2963–2972. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Bacher, U.; Kern, W.; Alpermann, T.; Haferlach, C.; Haferlach, T. Prognostic impact of FLT3-ITD load in NPM1 mutated acute myeloid leukemia. Leukemia 2011, 25, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Macijewski, K.; Weiss, T.; Bacher, U.; Schnittger, S.; Kern, W.; Kohlmann, A.; Klein, H.U.; Vignetti, M.; Piciocchi, A.; et al. Multilineage dysplasia has no impact on biologic, clinicopathologic, and prognostic features of aml with mutated nucleophosmin (NPM1). Blood 2010, 115, 3776–3786. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, C.; Mecucci, C.; Schnittger, S.; Kohlmann, A.; Mancini, M.; Cuneo, A.; Testoni, N.; Rege-Cambrin, G.; Santucci, A.; Vignetti, M.; et al. AML with mutated NPM1 carrying a normal or aberrant karyotype show overlapping biologic, pathologic, immunophenotypic, and prognostic features. Blood 2009, 114, 3024–3032. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Deng, D.H.; Huang, Y.; Ye, F.H.; Huang, L.L.; Xiao, Q.; Zhang, B.; Ye, B.B.; Lai, Y.R.; Mo, Z.N.; et al. Favorable prognosis of biallelic CEBPA gene mutations in acute myeloid leukemia patients: A meta-analysis. Eur. J. Haematol. 2014, in press. [Google Scholar]
- Pastore, F.; Kling, D.; Hoster, E.; Dufour, A.; Konstandin, N.P.; Schneider, S.; Sauerland, M.C.; Berdel, W.E.; Buechner, T.; Woermann, B.; et al. Long-term follow-up of cytogenetically normal CEBPA-mutated AML. J. Hematol. Oncol. 2014, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Wouters, B.J.; Löwenberg, B.; Erpelinck-Verschueren, C.A.; van Putten, W.L.; Valk, P.J.; Delwel, R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009, 113, 3088–3091. [Google Scholar] [CrossRef] [PubMed]
- Mueller, B.U.; Pabst, T. C/EBPalpha and the pathophysiology of acute myeloid leukemia. Curr. Opin. Hematol. 2006, 13, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Nerlov, C. C/EBPalpha mutations in acute myeloid leukaemias. Nat. Rev. Cancer 2004, 4, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Pabst, T.; Mueller, B.U.; Zhang, P.; Radomska, H.S.; Narravula, S.; Schnittger, S.; Behre, G.; Hiddemann, W.; Tenen, D.G. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (c/EBPalpha), in acute myeloid leukemia. Nat. Genet. 2001, 27, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.P.; Gönen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhu, Y.M.; Fan, X.; Shi, J.Y.; Wang, Q.R.; Yan, X.J.; Gu, Z.H.; Wang, Y.Y.; Chen, B.; Jiang, C.L.; et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood 2011, 118, 5593–5603. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Xu, Y.; Yin, J.; Tian, H.; Chen, S.; Wu, D.; Sun, A. TET2 gene mutation is unfavorable prognostic factor in cytogenetically normal acute myeloid leukemia patients with NPM1+ and FLT3-ITD- mutations. Int. J. Hematol. 2014, 100, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Care, R.S.; Valk, P.J.; Goodeve, A.C.; Abu-Duhier, F.M.; Geertsma-Kleinekoort, W.M.; Wilson, G.A.; Gari, M.A.; Peake, I.R.; Löwenberg, B.; Reilly, J.T. Incidence and prognosis of c-KIT and FLT3 mutations in core binding factor (CBF) acute myeloid leukaemias. Br. J. Haematol. 2003, 121, 775–777. [Google Scholar] [CrossRef] [PubMed]
- Beghini, A.; Ripamonti, C.B.; Cairoli, R.; Cazzaniga, G.; Colapietro, P.; Elice, F.; Nadali, G.; Grillo, G.; Haas, O.A.; Biondi, A.; et al. KIT activating mutations: Incidence in adult and pediatric acute myeloid leukemia, and identification of an internal tandem duplication. Haematologica 2004, 89, 920–925. [Google Scholar] [PubMed]
