The Impact of OXTR, COMT, and GRIN2B Polymorphisms on Brain Development in Preterm Infants
Abstract
1. Introduction
2. Materials and Methods
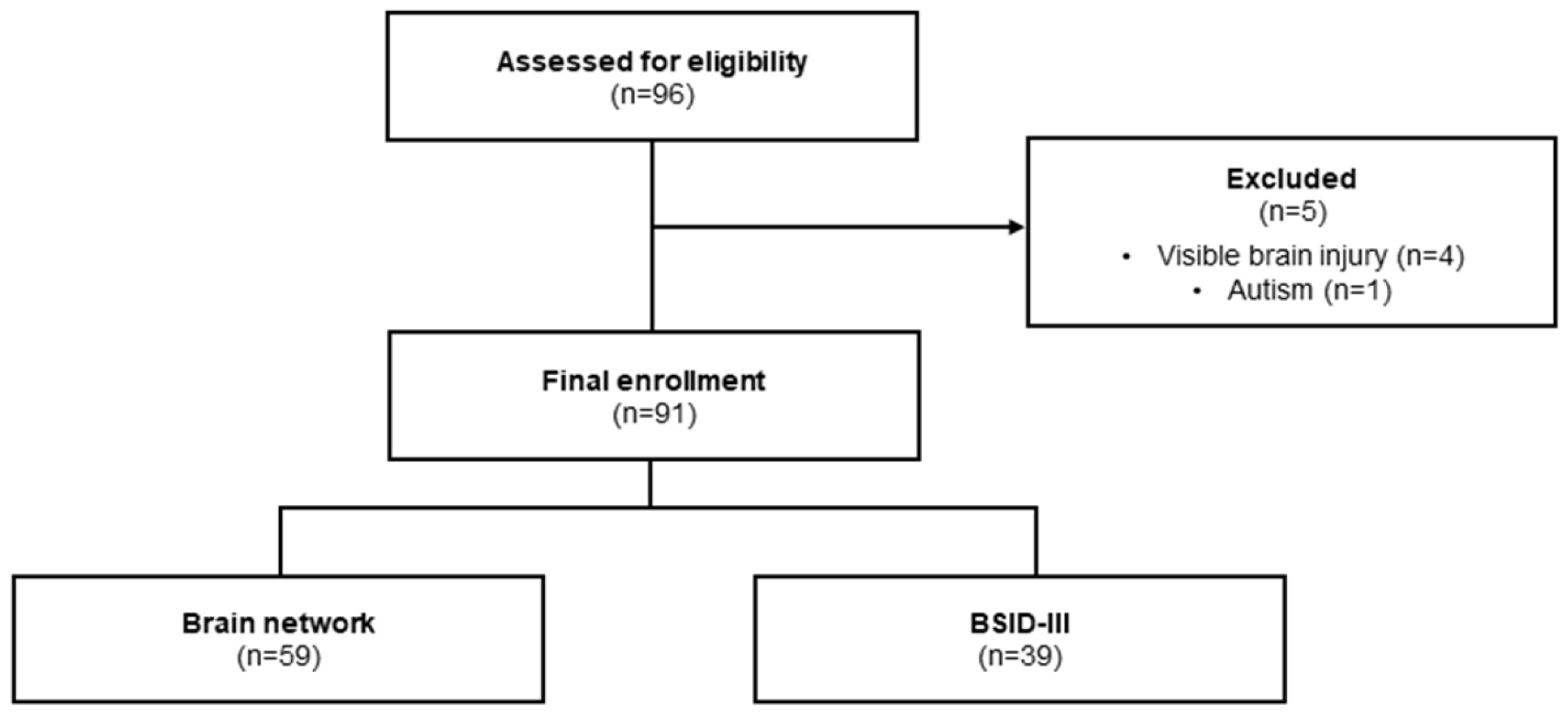
2.1. Study Design and Participants
2.2. DNA Sample Collection
2.3. MRI Acquisitions
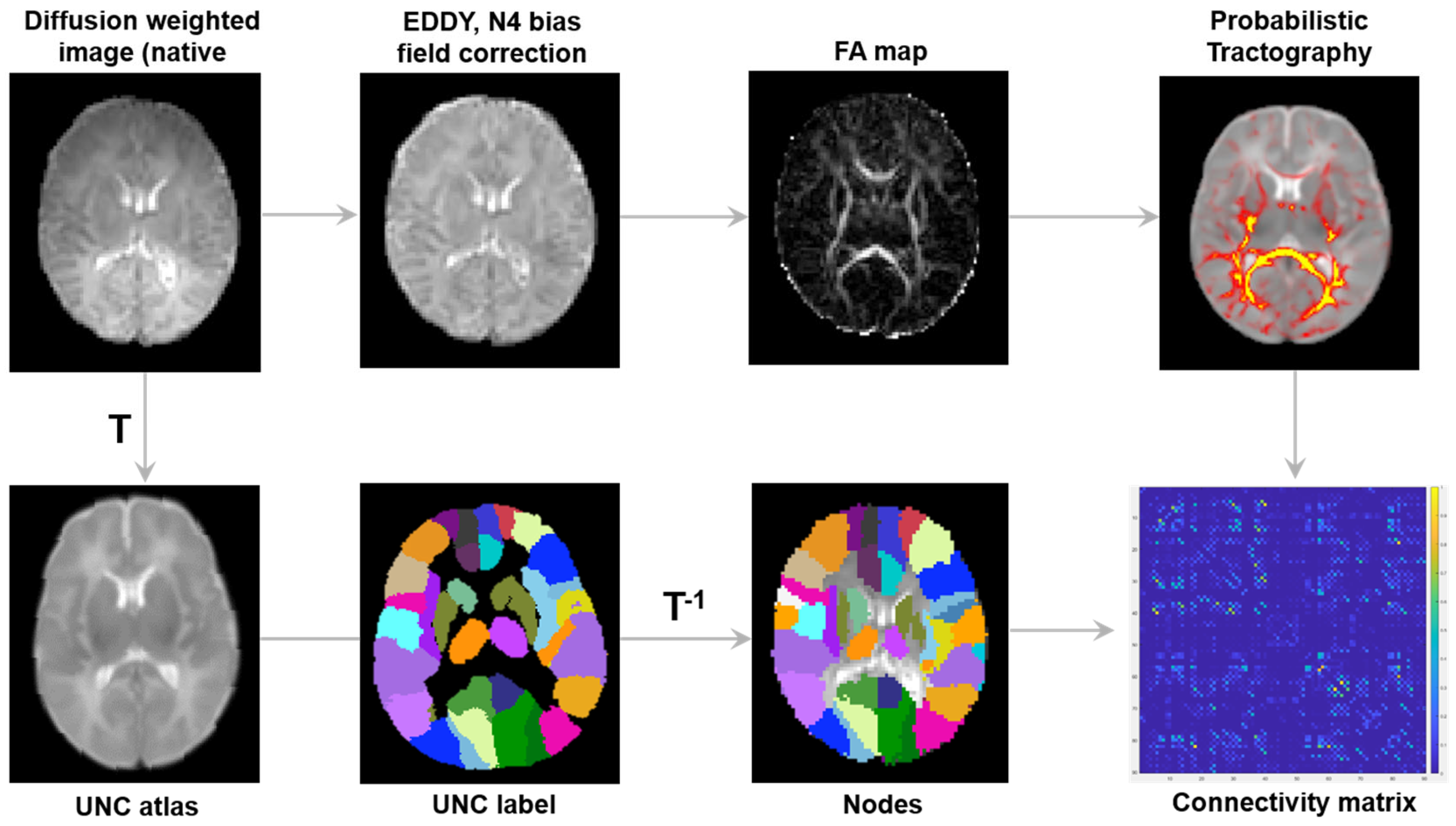
2.4. Image Processing
2.5. Network Construction
2.6. Global Network Analysis
2.7. Volumetric Analysis
2.8. Neurodevelopmental Assessment
2.9. Statistical Analysis
3. Results
3.1. Allelic and Genotypic Distributions
3.2. Association Between Minor Allele Frequencies and Neurodevelopmental Outcomes in Preterm Infants
3.3. Association Between Minor Allele Frequencies and Brain Network in Preterm Infants
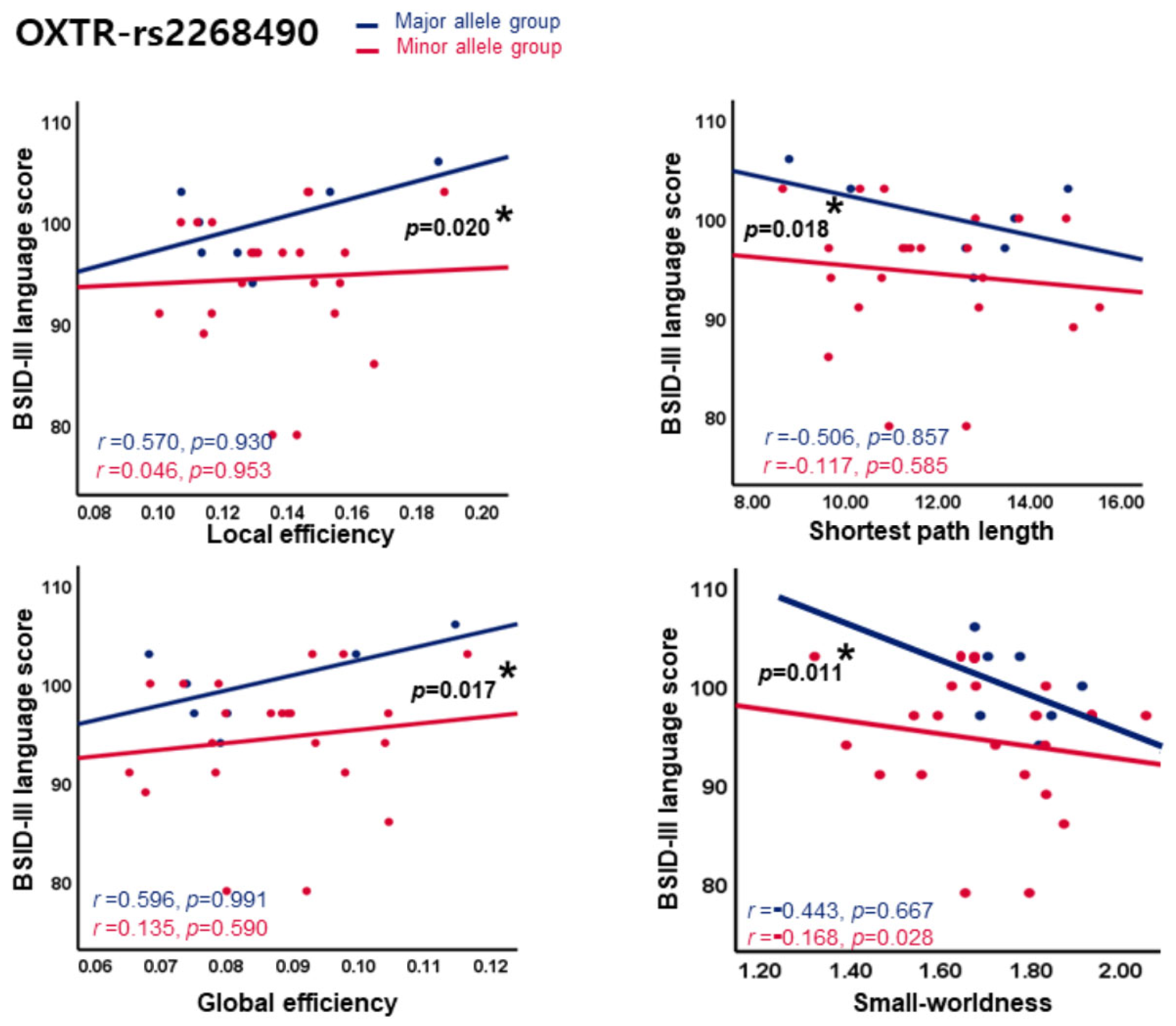
3.4. Relationship Between Brain Quantitative Values and BSID-III Scores According to Allele Group in Preterm Infants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Volpe, J.J. Systemic Inflammation, Oligodendroglial Maturation, and the Encephalopathy of Prematurity; Wiley Online Library: Hoboken, NJ, USA, 2011; pp. 525–529. [Google Scholar]
- Krishnan, M.L.; Wang, Z.; Aljabar, P.; Ball, G.; Mirza, G.; Saxena, A.; Counsell, S.J.; Hajnal, J.V.; Montana, G.; Edwards, A.D. Machine learning shows association between genetic variability in PPARG and cerebral connectivity in preterm infants. Proc. Natl. Acad. Sci. USA 2017, 114, 13744–13749. [Google Scholar] [CrossRef]
- Toga, A.W.; Thompson, P.M. Genetics of brain structure and intelligence. Annu. Rev. Neurosci. 2005, 28, 1–23. [Google Scholar] [CrossRef]
- Thompson, P.M.; Cannon, T.D.; Narr, K.L.; van Erp, T.; Poutanen, V.-P.; Huttunen, M.; Lönnqvist, J.; Standertskjöld-Nordenstam, C.-G.; Kaprio, J.; Khaledy, M.; et al. Genetic influences on brain structure. Nat. Neurosci. 2001, 4, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Prom-Wormley, E.C.; Perez, J.; Kubarych, T.; Styner, M.; Lin, W.; Neale, M.C.; Gilmore, J.H. White matter heritability using diffusion tensor imaging in neonatal brains. Twin Res. Hum. Genet. 2012, 15, 336–350. [Google Scholar] [CrossRef]
- Xie, X.; Zu, M.; Zhang, L.; Bai, T.; Wei, L.; Huang, W.; Ji, G.-J.; Qiu, B.; Hu, P.; Tian, Y. A common variant of the NOTCH4 gene modulates functional connectivity of the occipital cortex and its relationship with schizotypal traits. BMC Psychiatry 2020, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Feng, L.; Wang, S.; Lin, Z.; Li, T.; Zhao, B.; Zhu, H.; Zhang, H. Common genetic variants have associations with human cortical brain regions and risk of schizophrenia. Genet. Epidemiol. 2019, 43, 548–558. [Google Scholar] [CrossRef]
- Binder, E.B.; Gordon, J.A. Exploring and Exploiting Genetic Risk for Psychiatric Disorders; MIT Press: Cambridge, MA, USA, 2023. [Google Scholar]
