Voxel-Based Dose–Toxicity Modeling for Predicting Post-Radiotherapy Toxicity: A Critical Perspective
Abstract
1. Introduction
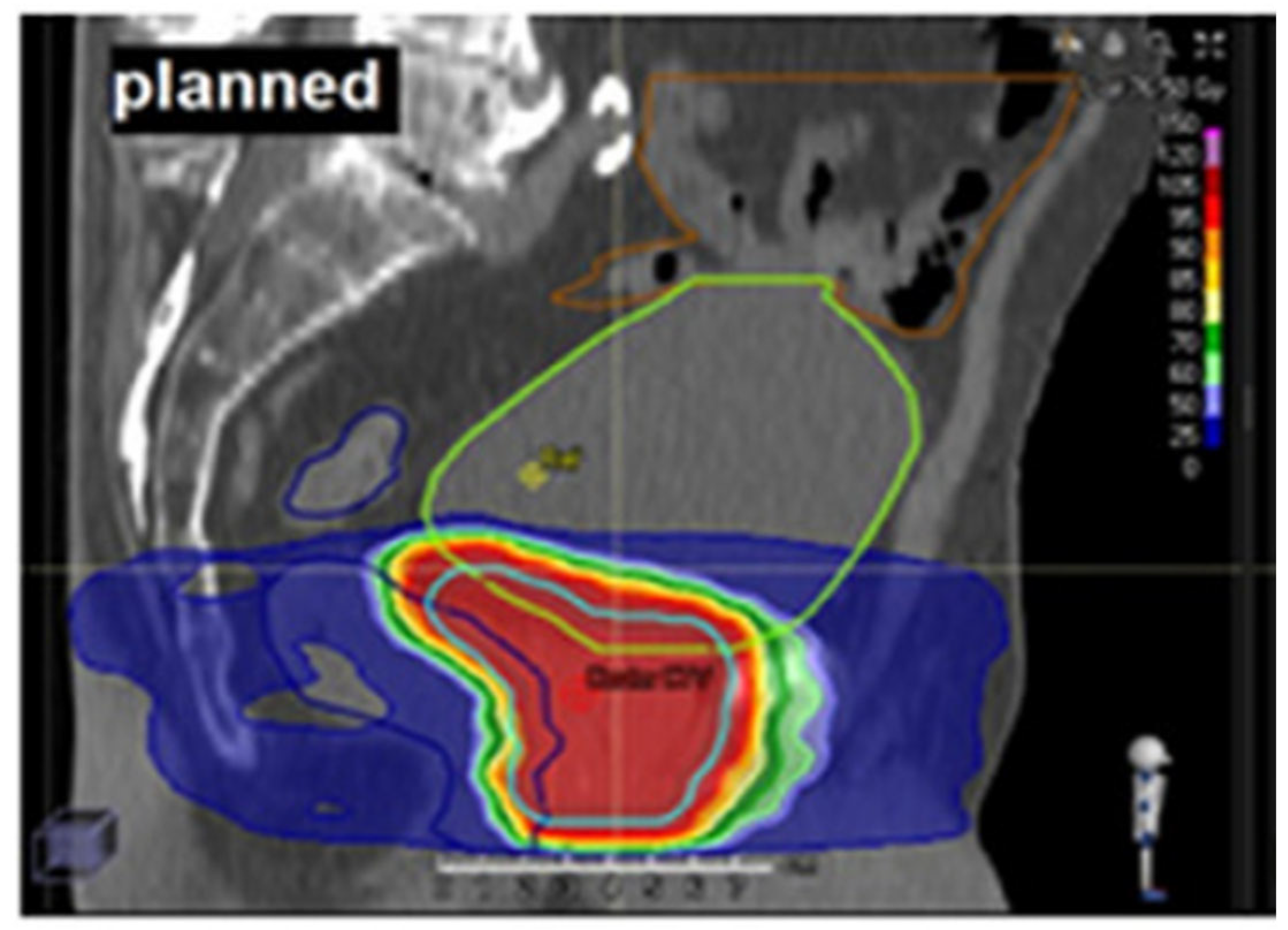
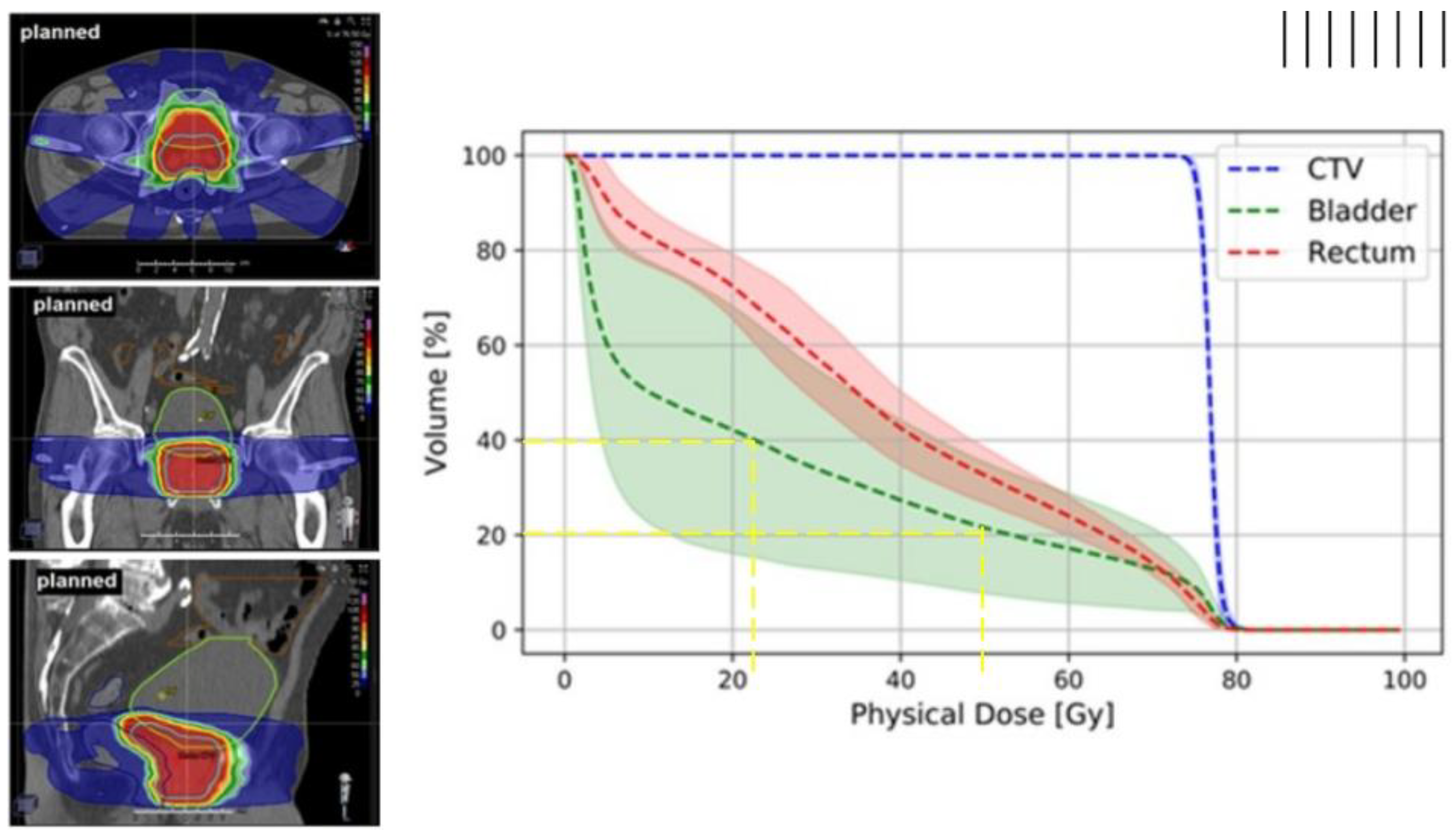
1.1. Dose–Volume Histograms
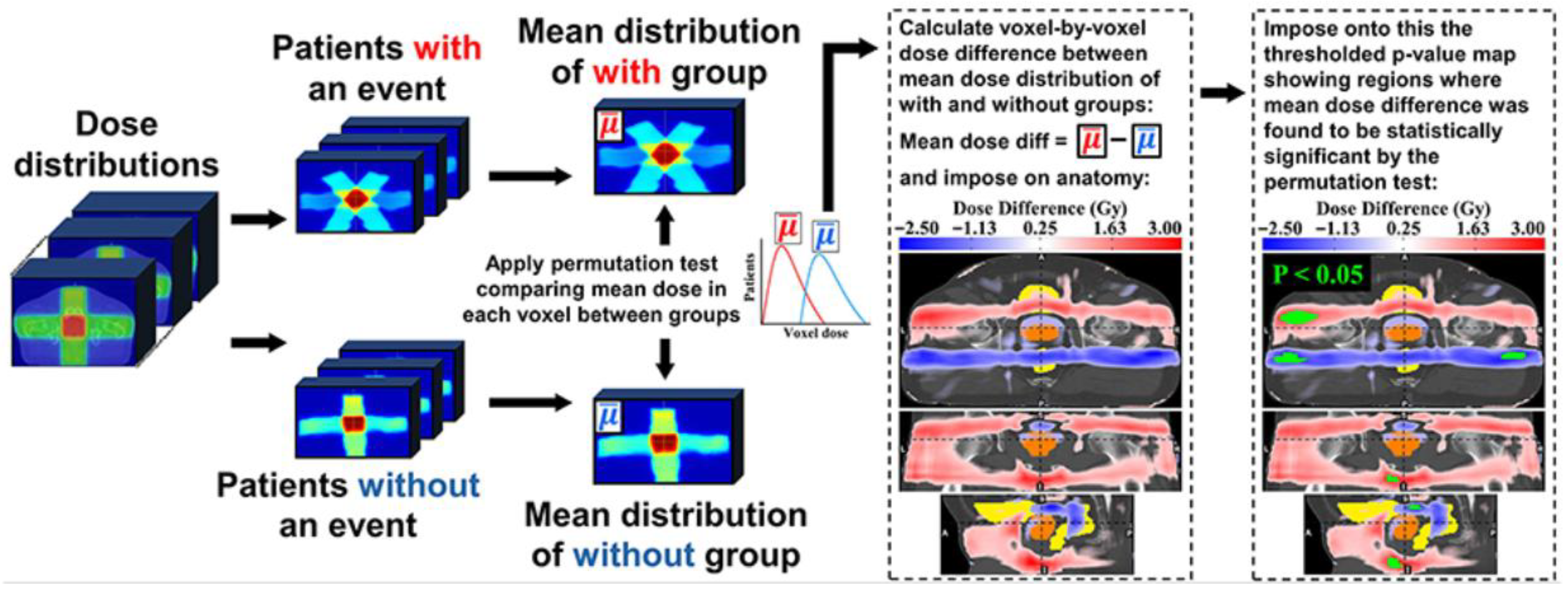
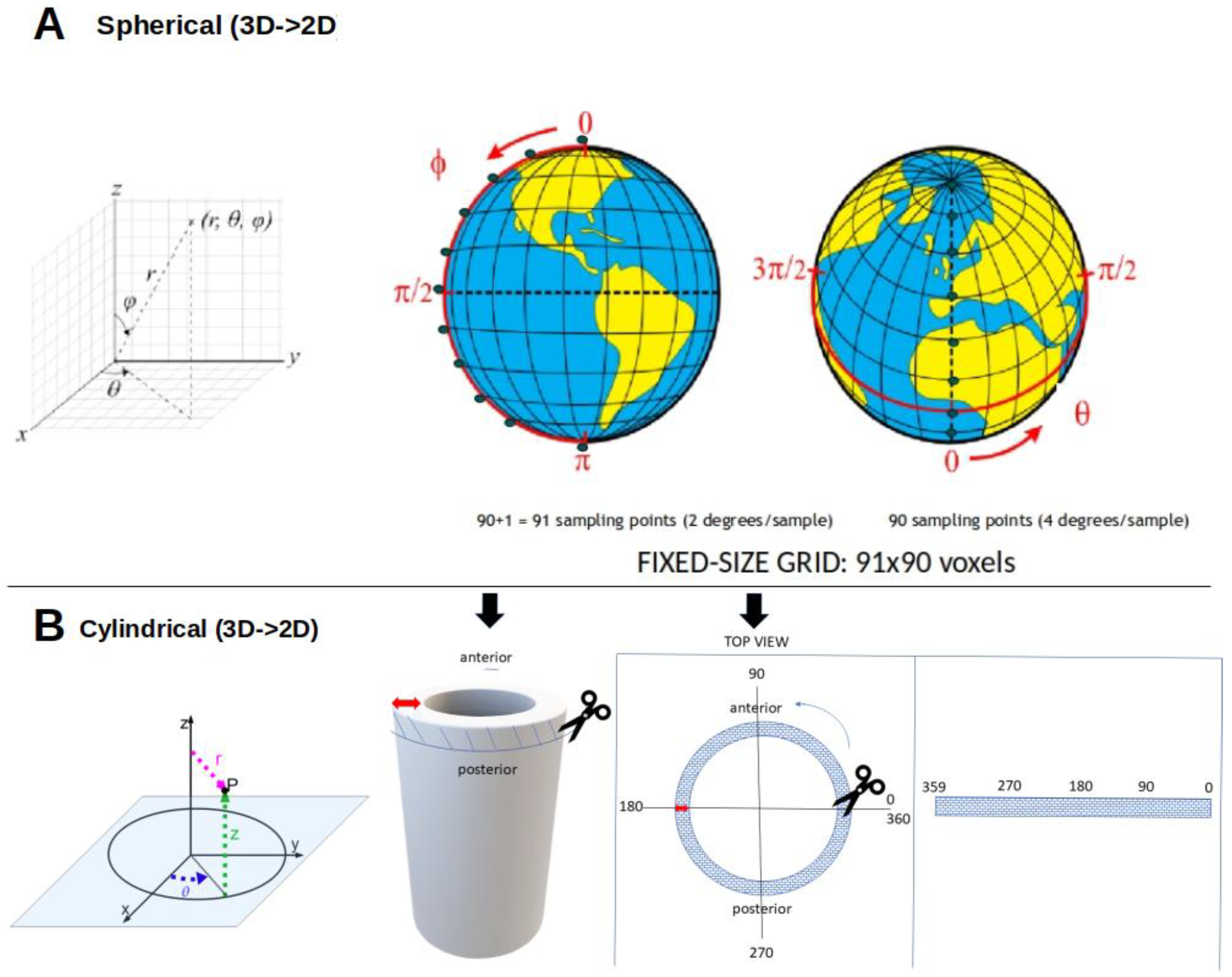
1.2. Voxel-Based Analysis (VBA) or Image-Based Data Mining (IBDM)
2. Problem
- (i)
- Lack of clinical validation of dose–toxicity models. This remains the most critical barrier because, without outcome-based validation demonstrating clinical benefit, statistical maps are hypothesis-generating only and cannot be directly translated into practice.
- (ii)
- Choice between parametric and non-parametric models, because model choice influences sensitivity to identify high-risk subregions within OARs, and robustness to assumption violations, thereby affecting reproducibility.
- (iii)
- Sensitivity of statistical models to uniform dose scaling, which can obscure clinically relevant differences when the dose scale changes.
- (iv)
- Choice of one-tailed versus two-tailed tests, which affects statistical power, false-positive rates, and interpretability.
- (v)
- Correct implementation of permutation testing to control the family-wise error rate (FWER). Improper use can inflate false positives or reduce interpretability.
- (vi)
- Use of dose-only predictors, which ignores patient, treatment, genomic, anatomical, and other clinical covariates that may confound dose–toxicity associations.
- (vii)
- Interpreting associations as causal without an appropriate framework, which risks misattribution of toxicity to dose when associations may instead be driven by confounding, bias, or artefacts.