- Mrózek, K.; Marcucci, G.; Paschka, P.; Bloomfield, C.D. Advances in molecular genetics and treatment of core-binding factor acute myeloid leukemia. Curr. Opin. Oncol. 2008, 20, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Paschka, P.; Marcucci, G.; Ruppert, A.S.; Mrózek, K.; Chen, H.; Kittles, R.A.; Vukosavljevic, T.; Perrotti, D.; Vardiman, J.W.; Carroll, A.J.; et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): A cancer and leukemia group B study. J. Clin. Oncol. 2006, 24, 3904–3911. [Google Scholar] [CrossRef] [PubMed]
- Cairoli, R.; Beghini, A.; Grillo, G.; Nadali, G.; Elice, F.; Ripamonti, C.B.; Colapietro, P.; Nichelatti, M.; Pezzetti, L.; Lunghi, M.; et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: An Italian retrospective study. Blood 2006, 107, 3463–3468. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Chi, H.S.; Min, S.K.; Park, B.G.; Jang, S.; Park, C.J. Prognostic impact of c-KIT mutations in core binding factor acute myeloid leukemia. Leuk. Res. 2011, 35, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Boissel, N.; Leroy, H.; Brethon, B.; Philippe, N.; de Botton, S.; Auvrignon, A.; Raffoux, E.; Leblanc, T.; Thomas, X.; Hermine, O.; et al. Incidence and prognostic impact of c-KIT, FLT3, and RAS gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia 2006, 20, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Jourdan, E.; Boissel, N.; Chevret, S.; Delabesse, E.; Renneville, A.; Cornillet, P.; Blanchet, O.; Cayuela, J.M.; Recher, C.; Raffoux, E.; et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood 2013, 121, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Cairoli, R.; Beghini, A.; Turrini, M.; Bertani, G.; Nadali, G.; Rodeghiero, F.; Castagnola, C.; Lazzaroni, F.; Nichelatti, M.; Ferrara, F.; et al. Old and new prognostic factors in acute myeloid leukemia with deranged core-binding factor beta. Am. J. Hematol. 2013, 88, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the european leukemianet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef] [PubMed]
- Devillier, R.; Gelsi-Boyer, V.; Brecqueville, M.; Carbuccia, N.; Murati, A.; Vey, N.; Birnbaum, D.; Mozziconacci, M.J. Acute myeloid leukemia with myelodysplasia-related changes are characterized by a specific molecular pattern with high frequency of ASXL1 mutations. Am. J. Hematol. 2012, 87, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Mercado, M.; Yip, B.H.; Pellagatti, A.; Davies, C.; Larrayoz, M.J.; Kondo, T.; Pérez, C.; Killick, S.; McDonald, E.J.; Odero, M.D.; et al. Mutation patterns of 16 genes in primary and secondary acute myeloid leukemia (AML) with normal cytogenetics. PLoS One 2012, 7, e42334. [Google Scholar] [CrossRef] [PubMed]
- Metzeler, K.H.; Becker, H.; Maharry, K.; Radmacher, M.D.; Kohlschmidt, J.; Mrózek, K.; Nicolet, D.; Whitman, S.P.; Wu, Y.Z.; Schwind, S.; et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal aml within the eln favorable genetic category. Blood 2011, 118, 6920–6929. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Tiacci, E.; Holmes, A.B.; Kohlmann, A.; Martelli, M.P.; Kern, W.; Spanhol-Rosseto, A.; Klein, H.U.; Dugas, M.; Schindela, S.; et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 2011, 118, 6153–6163. [Google Scholar] [CrossRef] [PubMed]