- Hashem, S.; Nisar, S.; Bhat, A.A.; Yadav, S.K.; Azeem, M.W.; Bagga, P.; Fakhro, K.; Reddy, R.; Frenneaux, M.P.; Haris, M. Genetics of structural and functional brain changes in autism spectrum disorder. Transl. Psychiatry 2020, 10, 229. [Google Scholar] [CrossRef]
- Kaya, K.; Önal, D.; Kartal, Y.; Budak, M.T.; Karabulut, E.; Karlı Oğuz, K.; Pehlivanoğlu, B. Situating the oxytocin receptor gene polymorphisms in the context of structural and connectome-level substrates and association with endogenous oxytocin. bioRxiv 2023. bioRxiv:2023.08. 24.554569. [Google Scholar] [CrossRef]
- Francis, S.M.; Kim, S.-J.; Kistner-Griffin, E.; Guter, S.; Cook, E.H.; Jacob, S. ASD and genetic associations with receptors for oxytocin and vasopressin—AVPR1A, AVPR1B, and OXTR. Front. Neurosci. 2016, 10, 516. [Google Scholar] [CrossRef]
- Dutt, A.; Shaikh, M.; Ganguly, T.; Nosarti, C.; Walshe, M.; Arranz, M.; Rifkin, L.; McDonald, C.; Chaddock, C.A.; McGuire, P. COMT gene polymorphism and corpus callosum morphometry in preterm born adults. Neuroimage 2011, 54, 148–153. [Google Scholar] [CrossRef]
- Esmaiel, N.N.; Ashaat, E.A.; Mosaad, R.; Fayez, A.; Ibrahim, M.; Abdallah, Z.Y.; Issa, M.Y.; Salem, S.; Ramadan, A.; El Wakeel, M.A. The potential impact of COMT gene variants on dopamine regulation and phenotypic traits of ASD patients. Behav. Brain Res. 2020, 378, 112272. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ahn, J.-H.; Lee, J.Y.; Jang, Y.H.; Kim, Y.-E.; Kim, J.I.; Kim, B.-N.; Lee, H.J. Altered cerebral curvature in preterm infants is associated with the common genetic variation related to autism spectrum disorder and lipid metabolism. J. Clin. Med. 2022, 11, 3135. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Yoo, J.H.; Kim, D.; Jeong, B.; Kim, B.-N. The effects of GRIN2B and DRD4 gene variants on local functional connectivity in attention-deficit/hyperactivity disorder. Brain Imaging Behav. 2018, 12, 247–257. [Google Scholar] [CrossRef]
- Monyer, H.; Burnashev, N.; Laurie, D.J.; Sakmann, B.; Seeburg, P.H. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 1994, 12, 529–540. [Google Scholar] [CrossRef]
- Kim, J.I.; Kim, J.-W.; Park, S.; Hong, S.-B.; Lee, D.S.; Paek, S.H.; Han, D.H.; Cheong, J.H.; Kim, B.-N. The GRIN2B and GRIN2A gene variants are associated with continuous performance test variables in ADHD. J. Atten. Disord. 2020, 24, 1538–1546. [Google Scholar] [CrossRef]
- Gizer, I.R.; Ficks, C.; Waldman, I.D. Candidate gene studies of ADHD: A meta-analytic review. Hum. Genet. 2009, 126, 51–90. [Google Scholar] [CrossRef]