3. Discussion
3.1. Dose–Toxicity Modeling Lacking Clinical Validation
3.2. Choosing Between Parametric and Nonparametric Statistical Models
3.3. Statistical Invariance to Dose Scaling or Shift
3.3.1. Scale and Shift Invariance in Common Tests
3.3.2. Clinical Pitfalls of Scale Invariance
3.3.3. Mitigating Invariance at the Model Level
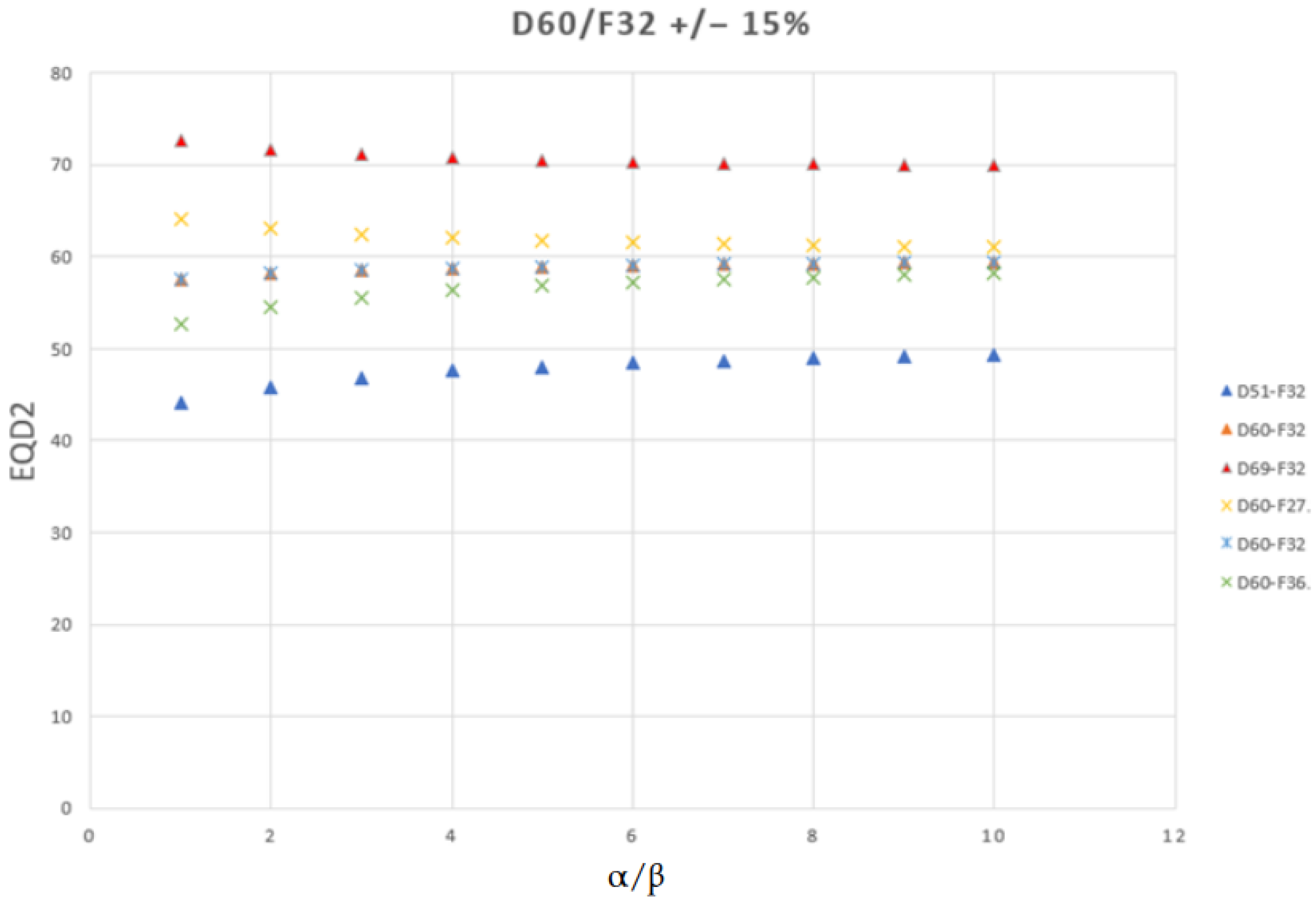
3.3.4. When Scale Invariance Is Appropriate: The EQD2 Context
3.4. Tail Choice in Hypothesis Testing
3.5. Error Types, Power, and Permutation Testing in VBA/IBDM
3.5.1. Permutation Testing
3.5.2. Variants of the Test Statistic
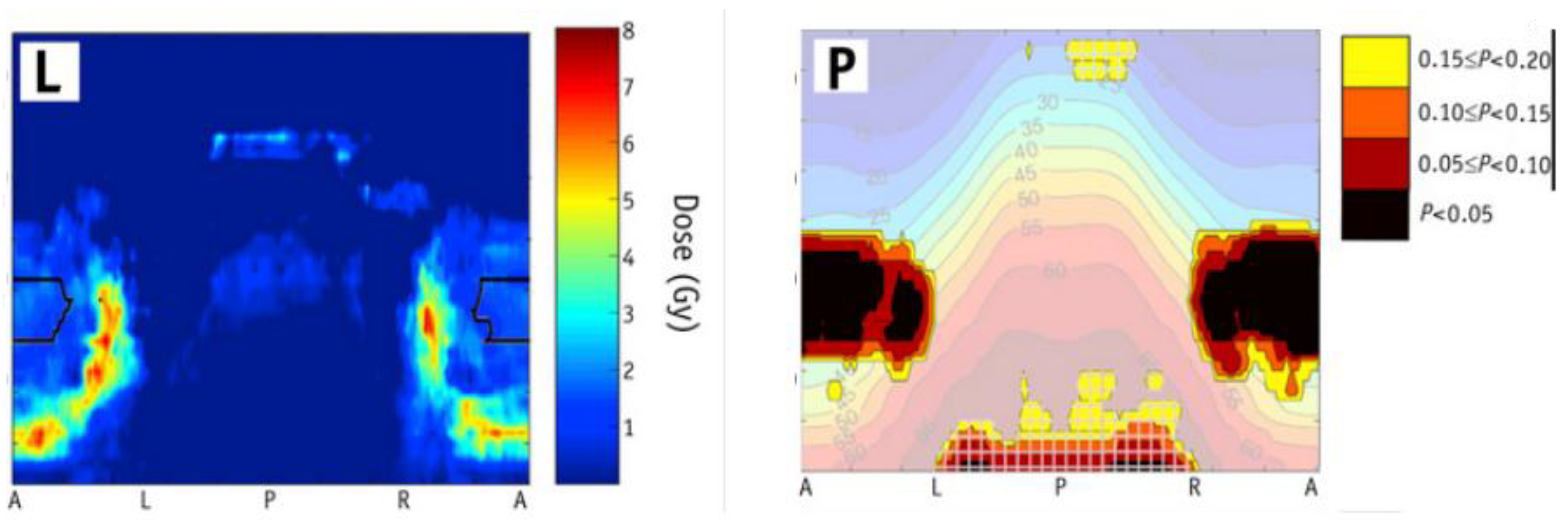
3.5.3. Spatial Dependence and Adaptive Methods
3.5.4. Enforcing Directionality
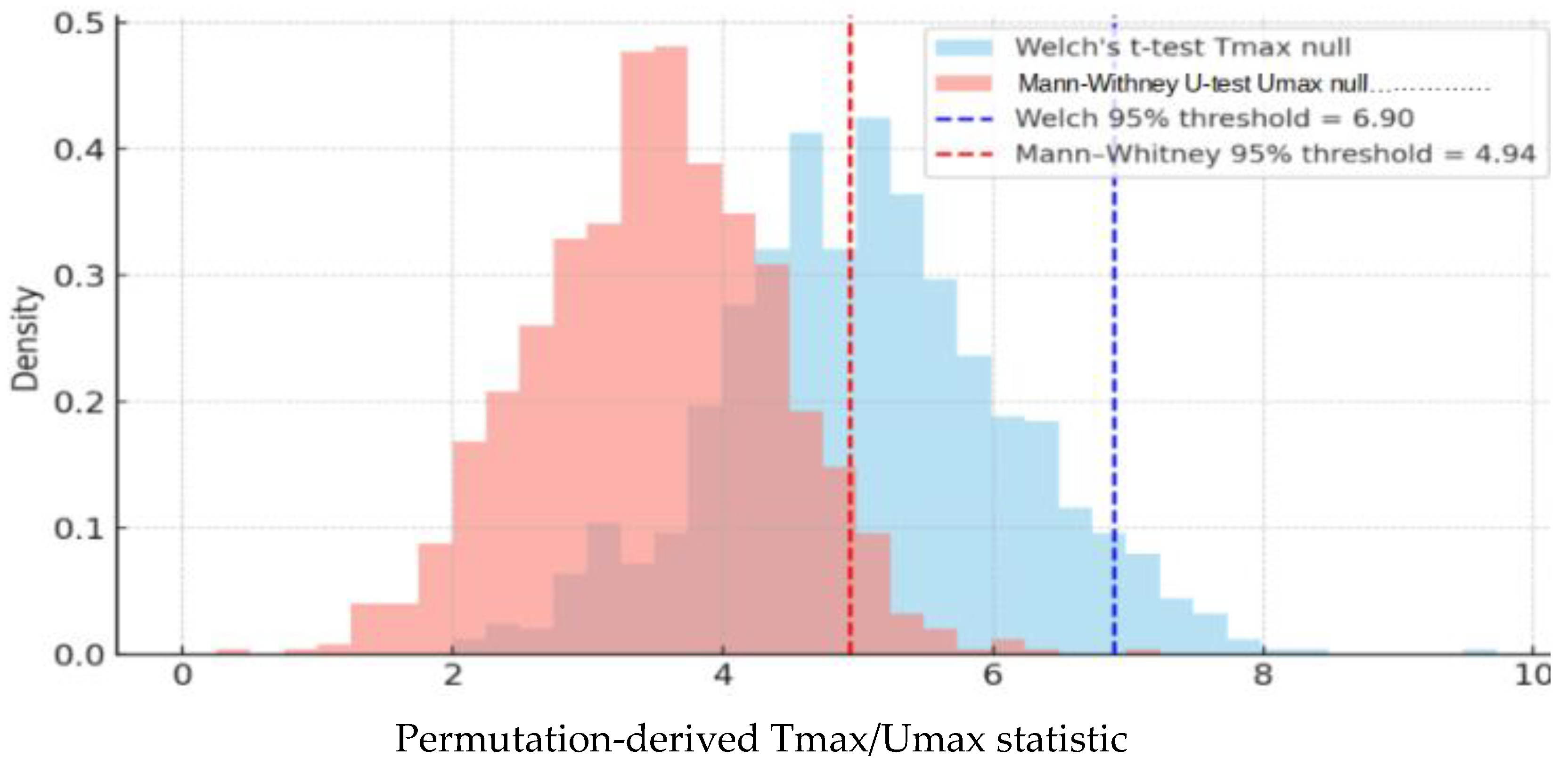
3.5.5. Power Calculations and Critical Perspectives
3.6. Using Dose-Only Data as the Predictor of Post-Radiotherapy Toxicity
3.7. Safeguards in Dose–Toxicity Modeling
3.8. Association Does Not Imply Causality
4. Why Is Dose–Survival Modeling Not Discussed?
5. Limitations
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vanherk, M. Errors and Margins in Radiotherapy. Semin. Radiat. Oncol. 2004, 14, 52–64. [Google Scholar] [CrossRef]
- Di Franco, R.; Borzillo, V.; Ravo, V.; Ametrano, G.; Falivene, S.; Cammarota, F.; Rossetti, S.; Romano, F.J.; D’Aniello, C.; Cavaliere, C.; et al. Rectal/Urinary Toxicity after Hypofractionated vs. Conventional Radiotherapy in Low/Intermediate Risk Localized Prostate Cancer: Systematic Review and Meta Analysis. Oncotarget 2017, 8, 17383–17395. [Google Scholar] [CrossRef]
- Dawson, L.A.; Sharpe, M.B. Image-Guided Radiotherapy: Rationale, Benefits, and Limitations. Lancet Oncol. 2006, 7, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Bostel, T.; Sachpazidis, I.; Splinter, M.; Bougatf, N.; Fechter, T.; Zamboglou, C.; Jäkel, O.; Huber, P.E.; Baltas, D.; Debus, J.; et al. Dosimetric Impact of Interfractional Variations in Prostate Cancer Radiotherapy—Implications for Imaging Frequency and Treatment Adaptation. Front. Oncol. 2019, 9, 940. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.X.; Wang, M.L.; Chen, L.X.; Liu, X. Evaluation of Dose-Volume Histogram Prediction for Organ-at Risk and Planning Target Volume Based on Machine Learning. Sci. Rep. 2021, 11, 3117. [Google Scholar] [CrossRef] [PubMed]
- Mirandola, A.; Russo, S.; Bonora, M.; Vischioni, B.; Camarda, A.M.; Ingargiola, R.; Molinelli, S.; Ronchi, S.; Rossi, E.; Vai, A.; et al. A Patient Selection Approach Based on NTCP Models and DVH Parameters for Definitive Proton Therapy in Locally Advanced Sinonasal Cancer Patients. Cancers 2022, 14, 2678. [Google Scholar] [CrossRef]
- Hanke, L.; Tang, H.; Schröder, C.; Windisch, P.; Kudura, K.; Shelan, M.; Buchali, A.; Bodis, S.; Förster, R.; Zwahlen, D. Dose-Volume Histogram Parameters and Quality of Life in Patients with Prostate Cancer Treated with Surgery and High-Dose Volumetric-Intensity-Modulated Arc Therapy to the Prostate Bed. Cancers 2023, 15, 3454. [Google Scholar] [CrossRef]
- Hu, J.; Schild, S.E.; Liu, W.; Li, J.; Fatyga, M. Improving Dose Volume Histogram (DVH) Based Analysis of Clinical Outcomes Using Modern Statistical Techniques: A Systematic Answer to Multiple Comparisons Concerns. Int. J. Radiat. Oncol. 2023, 117, S20. [Google Scholar] [CrossRef]
- Ting, J.Y.; Wu, X.; Fiedler, J.A.; Yang, C.; Watzich, M.L.; Markoe, A. Dose-Volume Histograms for Bladder and Rectum. Int. J. Radiat. Oncol. 1997, 38, 1105–1111. [Google Scholar] [CrossRef]
- Olsson, C.E.; Jackson, A.; Deasy, J.O.; Thor, M. A Systematic Post-QUANTEC Review of Tolerance Doses for Late Toxicity After Prostate Cancer Radiation Therapy. Int. J. Radiat. Oncol. 2018, 102, 1514–1532. [Google Scholar] [CrossRef]
- Moiseenko, V.; Van Dyk, J.; Battista, J.; Travis, E. Limitations in Using Dose-Volume Histograms for Radiotherapy Dose Optimization. In The Use of Computers in Radiation Therapy; Springer: Berlin/Heidelberg, Germany, 2000; pp. 239–241. ISBN 978-3-642-59758-9. [Google Scholar]
- McDonald, A.M.; Schneider, C.S.; Stahl, J.M.; Oster, R.A.; Popple, R.A.; Mayo, C.S. A Focused Review of Statistical Practices for Relating Radiation Dose-Volume Exposure and Toxicity. Radiat. Oncol. 2023, 18, 57. [Google Scholar] [CrossRef]
- Landoni, V.; Fiorino, C.; Cozzarini, C.; Sanguineti, G.; Valdagni, R.; Rancati, T. Predicting Toxicity in Radiotherapy for Prostate Cancer. Phys. Med. 2016, 32, 521–532. [Google Scholar] [CrossRef]
- Acosta, O.; Drean, G.; Ospina, J.D.; Simon, A.; Haigron, P.; Lafond, C.; de Crevoisier, R. Voxel-Based Population Analysis for Correlating Local Dose and Rectal Toxicity in Prostate Cancer Radiotherapy. Phys. Med. Biol. 2013, 58, 2581. [Google Scholar] [CrossRef]
- Wortel, R.C.; Witte, M.G.; Van Der Heide, U.A.; Pos, F.J.; Lebesque, J.V.; Van Herk, M.; Incrocci, L.; Heemsbergen, W.D. Dose–Surface Maps Identifying Local Dose–Effects for Acute Gastrointestinal Toxicity after Radiotherapy for Prostate Cancer. Radiother. Oncol. 2015, 117, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Palma, G.; Monti, S.; D’Avino, V.; Conson, M.; Liuzzi, R.; Pressello, M.C.; Donato, V.; Deasy, J.O.; Quarantelli, M.; Pacelli, R.; et al. A Voxel-Based Approach to Explore Local Dose Differences Associated With Radiation-Induced Lung Damage. Int. J. Radiat. Oncol. 2016, 96, 127–133. [Google Scholar] [CrossRef]
- Marcello, M.; Denham, J.W.; Kennedy, A.; Haworth, A.; Steigler, A.; Greer, P.B.; Holloway, L.C.; Dowling, J.A.; Jameson, M.G.; Roach, D.; et al. Increased Dose to Organs in Urinary Tract Associates With Measures of Genitourinary Toxicity in Pooled Voxel-Based Analysis of 3 Randomized Phase III Trials. Front. Oncol. 2020, 10, 1174. [Google Scholar] [CrossRef] [PubMed]
- Puri, T.; Rancati, T.; Seibold, P.; Webb, A.; Osorio, E.V.; Green, A.; Gioscio, E.; Azria, D.; Farcy-Jacquet, M.-P.; Chang-Claude, J.; et al. Dose-Response Mapping of Bladder and Rectum in Prostate Cancer Patients Undergoing Radiotherapy with and without Baseline Toxicity Correction. Phys. Imaging Radiat. Oncol. 2025, 35, 100805. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Moore, D.F.; Shih, W.; Lin, Y.; Li, H.; Shao, Y.-H.; Shen, S.; Lu-Yao, G.L. Severe Genitourinary Toxicity Following Radiation Therapy for Prostate Cancer—How Long Does It Last? J. Urol. 2013, 189, 116–121. [Google Scholar] [CrossRef]
- Seibold, P.; Webb, A.; Aguado-Barrera, M.E.; Azria, D.; Bourgier, C.; Brengues, M.; Briers, E.; Bultijnck, R.; Calvo-Crespo, P.; Carballo, A.; et al. REQUITE: A Prospective Multicentre Cohort Study of Patients Undergoing Radiotherapy for Breast, Lung or Prostate Cancer. Radiother. Oncol. 2019, 138, 59–67. [Google Scholar] [CrossRef]
