Impact of Immunosuppression on Immune Cell Dynamics in COVID-19: A Serial Comparison of Leukocyte Data in Healthy and Immunocompromised Patients Before and After Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
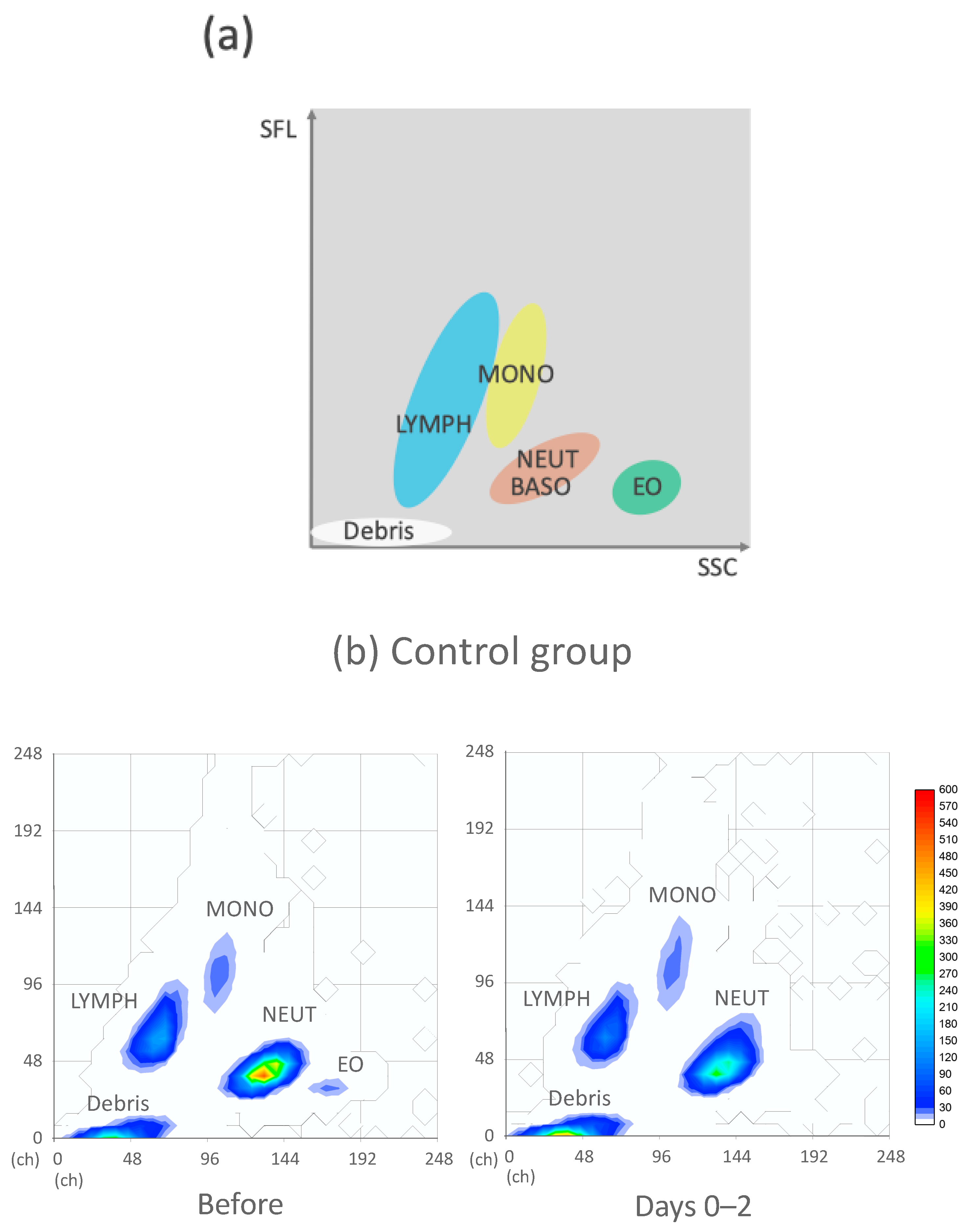
2.2. CPD and WBC Differential Fluorescence (WDF) Scattergram
2.3. Statistical Analysis
3. Results
3.1. Study Populations
3.2. Baseline Characteristics Between the Control and IST Groups
3.3. Changes in CBC and CPD in the Control Group
3.4. Changes in CBC and CPD in the IST Group
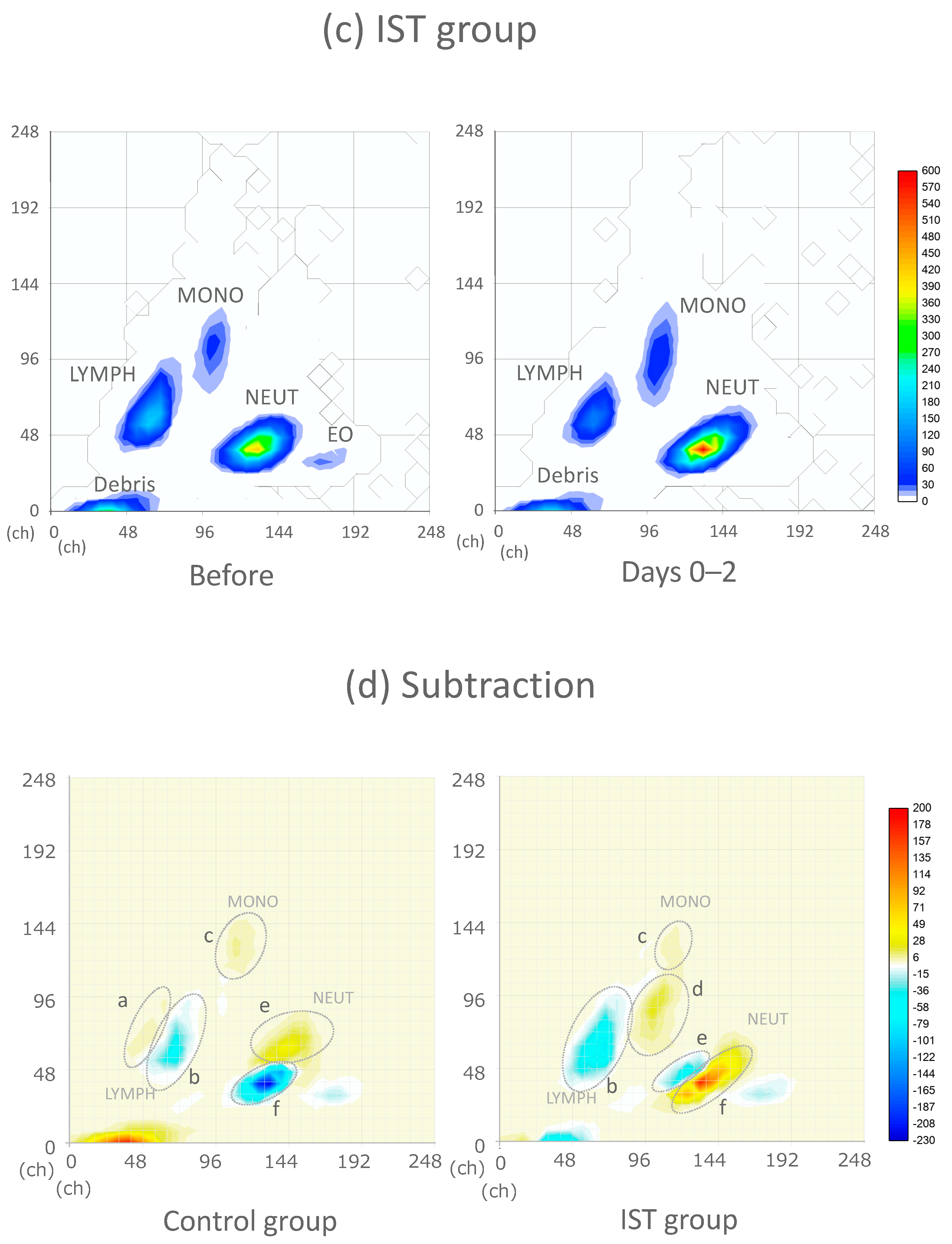
3.5. WDF Scattergram of the Control and IST Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMI | body mass index |
| COPD | chronic obstructive pulmonary disease |
| WBC | white blood cell count |
| HGB | hemoglobin concentration |
| HCT | hematocrit |
| PLT | absolute number of thrombocytes |
| NEUT# | neutrophil count |
| LYMPH# | lymphocyte count |
| MONO# | monocyte count |
| EO# | eosinophilic count |
| BASO# | basophilic count |
| NE-SSC | neutrophil complexity |
| NE-SFL | neutrophil fluorescence |
| NE-FSC | neutrophil size |
| NE-WX | width of dispersion of neutrophil complexity |
| NE-WY | width of dispersion of neutrophil fluorescence |
| NE-WZ | width of dispersion of neutrophil size |
| LY-X | lymphocyte complexity |
| LY-Y | lymphocyte fluorescence |
| LY-Z | lymphocyte size |
| LY-WX | width of dispersion of lymphocyte complexity |
| LY-WY | width of dispersion of lymphocyte fluorescence |
| LY-WZ | width of dispersion of lymphocyte size |
| MO-X | monocyte complexity |
| MO-Y | monocyte fluorescence |
| MO-Z | monocyte size |
| MO-WX | width of dispersion of monocyte complexity |
| MO-WY | width of dispersion of monocyte fluorescence |
| MO-WZ | width of dispersion of monocyte size |
| R-CHOP | rituximab, doxorubicin, vincristine, cyclophosphamide, prednisolone |
| S-1 | tegaful/gimeracil/oteracil potassium |
| CRP | C-reactive protein |
| LD | lactate dehydrogenase |
| AST | aspartate transaminase |
| ALT | alanine transaminase |
| eGFR | estimated glomerular filtration rate |
References
- Ketkar, A.; Willey, V.; Pollack, M.; Glasser, L.; Dobie, C.; Wenziger, C.; Teng, C.C.; Dube, C.; Cunningham, D.; Verduzco-Gutierrez, M. Assessing the risk and costs of COVID-19 in immunocompromised populations in a large United States commercial insurance health plan: The EPOCH-US Study. Curr. Med. Res. Opin. 2023, 39, 1103–1118. [Google Scholar] [CrossRef]
- Razonable, R.R. Protecting the vulnerable: Addressing the COVID-19 care needs of people with compromised immunity. Front. Immunol. 2024, 15, 1397040. [Google Scholar] [CrossRef]