- Loh, K.; Ramli, N.; Tan, L.; Roziah, M.; Rahmat, K.; Ariffin, H. Quantification of diffusion tensor imaging in normal white matter maturation of early childhood using an automated processing pipeline. Eur. Radiol. 2012, 22, 1413–1426. [Google Scholar] [CrossRef]
- Sporns, O. The human connectome: A complex network. Ann. N. Y. Acad. Sci. 2011, 1224, 109–125. [Google Scholar] [CrossRef]
- Potharst, E.S.; Houtzager, B.A.; Van Sonderen, L.; Tamminga, P.; Kok, J.H.; Last, B.F.; Van Wassenaer, A.G. Prediction of cognitive abilities at the age of 5 years using developmental follow-up assessments at the age of 2 and 3 years in very preterm children. Dev. Med. Child Neurol. 2012, 54, 240–246. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, E.S.L.; de Kieviet, J.F.; Königs, M.; van Elburg, R.M.; Oosterlaan, J. Predictive value of the Bayley scales of infant development on development of very preterm/very low birth weight children: A meta-analysis. Early Hum. Dev. 2013, 89, 487–496. [Google Scholar] [CrossRef]
- Han, S.; Kim, O.; Yoo, C.; Heo, J.S.; Lee, H.-S.; Jeon, J. Neurodevelopmental Correlations Between the Korean Developmental Screening Test and Bayley Scale III in Very-Low-Birth-Weight Infants. Neonatal Med. 2020, 27, 167–173. [Google Scholar] [CrossRef]
- Chung, H.J.; Yang, D.; Kim, G.-H.; Kim, S.K.; Kim, S.W.; Kim, Y.K.; Kim, Y.A.; Kim, J.S.; Kim, J.K.; Kim, C. Development of the Korean developmental screening test for infants and children (K-DST). Clin. Exp. Pediatr. 2020, 63, 438. [Google Scholar] [CrossRef] [PubMed]
- Behnia, F.; Parets, S.E.; Kechichian, T.; Yin, H.; Dutta, E.H.; Saade, G.R.; Smith, A.K.; Menon, R. Fetal DNA methylation of autism spectrum disorders candidate genes: Association with spontaneous preterm birth. Am. J. Obstet. Gynecol. 2015, 212, 533. e531–533. e539. [Google Scholar] [PubMed]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M. Fsl. Neuroimage 2012, 62, 782–790. [Google Scholar] [CrossRef]
- Andersson, J.L.; Sotiropoulos, S.N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 2016, 125, 1063–1078. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef]
- Shi, F.; Wang, L.; Wu, G.; Li, G.; Gilmore, J.H.; Lin, W.; Shen, D. Neonatal atlas construction using sparse representation. Hum. Brain Mapp. 2014, 35, 4663–4677. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Guerrero, G.D.; Cecilia, J.M.; García, J.M.; Inuggi, A.; Jbabdi, S.; Behrens, T.E.; Sotiropoulos, S.N. Accelerating fibre orientation estimation from diffusion weighted magnetic resonance imaging using GPUs. PLoS ONE 2013, 8, e61892. [Google Scholar] [CrossRef]
- Booth, T.; Dykiert, D.; Corley, J.; Gow, A.J.; Morris, Z.; Muñoz Maniega, S.; Royle, N.A.; Valdés Hernández, M.d.C.; Starr, J.M.; Penke, L. Reaction time variability and brain white matter integrity. Neuropsychology 2019, 33, 642. [Google Scholar] [CrossRef]