- Jaikuna, T.; Wilson, F.; Azria, D.; Chang-Claude, J.; De Santis, M.C.; Gutiérrez-Enríquez, S.; Van Herk, M.; Hoskin, P.; Kotzki, L.; Lambrecht, M.; et al. Optimising Inter-Patient Image Registration for Image-Based Data Mining in Breast Radiotherapy. Phys. Imaging Radiat. Oncol. 2024, 32, 100635. [Google Scholar] [CrossRef]
- Snyder, J.P. Map Projections: A Working Manual; U.S. Geological Survey Professional Paper; U.S. Government Printing Office: Washington, DC, USA, 1987. [Google Scholar]
- McMahon, S.J. The Linear Quadratic Model: Usage, Interpretation and Challenges. Phys. Med. Biol. 2018, 64, 01TR01. [Google Scholar] [CrossRef] [PubMed]
- Fowler, J.F. The Linear-Quadratic Model and Progress in Fractionated Radiotherapy. Br. J. Radiol. 1989, 62, 679–694. [Google Scholar] [CrossRef] [PubMed]
- Thames, H.D.; Withers, H.R.; Peters, L.J.; Fletcher, G.H. Changes in Early and Late Radiation Responses with Altered Dose Fractionation: Implications for Dose-Survival Relationships. Int. J. Radiat. Oncol. Biol. Phys. 1982, 8, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Brand, D.H.; Brüningk, S.C.; Wilkins, A.; Fernandez, K.; Naismith, O.; Gao, A.; Syndikus, I.; Dearnaley, D.P.; Tree, A.C.; Van As, N.; et al. Estimates of Alpha/Beta (α/β) Ratios for Individual Late Rectal Toxicity Endpoints: An Analysis of the CHHiP Trial. Int. J. Radiat. Oncol. 2021, 110, 596–608. [Google Scholar] [CrossRef]
- Brand, D.H.; Brüningk, S.C.; Wilkins, A.; Naismith, O.; Gao, A.; Syndikus, I.; Dearnaley, D.P.; Van As, N.; Hall, E.; Gulliford, S.; et al. The Fraction Size Sensitivity of Late Genitourinary Toxicity: Analysis of Alpha/Beta (α/β) Ratios in the CHHiP Trial. Int. J. Radiat. Oncol. 2023, 115, 327–336. [Google Scholar] [CrossRef]
- Fiorino, C.; Cozzarini, C.; Rancati, T.; Briganti, A.; Cattaneo, G.M.; Mangili, P.; Di Muzio, N.G.; Calandrino, R. Modelling the Impact of Fractionation on Late Urinary Toxicity After Postprostatectomy Radiation Therapy. Int. J. Radiat. Oncol. 2014, 90, 1250–1257. [Google Scholar] [CrossRef]
- Cui, M.; Gao, X.-S.; Li, X.; Ma, M.; Qi, X.; Shibamoto, Y. Variability of α/β Ratios for Prostate Cancer with the Fractionation Schedule: Caution against Using the Linear-Quadratic Model for Hypofractionated Radiotherapy. Radiat. Oncol. 2022, 17, 54. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591. [Google Scholar] [CrossRef]
- Welch, B.L. The Generalization of ‘Student’s’ Problem When Several Different Population Variances Are Involved. Biometrika 1947, 34, 28. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether One of Two Random Variables Is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Chen, C.; Witte, M.; Heemsbergen, W.; Herk, M.V. Multiple Comparisons Permutation Test for Image Based Data Mining in Radiotherapy. Radiat. Oncol. 2013, 8, 293. [Google Scholar] [CrossRef]
- Holmes, A.P.; Blair, R.C.; Watson, J.D.G.; Ford, I. Nonparametric Analysis of Statistic Images from Functional Mapping Experiments. J. Cereb. Blood Flow Metab. 1996, 16, 7–22. [Google Scholar] [CrossRef]
- Thomas, G.E.; Westfall, P.H.; Young, S.S. Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment. J. R. Stat. Soc. Ser. D 1994, 43, 347. [Google Scholar] [CrossRef]
- Puri, T.; Gioscio, E.; van Herk, M. Comparison of Two Independently Developed Voxel-Based Dose-Response Mapping Algorithms at Two Different Centres for Bladder and Rectum in Prostate Cancer Patients Undergoing Radical Radiotherapy; AAPM: Los Angeles, CA, USA, 2024. [Google Scholar]
- Gioscio, E.; Cicchetti, A.; Iacovacci, J.; Spampinato, S.; Waskiewicz, J.M.; Avuzzi, B.; Garibaldi, E.; Villa, E.; Magli, A.; Cante, D.; et al. PO-2099 Bladder Dose Surface Maps Identify Subregions Associated to Late Toxicities after Prostate Cancer RT. Radiother. Oncol. 2023, 182, S1882–S1883. [Google Scholar] [CrossRef]
- Nichols, T.E.; Holmes, A.P. Nonparametric Permutation Tests for Functional Neuroimaging: A Primer with Examples. Hum. Brain Mapp. 2001, 15, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Winkler, A.M.; Ridgway, G.R.; Webster, M.A.; Smith, S.M.; Nichols, T.E. Permutation Inference for the General Linear Model. NeuroImage 2014, 92, 381–397. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, A.; Palma, G.; Abravan, A.; Acosta, O.; Appelt, A.; Aznar, M.; Monti, S.; Onjukka, E.; Panettieri, V.; Placidi, L.; et al. Voxel-Based Analysis: Roadmap for Clinical Translation. Radiother. Oncol. 2023, 188, 109868. [Google Scholar] [CrossRef] [PubMed]
- Craddock, M.; Nestle, U.; Koenig, J.; Schimek-Jasch, T.; Kremp, S.; Lenz, S.; Banfill, K.; Davey, A.; Price, G.; Salem, A.; et al. Cardiac Function Modifies the Impact of Heart Base Dose on Survival: A Voxel-Wise Analysis of Patients With Lung Cancer From the PET-Plan Trial. J. Thorac. Oncol. 2023, 18, 57–66. [Google Scholar] [CrossRef]
- Vasquez Osorio, E.; Abravan, A.; Green, A.; Van Herk, M.; Lee, L.W.; Ganderton, D.; McPartlin, A. Dysphagia at 1 Year Is Associated With Mean Dose to the Inferior Section of the Brain Stem. Int. J. Radiat. Oncol. 2023, 117, 903–913. [Google Scholar] [CrossRef]
- Shelley, L.E.A.; Sutcliffe, M.P.F.; Thomas, S.J.; Noble, D.J.; Romanchikova, M.; Harrison, K.; Bates, A.M.; Burnet, N.G.; Jena, R. Associations between Voxel-Level Accumulated Dose and Rectal Toxicity in Prostate Radiotherapy. Phys. Imaging Radiat. Oncol. 2020, 14, 87–94. [Google Scholar] [CrossRef]
- Shelley, L.E.A.; Scaife, J.E.; Romanchikova, M.; Harrison, K.; Forman, J.R.; Bates, A.M.; Noble, D.J.; Jena, R.; Parker, M.A.; Sutcliffe, M.P.F.; et al. Delivered Dose Can Be a Better Predictor of Rectal Toxicity than Planned Dose in Prostate Radiotherapy. Radiother. Oncol. 2017, 123, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Puri, T.; Rancati, T.; Seibold, P.; Webb, A.; Osorio, E.V.; Azria, D.; Farcy-Jacquet, M.-P.; Chang-Claude, J.; Dunning, A.; Lambrecht, M.; et al. Sensitivity Analysis of Dose-Response Model Parameters for the Bladder in Prostate Cancer Radiotherapy. Phys. Med. 2025, 139, 105178. [Google Scholar] [CrossRef] [PubMed]
- El Naqa, I. Perspectives on Making Big Data Analytics Work for Oncology. Methods 2016, 111, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; McShane, L.M.; Michiels, S.; Ransohoff, D.F.; Altman, D.G.; Reis-Filho, J.S.; Hayes, D.F.; Pusztai, L. Biomarker Studies: A Call for a Comprehensive Biomarker Study Registry. Nat. Rev. Clin. Oncol. 2011, 8, 171–176. [Google Scholar] [CrossRef]