- Belsky, J.A.; Tullius, B.P.; Lamb, M.G.; Sayegh, R.; Stanek, J.R.; Auletta, J.J. COVID-19 in immunocompromised patients: A systematic review of cancer, hematopoietic cell and solid organ transplant patients. J. Infect. 2021, 82, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Schapkaitz, E.; Raburabu, S. Performance evaluation of the new measurement channels on the automated Sysmex XN-9000 hematology analyzer. Clin. Biochem. 2018, 53, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Osman, J.; Lambert, J.; Temple, M.; Devaux, F.; Favre, R.; Flaujac, C.; Bridoux, D.; Marque-Juillet, S.; Bruneel, F.; Mignon, F.; et al. Rapid screening of COVID-19 patients using white blood cell scattergrams, a study on 381 patients. Br. J. Haematol. 2020, 190, 718–722. [Google Scholar] [CrossRef]
- Lapic, I.; Brencic, T.; Rogic, D.; Lukic, M.; Lukic, I.; Kovacic, M.; Honovic, L.; Seric, V. Cell population data: Could a routine hematology analyzer aid in the differential diagnosis of COVID-19? Int. J. Lab. Hematol. 2021, 43, e64–e67. [Google Scholar] [CrossRef] [PubMed]
- Ono, D.; Ohno, Y.; Izumida, Y.; Ohno, H.; Oka, H.; Takeshita, K. Inflammation as an exacerbation marker and target for prophylaxis against Coronavirus Disease 2019-related thrombosis. Int. J. Med. Sci. 2023, 20, 136–141. [Google Scholar] [CrossRef]
- Briggs, C.; Longair, I.; Kumar, P.; Singh, D.; Machin, S.J. Performance evaluation of the Sysmex haematology XN modular system. J. Clin. Pathol. 2012, 65, 1024–1030. [Google Scholar] [CrossRef]
- Introcaso, G.; Galotta, A.; Salvini, L.; Faioni, E.M.; Bonomi, A.; Assanelli, E.; Biondi, M.L. Leukocyte cell population data as potential markers of COVID-19 disease characterization. J. Med. Biochem. 2023, 42, 454–459. [Google Scholar] [CrossRef]
- Bertini, C.D., Jr.; Khawaja, F.; Sheshadri, A. Coronavirus Disease-2019 in the Immunocompromised Host. Clin. Chest Med. 2023, 44, 395–406. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Hinedi, K.; Abbasi, S.; Babiker, M.; Sunji, A.; Eltigani, M. Hematologic, hepatic, and renal function changes in hospitalized patients with Middle East respiratory syndrome coronavirus. Int. J. Lab. Hematol. 2017, 39, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zeng, H.; Chen, H.; Fan, L.; Xu, C.; Huang, H.; Tang, T.; Li, M. Current knowledge of thrombocytopenia in sepsis and COVID-19. Front. Immunol. 2023, 14, 1213510. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Nair, M.G.; Jaroszewski, L.; Godzik, A. Deciphering Abnormal Platelet Subpopulations in COVID-19, Sepsis and Systemic Lupus Erythematosus through Machine Learning and Single-Cell Transcriptomics. Int. J. Mol. Sci. 2024, 25, 5941. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Maccaferri, M.; Ruini, C.; Tomasi, A.; Ozben, T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 2020, 57, 389–399. [Google Scholar] [CrossRef]