- Bordier, C.; Nicolini, C.; Bifone, A. Graph analysis and modularity of brain functional connectivity networks: Searching for the optimal threshold. Front. Neurosci. 2017, 11, 441. [Google Scholar] [CrossRef]
- Van Den Heuvel, M.P.; Fornito, A. Brain networks in schizophrenia. Neuropsychol. Rev. 2014, 24, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.H.; Kim, H.; Lee, J.Y.; Ahn, J.-H.; Chung, A.W.; Lee, H.J. Altered development of structural MRI connectome hubs at near-term age in very and moderately preterm infants. Cereb. Cortex 2023, 33, 5507–5523. [Google Scholar] [CrossRef]
- Jang, Y.H.; Ham, J.; Kasani, P.H.; Kim, H.; Lee, J.Y.; Lee, G.Y.; Han, T.H.; Kim, B.N.; Lee, H.J. Predicting two-year neurodevelopmental outcomes in preterm infants using multimodal structural brain MRI with local connectivity. Sci. Rep. 2024, 14, 9331. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Xia, M.; Liao, X.; Evans, A.; He, Y. GRETNA: A graph theoretical network analysis toolbox for imaging connectomics. Front. Hum. Neurosci. 2015, 9, 386. [Google Scholar]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 2010, 52, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Beare, R.J.; Chen, J.; Kelly, C.E.; Alexopoulos, D.; Smyser, C.D.; Rogers, C.E.; Loh, W.Y.; Matthews, L.G.; Cheong, J.L.; Spittle, A.J. Neonatal brain tissue classification with morphological adaptation and unified segmentation. Front. Neuroinformatics 2016, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Yim, C.-H.; Kim, G.-H.; Eun, B.-L. Usefulness of the Korean Developmental Screening Test for Infants and Children for the Evaluation of Developmental Delay in Korean Infants and Children: A Single-Center Study. Korean J. Pediatr. 2017, 60, 312. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M. Genetic association studies: Design, analysis and interpretation. Brief. Bioinform. 2002, 3, 146–153. [Google Scholar] [CrossRef]
- Wade, M.; Hoffmann, T.J.; Wigg, K.; Jenkins, J.M. Association between the oxytocin receptor (OXTR) gene and children’s social cognition at 18 months. Genes Brain Behav. 2014, 13, 603–610. [Google Scholar] [CrossRef]
- Quan, J.; Ong, M.-L.; Bureau, J.-F.; Sim, L.W.; Sanmugam, S.; Malik, A.B.A.; Wong, E.; Wong, J.; Chong, Y.-S.; Saw, S.M.; et al. The influence of CHRNA4, COMT, and maternal sensitivity on orienting and executive attention in 6-month-old infants. Brain Cogn. 2017, 116, 17–28. [Google Scholar] [CrossRef]