- Lake, P.; Drake, R. Information Systems Management in the Big Data Era; Advanced Information and Knowledge Processing; Springer International Publishing: Cham, Switzerland, 2014; ISBN 978-3-319-13502-1. [Google Scholar]
- Goodman, S.N.; Fanelli, D.; Ioannidis, J.P.A. What Does Research Reproducibility Mean? Sci. Transl. Med. 2016, 8, 341ps12. [Google Scholar] [CrossRef]
- Heinze, G.; Wallisch, C.; Dunkler, D. Variable Selection—A Review and Recommendations for the Practicing Statistician. Biom. J. 2018, 60, 431–449. [Google Scholar] [CrossRef]
- Open Science Collaboration Estimating the Reproducibility of Psychological Science. Science 2015, 349, aac4716. [CrossRef]
- Gundersen, O.E. The Fundamental Principles of Reproducibility. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2021, 379, 20200210. [Google Scholar] [CrossRef]
- Cagni, E.; Botti, A.; Orlandi, M.; Galaverni, M.; Iotti, C.; Iori, M.; Lewis, G.; Spezi, E. Evaluating the Quality of Patient-Specific Deformable Image Registration in Adaptive Radiotherapy Using a Digitally Enhanced Head and Neck Phantom. Appl. Sci. 2022, 12, 9493. [Google Scholar] [CrossRef]
- Lee, S.; Kim, H.; Ji, Y.; Cho, B.; Kim, S.S.; Jung, J.; Kwak, J.; Park, J.; Lee, S.; Kim, J.H.; et al. Evaluation of Hepatic Toxicity after Repeated Stereotactic Body Radiation Therapy for Recurrent Hepatocellular Carcinoma Using Deformable Image Registration. Sci. Rep. 2018, 8, 16224. [Google Scholar] [CrossRef]
- Rigaud, B.; Simon, A.; Castelli, J.; Lafond, C.; Acosta, O.; Haigron, P.; Cazoulat, G.; De Crevoisier, R. Deformable Image Registration for Radiation Therapy: Principle, Methods, Applications and Evaluation. Acta Oncol. 2019, 58, 1225–1237. [Google Scholar] [CrossRef]
- Mir, R.; Kelly, S.M.; Xiao, Y.; Moore, A.; Clark, C.H.; Clementel, E.; Corning, C.; Ebert, M.; Hoskin, P.; Hurkmans, C.W.; et al. Organ at Risk Delineation for Radiation Therapy Clinical Trials: Global Harmonization Group Consensus Guidelines. Radiother. Oncol. 2020, 150, 30–39. [Google Scholar] [CrossRef]
- Noël, G.; Le Fèvre, C.; Antoni, D. Delineation of Organs at Risk. Cancer/Radiothérapie 2022, 26, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Segedin, B.; Petric, P. Uncertainties in Target Volume Delineation in Radiotherapy—Are They Relevant and What Can We Do about Them? Radiol. Oncol. 2016, 50, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Dou, X.; Liu, J.; Feng, R.; Zhang, X.; Yue, J. National Competition Reveals Common Errors in Target Delineation among Young Clinicians in Radiation Therapy. BMC Med. Educ. 2025, 25, 870. [Google Scholar] [CrossRef] [PubMed]
- Karimkhani, L.; Saeedzadeh, E.; Sardari, D.; Mahdavi, S.R. Evaluation of Monaco Dose Calculation Errors for Out-of-Field Regions in Intensity Modulated Radiotherapy of Nasopharyngeal Cancer. Int. J. Radiat. Res. 2025, 23, 225–232. [Google Scholar] [CrossRef]
- Williams, G.; Tobler, M.; Gaffney, D.; Moeller, J.; Leavitt, D.D. Dose Calculation Errors Due to Inaccurate Representation of Heterogeneity Correction Obtained from Computerized Tomography. Med. Dosim. 2002, 27, 275–278. [Google Scholar] [CrossRef]
- Chen, W.-Z. Impact of Dose Calculation Algorithm on Radiation Therapy. World J. Radiol. 2014, 6, 874. [Google Scholar] [CrossRef]
- van der Merwe, D.; Van Dyk, J.; Healy, B.; Zubizarreta, E.; Izewska, J.; Mijnheer, B.; Meghzifene, A. Accuracy Requirements and Uncertainties in Radiotherapy: A Report of the International Atomic Energy Agency. Acta Oncol. 2016, 56, 1–6. [Google Scholar] [CrossRef]
- Batenburg, M.C.T.; Bartels, M.; Maarse, W.; Witkamp, A.; Verkooijen, H.M.; van den Bongard, H.J.G.D. Factors Associated with Late Local Radiation Toxicity after Post-Operative Breast Irradiation. Breast J. 2022, 2022, 1–13. [Google Scholar] [CrossRef]
- Pignon, T.; Gregor, A.; Schaake Koning, C.; Roussel, A.; Van Glabbeke, M.; Scalliet, P. Age Has No Impact on Acute and Late Toxicity of Curative Thoracic Radiotherapy. Radiother. Oncol. 1998, 46, 239–248. [Google Scholar] [CrossRef]
- Holmqvist, A.; Lindahl, G.; Mikivier, R.; Uppungunduri, S. Age as a Potential Predictor of Acute Side Effects during Chemoradiotherapy in Primary Cervical Cancer Patients. BMC Cancer 2022, 22, 371. [Google Scholar] [CrossRef]
- Massi, M.C.; Gasperoni, F.; Ieva, F.; Paganoni, A.M.; Zunino, P.; Manzoni, A.; Franco, N.R.; Veldeman, L.; Ost, P.; Fonteyne, V.; et al. A Deep Learning Approach Validates Genetic Risk Factors for Late Toxicity After Prostate Cancer Radiotherapy in a REQUITE Multi-National Cohort. Front. Oncol. 2020, 10, 541281. [Google Scholar] [CrossRef]
- Palumbo, E.; Piotto, C.; Calura, E.; Fasanaro, E.; Groff, E.; Busato, F.; El Khouzai, B.; Rigo, M.; Baggio, L.; Romualdi, C.; et al. Individual Radiosensitivity in Oncological Patients: Linking Adverse Normal Tissue Reactions and Genetic Features. Front. Oncol. 2019, 9, 987. [Google Scholar] [CrossRef]
- van Rossum, P.S.N.; Wolfhagen, N.; van Bockel, L.W.; Coremans, I.E.M.; van Es, C.A.; van der Geest, A.M.; De Jaeger, K.E.A.; Wachters, B.; Knol, H.P.; Koppe, F.L.A.; et al. Real-World Acute Toxicity and 90-Day Mortality in Patients With Stage I NSCLC Treated With Stereotactic Body Radiotherapy. J. Thorac. Oncol. 2024, 19, 1550–1563. [Google Scholar] [CrossRef] [PubMed]
- van der Weijst, L.; Azria, D.; Berkovic, P.; Boisselier, P.; Briers, E.; Bultijnck, R.; Chang-Claude, J.; Choudhury, A.; Defraene, G.; Demontois, S.; et al. The Correlation between Pre-Treatment Symptoms, Acute and Late Toxicity and Patient-Reported Health-Related Quality of Life in Non-Small Cell Lung Cancer Patients: Results of the REQUITE Study. Radiother. Oncol. 2022, 176, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Lehrer, E.J.; Rosenberg, J.; Trifiletti, D.M.; Zaorsky, N.G. Toxicity after Radiotherapy in Patients with Historically Accepted Contraindications to Treatment (CONTRAD): An International Systematic Review and Meta-Analysis. Radiother. Oncol. 2019, 135, 147–152. [Google Scholar] [CrossRef]
- Lee, J.; Chang, C.-L.; Lin, J.-B.; Wu, M.-H.; Sun, F.-J.; Wu, C.-J.; Tai, H.-C.; Hsu, S.-M.; Chen, Y.-J. The Effect of Body Mass Index and Weight Change on Late Gastrointestinal Toxicity in Locally Advanced Cervical Cancer Treated With Intensity-Modulated Radiotherapy. Int. J. Gynecol. Cancer 2018, 28, 1377–1386. [Google Scholar] [CrossRef]