- Pozdnyakova, O.; Connell, N.T.; Battinelli, E.M.; Connors, J.M.; Fell, G.; Kim, A.S. Clinical Significance of CBC and WBC Morphology in the Diagnosis and Clinical Course of COVID-19 Infection. Am. J. Clin. Pathol. 2021, 155, 364–375. [Google Scholar] [CrossRef]
- Chan, S.S.W.; Christopher, D.; Tan, G.B.; Chong, V.C.L.; Fan, B.E.; Lin, C.Y.; Ong, K.H. Peripheral lymphocyte subset alterations in COVID-19 patients. Int. J. Lab. Hematol. 2020, 42, e199–e203. [Google Scholar] [CrossRef]
- Zini, G.; d’Onofrio, G. Coronavirus disease 2019 (COVID-19): Focus on peripheral blood cell morphology. Br. J. Haematol. 2023, 200, 404–419. [Google Scholar] [CrossRef]
- Harte, J.V.; Mykytiv, V. A panhaemocytometric approach to COVID-19: A retrospective study on the importance of monocyte and neutrophil population data on Sysmex XN-series analysers. Clin. Chem. Lab. Med. 2021, 59, e169–e172. [Google Scholar] [CrossRef]
- Martens, R.J.H.; van Adrichem, A.J.; Mattheij, N.J.A.; Brouwer, C.G.; van Twist, D.J.L.; Broerse, J.; Magro-Checa, C.; van Dongen, C.M.P.; Mostard, R.L.M.; Ramiro, S.; et al. Hemocytometric characteristics of COVID-19 patients with and without cytokine storm syndrome on the sysmex XN-10 hematology analyzer. Clin. Chem. Lab. Med. 2021, 59, 783–793. [Google Scholar] [CrossRef]
- Zhang, S.; Asquith, B.; Szydlo, R.; Tregoning, J.S.; Pollock, K.M. Peripheral T cell lymphopenia in COVID-19: Potential mechanisms and impact. Immunother. Adv. 2021, 1, ltab015. [Google Scholar] [CrossRef]
- Weinberg, S.E.; Behdad, A.; Ji, P. Atypical lymphocytes in peripheral blood of patients with COVID-19. Br. J. Haematol. 2020, 190, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, E.; Kwiecien, I.; Raniszewska, A.; Sokolowski, R.; Bednarek, J.; Jahnz-Rozyk, K.; Chcialowski, A.; Rzepecki, P. New Neutrophil Parameters in Diseases with Various Inflammatory Processes. Biomedicines 2024, 12, 2016. [Google Scholar] [CrossRef]
- Borczuk, A.C.; Yantiss, R.K. The pathogenesis of coronavirus-19 disease. J. Biomed. Sci. 2022, 29, 87. [Google Scholar] [CrossRef]
- Cappenberg, A.; Kardell, M.; Zarbock, A. Selectin-Mediated Signaling-Shedding Light on the Regulation of Integrin Activity in Neutrophils. Cells 2022, 11, 1310. [Google Scholar] [CrossRef]
- Hansen, P.B.; Knudsen, L.M.; Johnsen, H.E.; Hansen, N.E. Stimulation tests for the bone marrow neutrophil pool in malignancies. Leuk. Lymphoma 1995, 16, 237–246. [Google Scholar] [CrossRef]
- Drifte, G.; Dunn-Siegrist, I.; Tissieres, P.; Pugin, J. Innate immune functions of immature neutrophils in patients with sepsis and severe systemic inflammatory response syndrome. Crit. Care Med. 2013, 41, 820–832. [Google Scholar] [CrossRef]
- Urrechaga, E. Reviewing the value of leukocytes cell population data (CPD) in the management of sepsis. Ann. Transl. Med. 2020, 8, 953. [Google Scholar] [CrossRef] [PubMed]