- Nobile, M.; Maggioni, E.; Mauri, M.; Garzitto, M.; Piccin, S.; Bonivento, C.; Giorda, R.; Girometti, R.; Tomasino, B.; Molteni, M.; et al. Brain anatomical mediators of GRIN2B gene association with attention/hyperactivity problems: An integrated genetic-neuroimaging study. Genes 2021, 12, 1193. [Google Scholar] [CrossRef]
- Di Napoli, A.; Warrier, V.; Baron-Cohen, S.; Chakrabarti, B. Genetic variation in the oxytocin receptor (OXTR) gene is associated with Asperger Syndrome. Mol. Autism 2014, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.W.; Kang, J.I.; An, S.K.; Kim, S.J. Oxytocin receptor gene variants are associated with emotion recognition and resilience, but not with false-belief reasoning performance in healthy young Korean volunteers. CNS Neurosci. Ther. 2019, 25, 519–526. [Google Scholar] [CrossRef]
- Dadds, M.R.; Moul, C.; Cauchi, A.; Dobson-Stone, C.; Hawes, D.J.; Brennan, J.; Urwin, R.; Ebstein, R.E. Polymorphisms in the oxytocin receptor gene are associated with the development of psychopathy. Dev. Psychopathol. 2014, 26, 21–31. [Google Scholar] [CrossRef]
- Hirata, Y.; Zai, C.C.; Nowrouzi, B.; Beitchman, J.H.; Kennedy, J.L. Study of the Catechol-O-Methyltransferase (COMT) gene with high aggression in children. Aggress. Behav. 2013, 39, 45–51. [Google Scholar] [CrossRef]
- Madzarac, Z.; Tudor, L.; Sagud, M.; Nedic Erjavec, G.; Mihaljevic Peles, A.; Pivac, N. The associations between COMT and MAO-B genetic variants with negative symptoms in patients with schizophrenia. Curr. Issues Mol. Biol. 2021, 43, 618–636. [Google Scholar] [CrossRef]
- Li, W.J.; Kou, C.G.; Yu, Y.; Sun, S.; Zhang, X.; Kosten, T.R.; Zhang, X.Y. Association of Catechol-O-methyltransferase gene polymorphisms with schizophrenia and negative symptoms in a Chinese population. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2012, 159, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Pap, D.; Gonda, X.; Molnar, E.; Lazary, J.; Benko, A.; Downey, D.; Thomas, E.; Chase, D.; Toth, Z.G.; Mekli, K. Genetic variants in the catechol-o-methyltransferase gene are associated with impulsivity and executive function: Relevance for major depression. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2012, 159, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Boardman, J.P.; Walley, A.; Ball, G.; Takousis, P.; Krishnan, M.L.; Hughes-Carre, L.; Aljabar, P.; Serag, A.; King, C.; Merchant, N. Common genetic variants and risk of brain injury after preterm birth. Pediatrics 2014, 133, e1655–e1663. [Google Scholar] [CrossRef]
- Xia, K.; Zhang, J.; Ahn, M.; Jha, S.; Crowley, J.; Szatkiewicz, J.; Li, T.; Zou, F.; Zhu, H.; Hibar, D. Genome-wide association analysis identifies common variants influencing infant brain volumes. Transl. Psychiatry 2017, 7, e1188. [Google Scholar] [CrossRef]