- Vargas, C.; Martinez, A.; Kestin, L.L.; Yan, D.; Grills, I.; Brabbins, D.S.; Lockman, D.M.; Liang, J.; Gustafson, G.S.; Chen, P.Y.; et al. Dose-Volume Analysis of Predictors for Chronic Rectal Toxicity after Treatment of Prostate Cancer with Adaptive Image-Guided Radiotherapy. Int. J. Radiat. Oncol. 2005, 62, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Fellin, G.; Fiorino, C.; Rancati, T.; Vavassori, V.; Baccolini, M.; Bianchi, C.; Cagna, E.; Gabriele, P.; Mauro, F.; Menegotti, L.; et al. Clinical and Dosimetric Predictors of Late Rectal Toxicity after Conformal Radiation for Localized Prostate Cancer: Results of a Large Multicenter Observational Study. Radiother. Oncol. 2009, 93, 197–202. [Google Scholar] [CrossRef]
- Valdagni, R.; Vavassori, V.; Rancati, T.; Fellin, G.; Baccolini, M.; Bianchi, C.; Cagna, E.; Gabriele, P.; Mauro, F.; Menegotti, L.; et al. Increasing the Risk of Late Rectal Bleeding after High-Dose Radiotherapy for Prostate Cancer: The Case of Previous Abdominal Surgery. Results from a Prospective Trial. Radiother. Oncol. 2012, 103, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, A.S.; Zelefsky, M.J.; Lee, H.J.; Lombardi, D.; Fuks, Z.; Leibel, S.A. Long-Term Urinary Toxicity after 3-Dimensional Conformal Radiotherapy for Prostate Cancer in Patients with Prior History of Transurethral Resection. Int. J. Radiat. Oncol. 2000, 48, 643–647. [Google Scholar] [CrossRef] [PubMed]
- Herold, D.M.; Hanlon, A.L.; Hanks, G.E. Diabetes Mellitus: A Predictor for Late Radiation Morbidity. Int. J. Radiat. Oncol. 1999, 43, 475–479. [Google Scholar] [CrossRef] [PubMed]
- K Jensen, N.B.; Pötter, R.; Spampinato, S.; Fokdal, L.U.; Chargari, C.; Lindegaard, J.C.; Schmid, M.P.; Sturdza, A.; Jürgenliemk-Schulz, I.M.; Mahantshetty, U.; et al. Dose-Volume Effects and Risk Factors for Late Diarrhea in Cervix Cancer Patients After Radiochemotherapy With Image Guided Adaptive Brachytherapy in the EMBRACE I Study. Int. J. Radiat. Oncol. 2021, 109, 688–700. [Google Scholar] [CrossRef]
- Fokdal, L.; Tanderup, K.; Pötter, R.; Sturdza, A.; Kirchheiner, K.; Chargari, C.; Jürgenliemk-Schulz, I.M.; Segedin, B.; Tan, L.-T.; Hoskin, P.; et al. Risk Factors for Ureteral Stricture After Radiochemotherapy Including Image Guided Adaptive Brachytherapy in Cervical Cancer: Results From the EMBRACE Studies. Int. J. Radiat. Oncol. 2019, 103, 887–894. [Google Scholar] [CrossRef]
- Eifel, P.J.; Jhingran, A.; Bodurka, D.C.; Levenback, C.; Thames, H. Correlation of Smoking History and Other Patient Characteristics With Major Complications of Pelvic Radiation Therapy for Cervical Cancer. J. Clin. Oncol. 2002, 20, 3651–3657. [Google Scholar] [CrossRef]
- Barnett, G.C.; De Meerleer, G.; Gulliford, S.L.; Sydes, M.R.; Elliott, R.M.; Dearnaley, D.P. The Impact of Clinical Factors on the Development of Late Radiation Toxicity: Results from the Medical Research Council RT01 Trial (ISRCTN47772397). Clin. Oncol. 2011, 23, 613–624. [Google Scholar] [CrossRef]
- Thomas, R.J.; Holm, M.; Williams, M.; Bowman, E.; Bellamy, P.; Andreyev, J.; Maher, J. Lifestyle Factors Correlate with the Risk of Late Pelvic Symptoms after Prostatic Radiotherapy. Clin. Oncol. 2013, 25, 246–251. [Google Scholar] [CrossRef]
- Laan, J.J.; Van Lonkhuijzen, L.R.C.W.; Van Os, R.M.; Tytgat, K.M.; Dávila Fajardo, R.; Pieters, B.R.; Stalpers, L.J.A.; Westerveld, G.H. Socioeconomic Status as an Independent Risk Factor for Severe Late Bowel Toxicity after Primary Radiotherapy for Cervical Cancer. Gynecol. Oncol. 2017, 147, 684–689. [Google Scholar] [CrossRef]
- Bentzen, S.M. Preventing or Reducing Late Side Effects of Radiation Therapy: Radiobiology Meets Molecular Pathology. Nat. Rev. Cancer 2006, 6, 702–713. [Google Scholar] [CrossRef]
- Ching, J.C.F.; Liu, K.C.K.; Pang, I.K.H.; Nicol, A.J.; Leung, V.W.S.; Cai, J.; Lee, S.W.Y. Predictive Factors for Gastrointestinal and Genitourinary Toxicities in Prostate Cancer External Beam Radiotherapy: A Scoping Review. Diagnostics 2025, 15, 1331. [Google Scholar] [CrossRef]
- Rades, D.; Cremers, F.; Ziemann, C.; Splettstösser, L.; Narvaez-Wolf, C.A.; Al-Salool, A.; Von Staden, M.; Küter, J.-D.; Janssen, S.; Timke, C. Changes in Bladder Volume During Radiotherapy for Prostate Cancer. Anticancer Res. 2024, 44, 4457–4463. [Google Scholar] [CrossRef] [PubMed]
- Joyce, R.; Murphy, J.; O’Neill, E.; Lyons, C.A. Bladder Volume Variation in Hypofractionated Prostate Radiotherapy. J. Clin. Oncol. 2022, 40, 234. [Google Scholar] [CrossRef]
- Willigenburg, T.; Van Der Velden, J.M.; Zachiu, C.; Teunissen, F.R.; Lagendijk, J.J.W.; Raaymakers, B.W.; De Boer, J.C.J.; Van Der Voort Van Zyp, J.R.N. Accumulated Bladder Wall Dose Is Correlated with Patient-Reported Acute Urinary Toxicity in Prostate Cancer Patients Treated with Stereotactic, Daily Adaptive MR-Guided Radiotherapy. Radiother. Oncol. 2022, 171, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, A.N.; Yorke, E.D.; Marks, L.B.; Eifel, P.J.; Shipley, W.U. Radiation Dose–Volume Effects of the Urinary Bladder. Int. J. Radiat. Oncol. 2010, 76, S116–S122. [Google Scholar] [CrossRef]
- Vinnikov, V.; Hande, M.P.; Wilkins, R.; Wojcik, A.; Zubizarreta, E.; Belyakov, O. Prediction of the Acute or Late Radiation Toxicity Effects in Radiotherapy Patients Using Ex Vivo Induced Biodosimetric Markers: A Review. J. Pers. Med. 2020, 10, 285. [Google Scholar] [CrossRef]
- Ozyigit, G.; Hurmuz, P.; Bayatfard, P.; Tilki, B.; Yedekci, Y.; Yilmaz, M.T. Predicting Acute and Late Toxicity in Prostate Cancer Stereotactic Ablative Radiotherapy: The Role of Dosimetric Parameters and Prostate Volume. Strahlenther. Onkol. 2025, 201, 788–798. [Google Scholar] [CrossRef]
- Rancati, T. Dose-Response Mapping at the Bladder in Prostate Cancer Patients Undergoing Radiotherapy: Success, Challenges, and Future Direction. Aarhus University Hospital: Aarhus, Denmark, October 2024. [Google Scholar]
- Rancati, T. Genetically-Based Analysis of Dose Surface Maps for Assessing Toxicity Post-RT. In Proceedings of the XXth International Conference on the use of Computers in Radiation therapy, Lyon, France, 8 July 2024. [Google Scholar]
- Sidebotham, D.; Barlow, C.J. The Central Limit Theorem: The Remarkable Theory That Explains All of Statistics. Anaesthesia 2024, 79, 1117–1121. [Google Scholar] [CrossRef]
- Kwak, S.G.; Kim, J.H. Central Limit Theorem: The Cornerstone of Modern Statistics. Korean J. Anesthesiol. 2017, 70, 144. [Google Scholar] [CrossRef]
- Sawada, T. Conditions of the Central-Limit Theorem Are Rarely Satisfied in Empirical Psychological Studies. Front. Psychol. 2021, 12, 762418. [Google Scholar] [CrossRef]
- Permutation, Parametric and Bootstrap Tests of Hypotheses; Springer Series in Statistics; Springer: New York, NY, USA, 2005; ISBN 978-0-387-20279-2.