- Perez, I.; Redin, M.E. Stability of leukocyte research parameters over time on the Sysmex XN: How to quantify the changes in cell morphology. Int. J. Lab. Hematol. 2018, 40, 569–576. [Google Scholar] [CrossRef]
- Seghezzi, M.; Buoro, S.; Previtali, G.; Moioli, V.; Manenti, B.; Simon-Lopez, R.; Ottomano, C.; Lippi, G. A Preliminary Proposal for Quality Control Assessment and Harmonization of Leukocytes Morphology-structural Parameters (cell Population Data Parameters). J. Med. Biochem. 2018, 37, 486–498. [Google Scholar] [CrossRef]


| No. | Background Diseases | Immunosuppressive Therapy |
|---|---|---|
| 1 | Myasthenia gravis | Prednisolone |
| 2 | Neuro-sweet disease | Prednisolone |
| 3 | Interstitial lung disease | Prednisolone |
| 4 | Encephalitis | Prednisolone |
| 5 | Eosinophilic granulomatosis with polyangiitis | Prednisolone |
| 6 | Diffuse large B cell lymphoma | R-CHOP |
| 7 | Multiple myeloma | Bortezomib, lenalidomide |
| 8 | Multiple myeloma | Bortezomib, dexamethasone |
| 9 | POEMS syndrome | Dexamethasone, lenalidomide |
| 10 | Essential thrombocythemia | Ruxolitinib |
| 11 | Rheumatoid arthritis | Infliximab, methotrexate |
| 12 | Microscopic polyangiitis, rapid progressive glomerulonephritis | Azathioprine, prednisolone |
| 13 | Crohn’s disease | Infliximab |
| 14 | Crohn’s disease | Azathioprine, infliximab, mesalazine |
| 15 | Ulcerative colitis | Betamethasone (enema), mesalazine |
| 16 | Pancreatic cancer | S-1 |
| 17 | Pancreatic cancer | S-1 |
| 18 | Lung cancer | Durvalumab, prednisolone |
| 19 | After liver transplantation | Cyclosporine, everolimus, methylprednisolone |
| Control Group | Immunosuppressive Treatment (IST) Group | p-Value | |||
|---|---|---|---|---|---|
| n = 29 | n = 19 | ||||
| Age (mean, year) | 65.7 ± 13.17 | 60.3 ± 17.11 | 0.226 | ||
| Male sex—no. (%) | 20 (69.0%) | 10 (52.6%) | 0.362 | ||
| BMI (mean, kg/m2) | 25.4 ± 4.90 | 23.9 ± 3.54 | 0.27 | ||
| Comorbidities, at least one, number (%) | |||||
| Hypertension | 15 (51.7%) | 8 (42.1%) | 0.566 | ||
| Diabetes mellitus | 10 (34.5%) | 5 (26.3%) | 0.751 | ||
| Obesity (BMI > 25 kg/m2) | 13 (46.4%) | 7 (36.8%) | 0.561 | ||
| COPD | 4 (13.8%) | 1 (5.3%) | 0.635 | ||
| Asthma | 0 (0%) | 2 (10.5%) | 0.152 | ||
| Chronic heart disease | 3 (10.3%) | 1 (5.3%) | 1 | ||
| Chronic kidney disease | 9 (31.0%) | 3 (15.8%) | 0.316 | ||
| Baseline complete blood count (CBC) and cell population data (CPD) before COVID-19 onset—median [25th–75th percentile] | |||||
| WBC count (103/μL) | 5.68 | [4.43–5.96] | 5.11 | [4.73–7.64] | 0.706 |
| HGB level (g/dL) | 13.0 | [12.4–13.9] | 12.7 | [12.0–13.3] | 0.333 |
| HCT count (%) | 38.8 | [37.8–40.1] | 39.5 | [34.9–40.3] | 0.706 |
| PLT count (104/μL) | 174 | [135–224] | 208 | [126–274] | 0.568 |