- Hermundstad, A.M.; Bassett, D.S.; Brown, K.S.; Aminoff, E.M.; Clewett, D.; Freeman, S.; Frithsen, A.; Johnson, A.; Tipper, C.M.; Miller, M.B. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc. Natl. Acad. Sci. USA 2013, 110, 6169–6174. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Zhu, D.; Yang, Z.; Liu, F.; Qin, W.; Zhu, J.; Liu, B.; Jiang, T.; Yu, C. A common variant in OXTR rs53576 impacts topological patterns of brain functional networks. Eur. Child Adolesc. Psychiatry 2020, 29, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L. Exploring Heterogeneity in Autism Neuroendophenotypes: Effects of Genetic Risk, Gender, and Behavioral Symptomatology; UCLA: Los Angeles, CA, USA, 2018. [Google Scholar]
- Schneiderman, I.; Kanat-Maymon, Y.; Ebstein, R.P.; Feldman, R. Cumulative risk on the oxytocin receptor gene (OXTR) underpins empathic communication difficulties at the first stages of romantic love. Soc. Cogn. Affect. Neurosci. 2014, 9, 1524–1529. [Google Scholar] [CrossRef]
- Sporns, O. Network attributes for segregation and integration in the human brain. Curr. Opin. Neurobiol. 2013, 23, 162–171. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, J.S.; Meskaldji, D.-E.; Loukas, S.; Lordier, L.; Gui, L.; Lazeyras, F.; Hüppi, P.S. Preterm birth leads to impaired rich-club organization and fronto-paralimbic/limbic structural connectivity in newborns. NeuroImage 2021, 225, 117440. [Google Scholar] [CrossRef]
- Feldman, R.; Monakhov, M.; Pratt, M.; Ebstein, R.P. Oxytocin pathway genes: Evolutionary ancient system impacting on human affiliation, sociality, and psychopathology. Biol. Psychiatry 2016, 79, 174–184. [Google Scholar] [CrossRef]
| Gene | SNP Name | Chromosome | Coordinate (Position) | Source | Variant (Major/Minor) | Korean MAF | Population MAF | HWE Unaffected, p |
|---|---|---|---|---|---|---|---|---|
| OXTR | rs1042778 | 3 | 8794545 | dbSNP | G/T | 0.087 | 0.076 | 0.504 |
| OXTR | rs2268490 | 3 | 8797085 | dbSNP | C/T | 0.494 | 0.492 | 0.856 |
| OXTR | rs2268493 | 3 | 8800840 | 1000_genomes | T/C | 0.161 | 0.139 | 0.465 |
| GRIN2B | rs2268116 | 12 | 13870080 | 1000_genomes | A/G | 0.368 | 0.349 | 0.687 |
| GRIN2B | rs2284411 | 12 | 13866172 | dbSNP | C/T | 0.186 | 0.164 | 0.736 |
| COMT | rs174690 | 22 | 19939432 | 1000_genomes | G/A | 0.283 | 0.290 | 0.824 |
| COMT | rs4818 | 22 | 19951207 | dbSNP | C/G | 0.334 | 0.277 | 0.106 |
| COMT | rs740603 | 22 | 19945177 | dbSNP | A/G | 0.423 | 0.357 | 0.548 |
| Characteristics | Preterm (n = 91) |
|---|---|
| Male sex | 43 (47.3) |
| Gestational age, mean ± SD | 31.32 ± 3.68 |
| Birth weight, mean ± SD | 1722.89 ± 698.96 |
| Scan age, mean ±SD | 37.95 ± 2.18 (59/91) |
| Moderate to severe BPD | 16 (17.6) |
| Stage II to III ROP | 6 (6.6) |
| K-DST | |