- Paschka, P.; Schlenk, R.F.; Gaidzik, V.I.; Habdank, M.; Krönke, J.; Bullinger, L.; Späth, D.; Kayser, S.; Zucknick, M.; Götze, K.; et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010, 28, 3636–3643. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Maharry, K.; Wu, Y.Z.; Radmacher, M.D.; Mrózek, K.; Margeson, D.; Holland, K.B.; Whitman, S.P.; Becker, H.; Schwind, S.; et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A cancer and leukemia group Bstudy. J. Clin. Oncol. 2010, 28, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Damm, F.; Wagner, K.; Göhring, G.; Schlegelberger, B.; Hoelzer, D.; Lübbert, M.; Heit, W.; Kanz, L.; Schlimok, G.; et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood 2010, 116, 614–616. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Lugthart, S.; Kavelaars, F.G.; Schelen, A.; Koenders, J.E.; Zeilemaker, A.; van Putten, W.J.; Rijneveld, A.W.; Löwenberg, B.; Valk, P.J. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: Prevalence and prognostic value. Blood 2010, 116, 2122–2126. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.N.; Gerbing, R.B.; Alonzo, T.A.; Ho, P.A.; Miller, K.; Hurwitz, C.; Heerema, N.A.; Hirsch, B.; Raimondi, S.C.; Lange, B.; et al. Prevalence and clinical implications of NRAS mutations in childhood AML: A report from the children's oncology group. Leukemia 2011, 25, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Bacher, U.; Haferlach, T.; Schoch, C.; Kern, W.; Schnittger, S. Implications of NRAS mutations in AML: A study of 2502 patients. Blood 2006, 107, 3847–3853. [Google Scholar] [CrossRef] [PubMed]
- Bowen, D.T.; Frew, M.E.; Hills, R.; Gale, R.E.; Wheatley, K.; Groves, M.J.; Langabeer, S.E.; Kottaridis, P.D.; Moorman, A.V.; Burnett, A.K.; et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood 2005, 106, 2113–2119. [Google Scholar] [CrossRef] [PubMed]
- Gaidzik, V.I.; Bullinger, L.; Schlenk, R.F.; Zimmermann, A.S.; Röck, J.; Paschka, P.; Corbacioglu, A.; Krauter, J.; Schlegelberger, B.; Ganser, A.; et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the AML study group. J. Clin. Oncol. 2011, 29, 1364–1372. [Google Scholar] [CrossRef] [PubMed]
- Gaidzik, V.I.; Paschka, P.; Späth, D.; Habdank, M.; Köhne, C.H.; Germing, U.; von Lilienfeld-Toal, M.; Held, G.; Horst, H.A.; Haase, D.; et al. TET2 mutations in acute myeloid leukemia (AML): Results from a comprehensive genetic and clinical analysis of the AML study group. J. Clin. Oncol. 2012, 30, 1350–1357. [Google Scholar] [CrossRef] [PubMed]
- Metzeler, K.H.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Margeson, D.; Becker, H.; Curfman, J.; Holland, K.B.; Schwind, S.; Whitman, S.P.; et al. TET2 mutations improve the new European leukemianet risk classification of acute myeloid leukemia: A cancer and leukemia group B study. J. Clin. Oncol. 2011, 29, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Krauth, M.T.; Alpermann, T.; Bacher, U.; Eder, C.; Dicker, F.; Ulke, M.; Kuznia, S.; Nadarajah, N.; Kern, W.; Haferlach, C.; et al. WT1 mutations are secondary events in AML, show varying frequencies and impact on prognosis between genetic subgroups. Leukemia 2014. [Google Scholar]
- Sano, H.; Shimada, A.; Tabuchi, K.; Taki, T.; Murata, C.; Park, M.J.; Ohki, K.; Sotomatsu, M.; Adachi, S.; Tawa, A.; et al. WT1 mutation in pediatric patients with acute myeloid leukemia: A report from the Japanese childhood AML cooperative study group. Int. J. Hematol. 2013, 98, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Eder, C.; Jeromin, S.; Alpermann, T.; Fasan, A.; Grossmann, V.; Kohlmann, A.; Illig, T.; Klopp, N.; Wichmann, H.E.; et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia 2013, 27, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.C.; Liu, H.C.; Yang, C.P.; Jaing, T.H.; Hung, I.J.; Yeh, T.C.; Chen, S.H.; Hou, J.Y.; Huang, Y.J.; Shih, Y.S.; et al. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood 2013, 121, 2988–2995. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawi, D.; Ali, A.; Evans, C.M.; Hills, R.K.; Burnett, A.K.; Linch, D.C.; Gale, R.E. ASXL1 mutations are infrequent in young patients with primary acute myeloid leukemia and their detection has a limited role in therapeutic risk stratification. Leuk. Lymphoma 2014, 55, 1326–1331. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.C.; Huang, H.H.; Hou, H.A.; Chen, C.Y.; Tang, J.L.; Yao, M.; Tsay, W.; Ko, B.S.; Wu, S.J.; Huang, S.Y.; et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood 2010, 116, 4086–4094. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.D.; Fischle, W.; Verdin, E.; Bardwell, V.J. BCOR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000, 14, 1810–1823. [Google Scholar] [PubMed]
- Herold, T.; Metzeler, K.H.; Vosberg, S.; Hartmann, L.; Röllig, C.; Stölzel, F.; Schneider, S.; Hubmann, M.; Zellmeier, E.; Ksienzyk, B.; et al. Isolated trisomy 13 defines a homogeneous aml subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood 2014, 124, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Thota, S.; Viny, A.D.; Makishima, H.; Spitzer, B.; Radivoyevitch, T.; Przychodzen, B.; Sekeres, M.A.; Levine, R.L.; Maciejewski, J.P. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 2014, 124, 1790–1798. [Google Scholar] [CrossRef] [PubMed]
- Ibrahem, L.; Mahfouz, R.; Elhelw, L.; Abdsalam, E.M.; Soliman, R. Prognostic significance of DNMT3A mutations in patients with acute myeloid leukemia. Blood Cells Mol. Dis. 2015, 54, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Cross, J.R.; Lu, C.; Weigert, O.; Abel-Wahab, O.; Levine, R.L.; Weinstock, D.M.; Sharp, K.A.; Thompson, C.B. Identification of additional IDH mutations associated with oncometabolite r(-)-2-hydroxyglutarate production. Oncogene 2012, 31, 2491–2498. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Evans, C.M.; Hills, R.K.; Burnett, A.K.; Linch, D.C.; Gale, R.E. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood 2010, 116, 2779–2782. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.A.; Kutny, M.A.; Alonzo, T.A.; Gerbing, R.B.; Joaquin, J.; Raimondi, S.C.; Gamis, A.S.; Meshinchi, S. Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: A report from the children’s oncology group. Pediatr. Blood Cancer 2011, 57, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Evans, C.M.; Zhao, L.; Hills, R.K.; Burnett, A.K.; Linch, D.C.; Gale, R.E. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 2011, 118, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, T.; Meyer, C.; Schwartz, S.; Hofmann, J.; Molkentin, M.; Kowarz, E.; Schneider, B.; Raff, T.; Reinhardt, R.; Gökbuget, N.; et al. The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia: Results from the gmall study group. Blood 2009, 113, 4011–4015. [Google Scholar] [CrossRef] [PubMed]
- Goemans, B.F.; Zwaan, C.M.; Miller, M.; Zimmermann, M.; Harlow, A.; Meshinchi, S.; Loonen, A.H.; Hählen, K.; Reinhardt, D.; Creutzig, U.; et al. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia 2005, 19, 1536–1542. [Google Scholar] [CrossRef] [PubMed]
- Todd, M.A.; Picketts, D.J. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J. Proteome Res. 2012, 11, 4326–4337. [Google Scholar] [CrossRef] [PubMed]