| NEUT# count (103/μL) | 3.42 | [2.25–4.46] | 2.67 | [2.55–3.54] | 0.716 |
| LYMPH# count (103/μL) | 1.15 | [1.07–1.85] | 1.79 | [1.36–1.98] | 0.303 |
| MONO# count 103/μL) | 0.30 | [0.19–0.35] | 0.31 | [0.30–0.37] | 0.220 |
| EO# count (103/μL) | 0.07 | [0.02–0.12] | 0.11 | [0.10–0.17] | 0.131 |
| BASO# count (103/μL) | 0.02 | [0.01–0.03] | 0.05 | [0.03–0.05] | 0.005 * |
| NE-SSC (ch) | 150 | [149–151] | 148 | [144–153] | 0.637 |
| NE-SFL (ch) | 47.9 | [46.2–48.3] | 47.2 | [46.0–51.1] | 0.608 |
| NE-FSC (ch) | 88.5 | [86.7–90.8] | 86.6 | [84.5–89.3] | 0.160 |
| NE-WX | 310 | [300–325] | 316 | [310–326] | 0.357 |
| NE-WY | 623 | [621–648] | 646 | [629–679] | 0.221 |
| NE-WZ | 685 | [631–834] | 669 | [603–794] | 0.457 |
| LY-X (ch) | 79.4 | [78.1–81.5] | 80.8 | [79.5–82.1] | 0.303 |
| LY-Y (ch) | 66.5 | [66.4–70.4] | 66.4 | [64.3–67.3] | 0.157 |
| LY-Z (ch) | 58.8 | [57.0–61.1] | 59.1 | [57.8–60.0] | 0.935 |
| LY-WX | 516 | [477–521] | 476 | [391–529] | 0.196 |
| LY-WY | 813 | [796–814] | 821 | [799–878] | 0.316 |
| LY-WZ | 608 | [538–634] | 533 | [484–646] | 0.237 |
| MO-X (ch) | 121 | [118–121] | 121 | [118–121] | 0.829 |
| MO-Y (ch) | 110 | [108–112] | 115 | [108–116] | 0.060 |
| MO-Z (ch) | 68.2 | [66.3–74.4] | 69.3 | [65.2–71.3] | 0.394 |
| MO-WX | 286 | [267–290] | 284 | [242–292] | 0.364 |
| MO-WY | 665 | [526–736] | 684 | [622–703] | 0.871 |
| MO-WZ | 632 | [609–633] | 606 | [571–695] | 0.526 |
| Control Group | Immunosuppressive Treatment (IST) Group | p-Value | |
|---|---|---|---|
| n = 29 | n = 19 | ||
| Severity—no. (%) | |||
| Mild | 21 (72.4%) | 13 (68.4%) | 1 |
| Severe | 8 (27.6%) | 6 (31.6%) | |
| Variable severity factors (including inflammatory variables)—median [25th–75th percentile] | |||
| CRP level (mg/dL) | 2.11 [1.24–4.19] | 0.77 [0.30–3.98] | 0.077 |
| Procalcitonin level (ng/mL) | 0.10 [0.10–0.20] | 0.10 [0.10–0.20] | 0.875 |
| Ferritin level (ng/mL) | 274 [126–499] | 312 [251–428] | 0.386 |
| LD level (units/L) | 272 [224–340] | 250 [192–339] | 0.424 |
| AST level (units/L) | 35.0 [25.8–45.5] | 31.5 [22.0–45.8] | 0.485 |
| ALT level (units/L) | 25.0 [16.0–43.0] | 24.0 [17.8–29.3] | 0.677 |
| eGFR (mL/min/1.73 m2) | 69.1 [51.3–80.0] | 66.9 [54.0–75.5] | 0.768 |
| Parameters | Parameter Description | Method Used to Analyze Clusters |
|---|---|---|
| NE-SSC (ch) | Complexity of the intracellular structure of the neutrophils (intracellular structure and granularity) | The laterally scattered light intensity of the neutrophil area on the WDF scattergram |
| NE-SFL (ch) | DNA/RNA content indicating cell immaturity or activation | The fluorescent light intensity of the neutrophil area on the WDF scattergram |
| NE-FSC (ch) | Size or volume of neutrophils | The forward-scattered light intensity of the neutrophil area on the WDF scattergram |