- Huber, P.J.; Ronchetti, E.M. Robust Statistics, Wiley Series in Probability and Statistics; 1st ed.; Wiley: Hoboken, NJ, USA, 2009; ISBN 978-0-470-12990-6. [Google Scholar]
- Davison, A.C.; Hinkley, D.V. Bootstrap Methods and Their Application, 1st ed.; Cambridge University Press: Cambridge, UK, 1997; ISBN 978-0-521-57391-7. [Google Scholar]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; Chapman and Hall/CRC: Boca Raton, FL, USA, 1994; ISBN 978-0-429-24659-3. [Google Scholar]
- Fay, M.P.; Proschan, M.A. Wilcoxon-Mann-Whitney or t-Test? On Assumptions for Hypothesis Tests and Multiple Interpretations of Decision Rules. Stat. Surv. 2010, 4, 1–39. [Google Scholar] [CrossRef]
- Pérez-Ortiz, M.F.; Lardy, T.; de Heide, R.; Grünwald, P. E-Statistics, Group Invariance and Anytime Valid Testing. arXiv 2023. [Google Scholar] [CrossRef]
- Hoff, P.D. Invariant Statistical Procedures [Lecture Notes]; Duke University: Durham, NC, USA, 2025. [Google Scholar]
- Monti, S.; Xu, T.; Mohan, R.; Liao, Z.; Palma, G.; Cella, L. Radiation-Induced Esophagitis in Non-Small-Cell Lung Cancer Patients: Voxel-Based Analysis and NTCP Modeling. Cancers 2022, 14, 1833. [Google Scholar] [CrossRef] [PubMed]
- El Naqa, I.; Bradley, J.; Blanco, A.I.; Lindsay, P.E.; Vicic, M.; Hope, A.; Deasy, J.O. Multivariable Modeling of Radiotherapy Outcomes, Including Dose–Volume and Clinical Factors. Int. J. Radiat. Oncol. 2006, 64, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Wang, G.; Qin, X.; Li, Y.; Sun, B.; Kong, F.-M.; Gao, Y.; Jin, J.-Y. A Statistical Approach to Assess Biological Equivalent Mean Organ Dose (MOD) for Different Fractionations in Thoracic Radiotherapy. Biomedicines 2025, 13, 1110. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Mattson, M.P. How Does Hormesis Impact Biology, Toxicology, and Medicine? Npj Aging Mech. Dis. 2017, 3, 13. [Google Scholar] [CrossRef]
- Luckey, T.D. Radiation Hormesis: The Good, the Bad, and the Ugly. Dose-Response 2006, 4. [Google Scholar] [CrossRef]
- Lau, Y.S.; Chew, M.T.; Alqahtani, A.; Jones, B.; Hill, M.A.; Nisbet, A.; Bradley, D.A. Low Dose Ionising Radiation-Induced Hormesis: Therapeutic Implications to Human Health. Appl. Sci. 2021, 11, 8909. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Li, A.; Barber, R.F. Multiple Testing with the Structure-Adaptive Benjamini–Hochberg Algorithm. J. R. Stat. Soc. Ser. B Stat. Methodol. 2019, 81, 45–74. [Google Scholar] [CrossRef]
- SMITH, S.; NICHOLS, T. Threshold-Free Cluster Enhancement: Addressing Problems of Smoothing, Threshold Dependence and Localisation in Cluster Inference. NeuroImage 2009, 44, 83–98. [Google Scholar] [CrossRef]
- Worsley, K.J.; Evans, A.C.; Marrett, S.; Neelin, P. A Three-Dimensional Statistical Analysis for CBF Activation Studies in Human Brain. J. Cereb. Blood Flow Amp Metab. 1992, 12, 900–918. [Google Scholar] [CrossRef]
- Fischer, L.; Ramdas, A. Sequential Monte Carlo Testing by Betting. J. R. Stat. Soc. Ser. B Stat. Methodol. 2025, 87, 1200–1220. [Google Scholar] [CrossRef]
- Schweiger, R.; Fisher, E.; Weissbrod, O.; Rahmani, E.; Müller-Nurasyid, M.; Kunze, S.; Gieger, C.; Waldenberger, M.; Rosset, S.; Halperin, E. Detecting Heritable Phenotypes without a Model Using Fast Permutation Testing for Heritability and Set-Tests. Nat. Commun. 2018, 9, 4919. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chu, A.; Xu, Z. Dose Uncertainty from Calculation Grid Resolution and Its Alignment with MLC. Med. Dosim. 2019, 44, e1–e7. [Google Scholar] [CrossRef] [PubMed]
- Saez, J.; Bar-Deroma, R.; Bogaert, E.; Cayez, R.; Chow, T.; Clark, C.H.; Esposito, M.; Feygelman, V.; Monti, A.F.; Garcia-Miguel, J.; et al. Universal Evaluation of MLC Models in Treatment Planning Systems Based on a Common Set of Dynamic Tests. Radiother. Oncol. 2023, 186, 109775. [Google Scholar] [CrossRef]
- Goodall, S.K.; Ebert, M.A. Recommended Dose Voxel Size and Statistical Uncertainty Parameters for Precision of Monte Carlo Dose Calculation in Stereotactic Radiotherapy. J. Appl. Clin. Med. Phys. 2020, 21, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Kubo, H.D.; Wilder, R.B.; Pappas, C.T.E. Impact of Collimator Leaf Width on Stereotactic Radiosurgery and 3D Conformal Radiotherapy Treatment Plans. Int. J. Radiat. Oncol. 1999, 44, 937–945. [Google Scholar] [CrossRef]
- Cheung, M.R.; Tucker, S.L.; Dong, L.; De Crevoisier, R.; Lee, A.K.; Frank, S.; Kudchadker, R.J.; Thames, H.; Mohan, R.; Kuban, D. Investigation of Bladder Dose and Volume Factors Influencing Late Urinary Toxicity after External Beam Radiotherapy for Prostate Cancer. Int. J. Radiat. Oncol. 2007, 67, 1059–1065. [Google Scholar] [CrossRef]
- Steike, D.R.; Troschel, F.M.; Roers, J.; Siats, J.J.; Kittel, C.; Pepper, N.B.; Gravemeyer, S.; Papavassilis, P.; Schrader, A.J.; Eich, H.T.; et al. Effect of Prior Transurethral Prostate Resection (TURP) or Laser Enucleation (ThuLEP) on Radiotherapy-Induced Toxicity and Quality of Life in Prostate Cancer Patients Undergoing Definitive Radiotherapy. Cancers 2024, 16, 3403. [Google Scholar] [CrossRef]
- Pinkawa, M.; Klotz, J.; Djukic, V.; Petz, D.; Holy, R.; Eble, M.J. Transurethral Resection of the Prostate after Radiotherapy for Prostate Cancer: Impact on Quality of Life. Int. J. Urol. 2014, 21, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Heemsbergen, W.D.; Al-Mamgani, A.; Witte, M.G.; Van Herk, M.; Pos, F.J.; Lebesque, J.V. Urinary Obstruction in Prostate Cancer Patients From the Dutch Trial (68 Gy vs. 78 Gy): Relationships With Local Dose, Acute Effects, and Baseline Characteristics. Int. J. Radiat. Oncol. 2010, 78, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Gioscio, E.; Massi, M.C.; Franco, N.R.; Seibold, P.; Avuzzi, B.; Cicchetti, A.; Rosenstein, B.; Azria, D.; Choudhury, A.; De Ruysscher, D.; et al. 1378: Genetically-Based Analysis of Dose Surface Maps for Assessing Toxicity Post-RT: An Innovative Method. Radiother. Oncol. 2024, 194, S5037–S5041. [Google Scholar] [CrossRef]
- Franco, N.R.; Massi, M.C.; Ieva, F.; Manzoni, A.; Paganoni, A.M.; Zunino, P.; Veldeman, L.; Ost, P.; Fonteyne, V.; Talbot, C.J.; et al. Development of a Method for Generating SNP Interaction-Aware Polygenic Risk Scores for Radiotherapy Toxicity. Radiother. Oncol. 2021, 159, 241–248. [Google Scholar] [CrossRef]
- Barnett, G.C.; Kerns, S.L.; Dorling, L.; Fachal, L.; Aguado-Barrera, M.E.; Martínez-Calvo, L.; Jandu, H.K.; Welsh, C.; Tyrer, J.; Coles, C.E.; et al. No Association Between Polygenic Risk Scores for Cancer and Development of Radiation Therapy Toxicity. Int. J. Radiat. Oncol. 2022, 114, 494–501. [Google Scholar] [CrossRef]
- Dorling, L.; Barnett, G.C.; Michailidou, K.; Coles, C.E.; Burnet, N.G.; Yarnold, J.; Elliott, R.M.; Dunning, A.M.; Pharoah, P.D.P.; West, C.M. Patients with a High Polygenic Risk of Breast Cancer Do Not Have An Increased Risk of Radiotherapy Toxicity. Clin. Cancer Res. 2016, 22, 1413–1420. [Google Scholar] [CrossRef]
- Farazi, M.; Yang, X.; Gehl, C.J.; Barnett, G.C.; Burnet, N.G.; Chang-Claude, J.; Parker, C.C.; Dunning, A.M.; Azria, D.; Choudhury, A.; et al. A Polygenic Risk Score for Late Bladder Toxicity Following Radiotherapy for Non-Metastatic Prostate Cancer. Cancer Epidemiol. Biomark. Prev. 2025, 34, 795–804. [Google Scholar] [CrossRef]
- Nuijens, A.C.; Oei, A.L.; Franken, N.A.P.; Rasch, C.R.N.; Stalpers, L.J.A. Towards Personalized Radiotherapy in Pelvic Cancer: Patient-Related Risk Factors for Late Radiation Toxicity. Curr. Oncol. 2025, 32, 47. [Google Scholar] [CrossRef]