| <−2 SD in any domain | 10/87 (11.5) |
| <−2 SD in gross motor domain | 4/87 (4.6) |
| <−2 SD in fine motor domain | 2/87 (2.3) |
| <−2 SD in cognition domain | 5/87 (5.7) |
| <−2 SD in language domain | 5/87 (5.7) |
| <−2 SD in sociality domain | 1/87 (1.2) |
| Gene | SNP Name | Variant | Preterm (n = 91) |
|---|---|---|---|
| OXTR | rs1042778 | G/G | 76 (84.4) |
| G/T | 13 (14.4) | ||
| T/T | 1 (1.1) | ||
| OXTR | rs2268490 | T/T | 21 (23.1) |
| C/T | 49 (53.8) | ||
| C/C | 21 (23.1) | ||
| OXTR | rs2268493 | T/T | 67 (73.6) |
| T/C | 24 (26.4) | ||
| C/C | 0 (0.0) | ||
| GRIN2B | rs2268116 | A/A | 41 (45.1) |
| A/G | 41 (45.1) | ||
| G/G | 9 (9.9) | ||
| GRIN2B | rs2284411 | C/C | 65 (71.4) |
| C/T | 24 (26.4) | ||
| T/T | 2 (2.2) | ||
| COMT | rs174690 | G/G | 47 (51.6) |
| G/A | 36 (39.6) | ||
| A/A | 8 (8.8) | ||
| COMT | rs4818 | C/C | 49 (53.8) |
| C/G | 34 (37.4) | ||
| G/G | 8 (8.8) | ||
| COMT | rs740603 | A/A | 39 (42.9) |
| A/G | 40 (44.0) | ||
| G/G | 12 (13.2) |
| BSID-III (n = 39) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP Name | Cognition | Language | Motor | Social–Emotional | Adaptive Behavior | |||||
| B (95% CI) | p | B (95% CI) | p | B (95% CI) | p | B (95% CI) | p | B (95% CI) | p | ||
| OXTR | rs1042778 | −5.881 (−13.25 to 1.488) | 0.125 | −2.05 (−8.939 to 4.839) | 0.563 | −2.643 (−12.17 to 6.883) | 0.589 | −0.842 (−13.97 to 12.28) | 0.900 | 7.171 (−3.501 to 17.84) | 0.194 |
| OXTR | rs2268490 | −2.605 (−9.212 to 4.002) | 0.444 | −6.51 (−12.1 to −0.924) | 0.027 | −4.03 (−12.2 to 4.139) | 0.339 | −1.446 (−12.87 to 9.982) | 0.805 | −10.23 (−18.95 to −1.515) | 0.026 |
| OXTR | rs2268493 | −2.073 (−8.215 to 4.069) | 0.512 | −2.455 (−7.874 to 2.964) | 0.379 | −0.197 (−7.852 to 7.459) | 0.960 | −2.566 (−13.15 to 8.021) | 0.637 | −1.675 (−10.2 to 6.846) | 0.702 |
| GRIN2B | rs2268116 | 0.121 (−6.017 to 6.258) | 0.970 | 1.205 (−4.219 to 6.63) | 0.665 | −3.544 (−11.09 to 4.004) | 0.362 | −0.083 (−10.64 to 10.47) | 0.988 | −1.371 (−9.853 to 7.11) | 0.753 |
| GRIN2B | rs2284411 | −0.862 (−7.34 to 5.616) | 0.795 | 2.396 (−3.303 to 8.095) | 0.414 | −2.652 (−10.66 to 5.355) | 0.519 | 2.338 (−8.791 to 13.47) | 0.682 | 4.966 (−3.888 to 13.82) | 0.277 |
| COMT | rs174690 | 2.583 (−3.372 to 8.539) | 0.400 | 0.043 (−5.272 to 5.357) | 0.988 | 5.654 (−1.615 to 12.92) | 0.134 | 6.722 (−3.419 to 16.86) | 0.200 | 0.232 (−8.069 to 8.534) | 0.957 |
| COMT | rs4818 | 4.516 (−1.333 to 10.36) | 0.137 | 4.032 (−1.145 to 9.209) | 0.134 | 3.679 (−3.677 to 11.04) | 0.332 | −12.25 (−21.94 to −2.568) | 0.017 | −3.309 (−11.54 to 4.922) | 0.435 |
| COMT | rs740603 | 3.836 (−2.084 to 9.756) | 0.210 | −0.086 (−5.418 to 5.246) | 0.975 | −4.197 (−11.57 to 3.177) | 0.270 | −11.55 (−21.36 to −1.734) | 0.026 | −7.07 (−15.15 to 1.009) | 0.093 |