| NE-WX | Dispersion of the NE-SSC signal of the neutrophils | The laterally scattered light distribution width index of the neutrophil area on the WDF scattergram |
| NE-WY | Dispersion of the NE-SFL signal of the neutrophils | The fluorescent light distribution width index of the neutrophil area on the WDF scattergram |
| NE-WZ | Dispersion of the NE-FSC signal of the neutrophils | The forward-scattered light distribution width index of the neutrophil area on the WDF scattergram |
| LY-X (ch) | Complexity of the intracellular structure of the lymphocytes (e.g., nuclear irregularities and vacuolation) | The laterally scattered light intensity of the lymphocyte area on the WDF scattergram |
| LY-Y (ch) | DNA/RNA content indicating cell immaturity or activation | The fluorescent light intensity of the lymphocyte area on the WDF scattergram |
| LY-Z (ch) | Size or volume of the lymphocytes | The forward-scattered light intensity of the lymphocyte area on the WDF scattergram |
| LY-WX | Dispersion of the LY-X signal of the lymphocytes | The laterally scattered light distribution width index of the lymphocyte area on the WDF scattergram |
| LY-WY | Dispersion of the LY-Y signal of the lymphocytes | The fluorescent light distribution width index of the lymphocyte area on the WDF scattergram |
| LY-WZ | Dispersion of the LY-Z signal of the lymphocytes | The forward-scattered light distribution width index of the lymphocyte area on the WDF scattergram |
| MO-X (ch) | Complexity of the intracellular structure of the monocytes (e.g., nuclear irregularities and vacuolation) | The laterally scattered light intensity of the monocyte area on the WDF scattergram |
| MO-Y (ch) | DNA/RNA content indicating cell immaturity or activation | The fluorescent light intensity of the monocyte area on the WDF scattergram |
| MO-Z (ch) | Size or volume of the monocytes | The forward-scattered light intensity of the monocyte area on the WDF scattergram |
| MO-WX | Dispersion of the MO-X signal of the monocytes | The laterally scattered light distribution width index of the monocyte area on the WDF scattergram |
| MO-WY | Dispersion of the MO-Y signal of the monocytes | The fluorescent light distribution width index of the monocyte area on the WDF scattergram |
| MO-WZ | Dispersion of the MO-Z signal of the monocytes | The forward-scattered light distribution width index of the monocyte area on the WDF scattergram |
| Control Group | Immunosuppressive Treatment (IST) Group | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median [25th–75th Percentile] | p-Value | Median [25th–75th Percentile] | p Value | |||||||||||||
| Before | Days 0–2 | Days 3–5 | Before | Days 0–2 | Before | Days 0–2 | Days 3–5 | Before | Days 0–2 | |||||||
| vs. | vs. | vs. | vs. | |||||||||||||