- Lower, K.M.; Turner, G.; Kerr, B.A.; Mathews, K.D.; Shaw, M.A.; Gedeon, A.K.; Schelley, S.; Hoyme, H.E.; White, S.M.; Delatycki, M.B.; et al. Mutations in PHF6 are associated with Börjeson-Forssman-Lehmann syndrome. Nat. Genet. 2002, 32, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Yoo, N.J.; Kim, Y.R.; Lee, S.H. Somatic mutation of PHF6 gene in T-cell acute lymphoblatic leukemia, acute myelogenous leukemia and hepatocellular carcinoma. Acta Oncol. 2012, 51, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Van Vlierberghe, P.; Patel, J.; Abdel-Wahab, O.; Lobry, C.; Hedvat, C.V.; Balbin, M.; Nicolas, C.; Payer, A.R.; Fernandez, H.F.; Tallman, M.S.; et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia 2011, 25, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Preudhomme, C.; Renneville, A.; Bourdon, V.; Philippe, N.; Roche-Lestienne, C.; Boissel, N.; Dhedin, N.; André, J.M.; Cornillet-Lefebvre, P.; Baruchel, A.; et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood 2009, 113, 5583–5587. [Google Scholar] [CrossRef] [PubMed]
- Aslanyan, M.G.; Kroeze, L.I.; Langemeijer, S.M.; Koorenhof-Scheele, T.N.; Massop, M.; van Hoogen, P.; Stevens-Linders, E.; van de Locht, L.T.; Tönnissen, E.; van der Heijden, A.; et al. Clinical and biological impact of TET2 mutations and expression in younger adult AML patients treated within the EORTC/GIMEMA AML-12 clinical trial. Ann. Hematol. 2014, 93, 1401–1412. [Google Scholar] [PubMed]
- Langemeijer, S.M.; Jansen, J.H.; Hooijer, J.; van Hoogen, P.; Stevens-Linders, E.; Massop, M.; Waanders, E.; van Reijmersdal, S.V.; Stevens-Kroef, M.J.; Zwaan, C.M.; et al. TET2 mutations in childhood leukemia. Leukemia 2011, 25, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Busque, L.; Patel, J.P.; Figueroa, M.E.; Vasanthakumar, A.; Provost, S.; Hamilou, Z.; Mollica, L.; Li, J.; Viale, A.; Heguy, A.; et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 2012, 44, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, Y.; Suarez Saiz, F.; Saurez Saiz, F.; Minden, M.D. A tumor suppressor and oncogene: The WT1 story. Leukemia 2007, 21, 868–876. [Google Scholar] [PubMed]
- Lyu, X.; Xin, Y.; Mi, R.; Ding, J.; Wang, X.; Hu, J.; Fan, R.; Wei, X.; Song, Y.; Zhao, R.Y. Overexpression of wilms tumor 1 gene as a negative prognostic indicator in acute myeloid leukemia. PLoS One 2014, 9, e92470. [Google Scholar] [CrossRef] [PubMed]
- Woehlecke, C.; Wittig, S.; Arndt, C.; Gruhn, B. Prognostic impact of WT1 expression prior to hematopoietic stem cell transplantation in children with malignant hematological diseases. J. Cancer Res. Clin. Oncol. 2014. [Google Scholar]
- Rossi, G.; Carella, A.M.; Minervini, M.M.; Savino, L.; Fontana, A.; Pellegrini, F.; Greco, M.M.; Merla, E.; Quarta, G.; Loseto, G.; et al. Minimal residual disease after allogeneic stem cell transplant: A comparison among multiparametric flow cytometry, wilms tumor 1 expression and chimerism status (complete chimerism versus low level mixed chimerism) in acute leukemia. Leuk. Lymphoma 2013, 54, 2660–2666. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Kim, H.J.; Shin, S.H.; Yahng, S.A.; Lee, S.E.; Cho, B.S.; Eom, K.S.; Kim, Y.J.; Lee, S.; Min, C.K.; et al. Serial measurement of WT1 expression and decrement ratio until hematopoietic cell transplantation as a marker of residual disease in patients with cytogenetically normal acute myelogenous leukemia. Biol. Blood Marrow Transpl. 2013, 19, 958–966. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yohe, S. Molecular Genetic Markers in Acute Myeloid Leukemia. J. Clin. Med. 2015, 4, 460-478. https://doi.org/10.3390/jcm4030460
Yohe S. Molecular Genetic Markers in Acute Myeloid Leukemia. Journal of Clinical Medicine. 2015; 4(3):460-478. https://doi.org/10.3390/jcm4030460
Chicago/Turabian StyleYohe, Sophia. 2015. "Molecular Genetic Markers in Acute Myeloid Leukemia" Journal of Clinical Medicine 4, no. 3: 460-478. https://doi.org/10.3390/jcm4030460
APA StyleYohe, S. (2015). Molecular Genetic Markers in Acute Myeloid Leukemia. Journal of Clinical Medicine, 4(3), 460-478. https://doi.org/10.3390/jcm4030460