| Brain Network (n = 59) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP Name | SW | GE | LE | LP | ||||
| B (95% CI) | p | B (95% CI) | p | B (95% CI) | p | B (95% CI) | p | ||
| OXTR | rs1042778 | 0.183 (0.050 to 0.316) | 0.009 | −0.014 (−0.027 to −0.002) | 0.030 | −0.020 (−0.041 to 0.000) | 0.058 | 2.101 (0.360 to 3.842) | 0.022 |
| OXTR | rs2268490 | −0.145 (−0.254 to −0.035) | 0.012 | 0.011 (−0.016 to 0.004) | 0.038 | 0.018 (0.001 to 0.034) | 0.042 | −1.912 (−3.331 to 0.512) | 0.010 |
| OXTR | rs2268493 | −0.061 (−0.167 to 0.0451) | 0.264 | −0.006 (0.001 to 0.022) | 0.241 | −0.010 (−0.026 to 0.006) | 0.226 | 0.747 (−0.622 to 2.116) | 0.290 |
| GRIN2B | rs2268116 | 0.010 (−0.093 to 0.112) | 0.854 | −0.000 (−0.010 to 0.009) | 0.942 | −0.008 (−0.019 to 0.012) | 0.664 | 0.187 (−1.129 to 1.502) | 0.782 |
| GRIN2B | rs2284411 | 0.052 (−0.055 to 0.160) | 0.342 | −0.005 (−0.015 to 0.005) | 0.369 | −0.003 (−0.024 to 0.008) | 0.308 | 0.557 (−0.827 to 1.941) | 0.434 |
| COMT | rs174690 | −0.046 (−0.148 to 0.056) | 0.380 | 0.004 (−0.005 to 0.014) | 0.386 | 0.008 (−0.008 to 0.023) | 0.334 | −0.824 (−2.128 to 0.481) | 0.221 |
| COMT | rs4818 | −0.005 (−0.107 to 0.097) | 0.923 | −0.002 (−0.011 to 0.008) | 0.699 | −0.002 (−0.017 to 0.013) | 0.785 | 0.495 (−0.815 to 1.806) | 0.462 |
| COMT | rs740603 | −0.084 (−0.183 to 0.016) | 0.105 | 0.004 (−0.006 to 0.013) | 0.426 | 0.006 (−0.009 to 0.021) | 0.459 | −0.226 (−1.536 to 1.084) | 0.737 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.Y.; Kim, H.; Jang, Y.H.; Hwang, W.; Hur, J.K.; Kim, Y.-E.; Lim, S.; Ye, D.-H.; Lee, H.J. The Impact of OXTR, COMT, and GRIN2B Polymorphisms on Brain Development in Preterm Infants. J. Clin. Med. 2025, 14, 8233. https://doi.org/10.3390/jcm14228233
Kim EY, Kim H, Jang YH, Hwang W, Hur JK, Kim Y-E, Lim S, Ye D-H, Lee HJ. The Impact of OXTR, COMT, and GRIN2B Polymorphisms on Brain Development in Preterm Infants. Journal of Clinical Medicine. 2025; 14(22):8233. https://doi.org/10.3390/jcm14228233
Chicago/Turabian StyleKim, Eon Yak, Hyuna Kim, Yong Hun Jang, Woochang Hwang, Junho K Hur, Young-Eun Kim, Sungmin Lim, Dong-Hye Ye, and Hyun Ju Lee. 2025. "The Impact of OXTR, COMT, and GRIN2B Polymorphisms on Brain Development in Preterm Infants" Journal of Clinical Medicine 14, no. 22: 8233. https://doi.org/10.3390/jcm14228233
APA StyleKim, E. Y., Kim, H., Jang, Y. H., Hwang, W., Hur, J. K., Kim, Y.-E., Lim, S., Ye, D.-H., & Lee, H. J. (2025). The Impact of OXTR, COMT, and GRIN2B Polymorphisms on Brain Development in Preterm Infants. Journal of Clinical Medicine, 14(22), 8233. https://doi.org/10.3390/jcm14228233