| Days 0–2 | Days 3–5 | Days 0–2 | Days 3–5 | |||||||||||||
| WBC count (103/μL) | 5.68 | [4.43–5.96] | 3.91 | [3.63–5.93] | 4.43 | [3.94–6.50] | 0.317 | 0.174 | 5.11 | [4.73–7.64] | 5.3 | [4.87–7.73] | 5.97 | [3.93–6.93] | 0.595 | 0.583 |
| NEUT count (103/μL) | 3.42 | [2.25–4.46] | 2.41 | [2.05–2.67] | 2.88 | [2.10–5.02] | 0.096 | 0.335 | 2.67 | [2.55–3.54] | 3.89 | [3.37–5.66] | 3.49 | [2.45–5.14] | 0.073 | 0.429 |
| LYMPH# count (103/μL) | 1.15 | [1.07–1.85] | 0.98 | [0.86–1.13] | 1.28 | [1.01–2.00] | 0.018 * | 0.082 | 1.79 | [1.36–1.98] | 0.73 | [0.54–1.23] | 0.76 | [0.65–1.21] | 0.004 * | 0.78 |
| MONO# count (103/μL) | 0.3 | [0.19–0.35] | 0.33 | [0.35–0.40] | 0.45 | [0.32–0.54] | 0.204 | 0.024 * | 0.31 | [0.30–0.37] | 0.42 | [0.35–0.65] | 0.47 | [0.25–0.56] | 0.105 | 0.653 |
| HGB level (g/dL) | 13 | [12.4–13.9] | 12.7 | [11.8–0.40] | 13.7 | [12.4–15.0] | 0.494 | 0.15 | 12.7 | [12.0–13.3] | 13.2 | [13.0–13.5] | 13.1 | [10.1–14.8] | 0.322 | 0.815 |
| HCT count (%) | 38.8 | [37.8–40.1] | 36.9 | [35.3–42.9] | 39.9 | [37.4–44.7] | 0.45 | 0.075 | 39.5 | [34.9–40.3] | 40.2 | [37.7–40.6] | 40.4 | [30.9–43.6] | 0.467 | 0.96 |
| PLT count (104/μL) | 174 | [135–224] | 189 | [119–224] | 137 | [112–181] | 0.963 | 0.037 * | 208 | [126–274] | 205 | [133–237] | 105 | [98.0–189] | 0.785 | 0.037 * |
| NE-SSC (ch) | 150 | [149–151] | 151 | [148–152] | 152 | [149–155] | 0.369 | 0.584 | 148 | [144–153] | 149 | [149–159] | 155 | [149–158] | 0.112 | 0.653 |
| NE-SFL (ch) | 47.9 | [46.2–48.3] | 47 | [46.0–48.4] | 49 | [46.9–51.1] | 0.919 | 0.353 | 47.2 | [46.0–51.1] | 47.1 | [44.0–51.6] | 50.4 | [48.1–52.2] | 0.784 | 0.125 |
| NE-FSC (ch) | 88.5 | [86.7–90.8] | 85.7 | [84.5–88.0] | 87.6 | [85.4–89.8] | 0.009 * | 0.167 | 86.6 | [84.5–89.3] | 88.3 | [86.4–90.5] | 88.2 | [84.9–90.4] | 0.356 | 0.78 |
| NE-WX | 310 | [300–325] | 307 | [304–318] | 325 | [317–334] | 0.433 | 0.025 * | 316 | [310–326] | 317 | [301–339] | 310 | [298–330] | 0.885 | 0.618 |
| NE-WY | 623 | [621–648] | 645 | [631–658] | 650 | [624–672] | 0.345 | 0.713 | 646 | [629–679] | 659 | [637–680] | 623 | [607–638] | 0.784 | 0.010 * |
| NE-WZ | 685 | [631–834] | 669 | [639–715] | 647 | [616–742] | 0.345 | 0.713 | 669 | [603–794] | 643 | [605–710] | 658 | [600–720] | 0.664 | 0.799 |
| LY-X (ch) | 79.4 | [78.1–81.5] | 80 | [78.2–81.7] | 77.3 | [74.5–79.9] | 0.812 | 0.059 | 80.8 | [79.5–82.1] | 77.5 | [75.9–81.2] | 79.3 | [76.2–83.7] | 0.137 | 0.246 |
| LY-Y (ch) | 66.5 | [66.4–70.4] | 68.7 | [66.0–69.7] | 67.3 | [65.0–69.7] | 0.533 | 0.382 | 66.4 | [64.3–67.3] | 64.1 | [62.4–67.1] | 68.4 | [65.2–70.6] | 0.447 | 0.020 * |
| LY-Z (ch) | 58.8 | [57.0–61.1] | 57.9 | [57.0–59.4] | 57.8 | [56.8–58.9] | 0.548 | 0.726 | 59.1 | [57.8–60.0] | 57.6 | [56.9–58.4] | 59.6 | [57.6–60.6] | 0.21 | 0.166 |
| LY-WX | 516 | [477–521] | 558 | [444–586] | 534 | [477–589] | 0.24 | 0.66 | 476 | [391–529] | 477 | [456–549] | 552 | [480–609] | 0.392 | 0.251 |
| LY-WY | 813 | [796–814] | 867 | [809–926] | 809 | [732–589] | 0.24 | 0.134 | 821 | [799–878] | 776 | [678–820] | 797 | [726–895] | 0.049 * | 0.312 |
| LY-WZ | 608 | [538–634] | 586 | [498–630] | 560 | [538–613] | 0.489 | 0.979 | 533 | [484–646] | 525 | [498–552 | 583 | [553–605] | 0.885 | 0.034 * |
| MO-X (ch) | 121 | [118–121] | 122 | [120–123] | 121 | [119–122] | 0.139 | 0.229 | 121 | [118–121] | 120 | [118–123] | 121 | [120–123] | 0.834 | 0.3 |
| MO-Y (ch) | 110 | [108–112] | 114 | [104–122] | 112 | [109–117] | 0.3 | 0.957 | 115 | [108–116] | 106 | [99.0–116] | 108 | [105–113] | 0.322 | 0.544 |
| MO-Z (ch) | 68.2 | [66.3–74.4] | 66.9 | [64.0–69.0] | 66.2 | [65.2–67.4] | 0.111 | 0.822 | 69.3 | [65.2–71.3] | 66.5 | [63.7–69.9] | 67.6 | [64.7–69.7] | 0.339 | 0.837 |
| MO-WX | 286 | [267–290] | 267 | [247–292] | 261 | [247–276] | 0.334 | 0.686 | 284 | [242–292] | 269 | [250–287] | 265 | [243–290] | 0.988 | 0.636 |
| MO-WY | 665 | [526–736] | 696 | [580–740] | 725 | [683–745] | 0.503 | 0.321 | 684 | [622–703] | 775 | [720–813] | 664 | [603–708] | 0.008 * | 0.017 * |
| MO-WZ | 632 | [609–633] | 620 | [561–650] | 668 | [581–709] | 0.563 | 0.226 | 606 | [571–695] | 605 | [595–642] | 617 | [586–694] | 0.688 | 0.904 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogawa, M.; Suzuki, Y.; Nishida, Y.; Ono, D.; Kataoka, H.; Takeshita, K. Impact of Immunosuppression on Immune Cell Dynamics in COVID-19: A Serial Comparison of Leukocyte Data in Healthy and Immunocompromised Patients Before and After Infection. J. Clin. Med. 2025, 14, 3223. https://doi.org/10.3390/jcm14093223
Ogawa M, Suzuki Y, Nishida Y, Ono D, Kataoka H, Takeshita K. Impact of Immunosuppression on Immune Cell Dynamics in COVID-19: A Serial Comparison of Leukocyte Data in Healthy and Immunocompromised Patients Before and After Infection. Journal of Clinical Medicine. 2025; 14(9):3223. https://doi.org/10.3390/jcm14093223
Chicago/Turabian StyleOgawa, Masumi, Yasufumi Suzuki, Yusuke Nishida, Daisuke Ono, Hiromi Kataoka, and Kyosuke Takeshita. 2025. "Impact of Immunosuppression on Immune Cell Dynamics in COVID-19: A Serial Comparison of Leukocyte Data in Healthy and Immunocompromised Patients Before and After Infection" Journal of Clinical Medicine 14, no. 9: 3223. https://doi.org/10.3390/jcm14093223
APA StyleOgawa, M., Suzuki, Y., Nishida, Y., Ono, D., Kataoka, H., & Takeshita, K. (2025). Impact of Immunosuppression on Immune Cell Dynamics in COVID-19: A Serial Comparison of Leukocyte Data in Healthy and Immunocompromised Patients Before and After Infection. Journal of Clinical Medicine, 14(9), 3223. https://doi.org/10.3390/jcm14093223