- He, Y.-Q.; Wang, T.-M.; Yang, D.-W.; Xue, W.-Q.; Deng, C.-M.; Li, D.-H.; Zhang, W.-L.; Liao, Y.; Xiao, R.-W.; Luo, L.-T.; et al. A Comprehensive Predictive Model for Radiation-Induced Brain Injury in Risk Stratification and Personalized Radiotherapy of Nasopharyngeal Carcinoma. Radiother. Oncol. 2024, 190, 109974. [Google Scholar] [CrossRef]
- Herskind, C.; Talbot, C.J.; Kerns, S.L.; Veldwijk, M.R.; Rosenstein, B.S.; West, C.M.L. Radiogenomics: A Systems Biology Approach to Understanding Genetic Risk Factors for Radiotherapy Toxicity? Cancer Lett. 2016, 382, 95–109. [Google Scholar] [CrossRef]
- Yahya, N.; Ebert, M.A.; House, M.J.; Kennedy, A.; Matthews, J.; Joseph, D.J.; Denham, J.W. Modeling Urinary Dysfunction After External Beam Radiation Therapy of the Prostate Using Bladder Dose-Surface Maps: Evidence of Spatially Variable Response of the Bladder Surface. Int. J. Radiat. Oncol. 2017, 97, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yang, Z.; Wang, J.; Hu, W. Dosimetric Impact of Different Bladder and Rectum Filling during Prostate Cancer Radiotherapy. Radiat. Oncol. 2016, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Padhani, A.R.; Khoo, V.S.; Suckling, J.; Husband, J.E.; Leach, M.O.; Dearnaley, D.P. Evaluating the Effect of Rectal Distension and Rectal Movement on Prostate Gland Position Using Cine MRI. Int. J. Radiat. Oncol. 1999, 44, 525–533. [Google Scholar] [CrossRef]
- Kievit, R.A.; Frankenhuis, W.E.; Waldorp, L.J.; Borsboom, D. Simpson’s Paradox in Psychological Science: A Practical Guide. Front. Psychol. 2013, 4, 513. [Google Scholar] [CrossRef]
- Simpson, E.H. The Interpretation of Interaction in Contingency Tables. J. R. Stat. Soc. Ser. B Methodol. 1951, 13, 238–241. [Google Scholar] [CrossRef]
- Tomasetti, C.; Vogelstein, B. Variation in Cancer Risk among Tissues Can Be Explained by the Number of Stem Cell Divisions. Science 2015, 347, 78–81. [Google Scholar] [CrossRef]
- Albini, A.; Cavuto, S.; Apolone, G.; Noonan, D.M. Strategies to Prevent “Bad Luck” in Cancer. J. Natl. Cancer Inst. 2015, 107, djv213. [Google Scholar] [CrossRef]
- Feuerriegel, S.; Frauen, D.; Melnychuk, V.; Schweisthal, J.; Hess, K.; Curth, A.; Bauer, S.; Kilbertus, N.; Kohane, I.S.; Van Der Schaar, M. Causal Machine Learning for Predicting Treatment Outcomes. Nat. Med. 2024, 30, 958–968. [Google Scholar] [CrossRef]
- Harber, P. Causal Inference Analysis for Poorly Soluble Low Toxicity Particles, Lung Function, and Malignancy. Front. Public Health 2022, 10, 863402. [Google Scholar] [CrossRef]
- Jenkins, A.; Osorio, E.V.; Green, A.; Van Herk, M.; Sperrin, M.; McWilliam, A. Methods of Causal Effect Estimation for High-dimensional Treatments: A Radiotherapy Simulation Study. Med. Phys. 2025, 52. [Google Scholar] [CrossRef]
- van der Laan, M.J.; Rose, S. Targeted Learning: Causal Inference for Observational and Experimental Data; Springer: New York, NY, USA, 2011; ISBN 978-1-4419-9782-1. [Google Scholar]
- Angrist, J.D.; Imbens, G.W.; Rubin, D.B. Identification of Causal Effects Using Instrumental Variables. J. Am. Stat. Assoc. 1996, 91, 444–455. [Google Scholar] [CrossRef]
- Baiocchi, M.; Cheng, J.; Small, D.S. Instrumental Variable Methods for Causal Inference: Instrumental Variable Methods for Causal Inference. Stat. Med. 2014, 33, 2297–2340. [Google Scholar] [CrossRef] [PubMed]
- Dunning, T. Natural Experiments in the Social Sciences: A Design-Based Approach; Cambridge University Press: Cambridge, UK, 2012; ISBN 978-1-139-08444-4. [Google Scholar]
- Rosenbaum, P.R. Design of Observational Studies; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; ISBN 978-3-030-46405-9. [Google Scholar]
- Hernan, M.A.; Robins, J.M. Causal Inference: What If, illustrated ed.; CRC Press: Boca Raton, FL, USA, 2020; ISBN 1-4200-7616-7. [Google Scholar]
- McWilliam, A.; Kennedy, J.; Hodgson, C.; Vasquez Osorio, E.; Faivre-Finn, C.; Van Herk, M. Radiation Dose to Heart Base Linked with Poorer Survival in Lung Cancer Patients. Eur. J. Cancer 2017, 85, 106–113. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, A.; Dootson, C.; Graham, L.; Banfill, K.; Abravan, A.; Van Herk, M. Dose Surface Maps of the Heart Can Identify Regions Associated with Worse Survival for Lung Cancer Patients Treated with Radiotherapy. Phys. Imaging Radiat. Oncol. 2020, 15, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Franzese, C.; Dei, D.; Lambri, N.; Teriaca, M.A.; Badalamenti, M.; Crespi, L.; Tomatis, S.; Loiacono, D.; Mancosu, P.; Scorsetti, M. Enhancing Radiotherapy Workflow for Head and Neck Cancer with Artificial Intelligence: A Systematic Review. J. Pers. Med. 2023, 13, 946. [Google Scholar] [CrossRef]
- Santoro, M.; Strolin, S.; Paolani, G.; Della Gala, G.; Bartoloni, A.; Giacometti, C.; Ammendolia, I.; Morganti, A.G.; Strigari, L. Recent Applications of Artificial Intelligence in Radiotherapy: Where We Are and Beyond. Appl. Sci. 2022, 12, 3223. [Google Scholar] [CrossRef]
- Piras, A.; Corso, R.; Benfante, V.; Ali, M.; Laudicella, R.; Alongi, P.; D’Aviero, A.; Cusumano, D.; Boldrini, L.; Salvaggio, G.; et al. Artificial Intelligence and Statistical Models for the Prediction of Radiotherapy Toxicity in Prostate Cancer: A Systematic Review. Appl. Sci. 2024, 14, 10947. [Google Scholar] [CrossRef]
- Morelli, I.; Banini, M.; Greto, D.; Visani, L.; Garlatti, P.; Loi, M.; Aquilano, M.; Valzano, M.; Salvestrini, V.; Bertini, N.; et al. Integrating Radiomics and Artificial Intelligence (AI) in Stereotactic Body Radiotherapy (SBRT)/Stereotactic Radiosurgery (SRS): Predictive Tools for Tailored Cancer Care. Cancers 2025, 17, 2906. [Google Scholar] [CrossRef]
- Lastrucci, A.; Wandael, Y.; Ricci, R.; Maccioni, G.; Giansanti, D. The Integration of Deep Learning in Radiotherapy: Exploring Challenges, Opportunities, and Future Directions through an Umbrella Review. Diagnostics 2024, 14, 939. [Google Scholar] [CrossRef]
- Deodato, F.; Macchia, G.; Duhanxhiu, P.; Mammini, F.; Cavallini, L.; Ntreta, M.; Zamfir, A.A.; Buwenge, M.; Cellini, F.; Ciabatti, S.; et al. A Multicenter Machine Learning-Based Predictive Model of Acute Toxicity in Prostate Cancer Patients Undergoing Salvage Radiotherapy (ICAROS Study). Cancers 2025, 17, 2142. [Google Scholar] [CrossRef]
- Najjar, R. Redefining Radiology: A Review of Artificial Intelligence Integration in Medical Imaging. Diagnostics 2023, 13, 2760. [Google Scholar] [CrossRef] [PubMed]
- Vendrame, A.; Cappelletto, C.; Chiovati, P.; Vinante, L.; Parvej, M.; Caroli, A.; Pirrone, G.; Barresi, L.; Drigo, A.; Avanzo, M. Artificial Intelligence-Based Patient Selection for Deep Inspiration Breath-Hold Breast Radiotherapy from Respiratory Signals. Appl. Sci. 2023, 13, 4962. [Google Scholar] [CrossRef]
- Ubeira-Gabellini, M.G.; Mori, M.; Palazzo, G.; Cicchetti, A.; Mangili, P.; Pavarini, M.; Rancati, T.; Fodor, A.; del Vecchio, A.; Di Muzio, N.G.; et al. Comparing Performances of Predictive Models of Toxicity after Radiotherapy for Breast Cancer Using Different Machine Learning Approaches. Cancers 2024, 16, 934. [Google Scholar] [CrossRef] [PubMed]
- Huhulea, E.N.; Huang, L.; Eng, S.; Sumawi, B.; Huang, A.; Aifuwa, E.; Hirani, R.; Tiwari, R.K.; Etienne, M. Artificial Intelligence Advancements in Oncology: A Review of Current Trends and Future Directions. Biomedicines 2025, 13, 951. [Google Scholar] [CrossRef]
- Suwanraksa, C.; Bridhikitti, J.; Liamsuwan, T.; Chaichulee, S. CBCT-to-CT Translation Using Registration-Based Generative Adversarial Networks in Patients with Head and Neck Cancer. Cancers 2023, 15, 2017. [Google Scholar] [CrossRef]
- Thor, M.; Bentzen, L.; Hysing, L.B.; Ekanger, C.; Helle, S.-I.; Karlsdóttir, Á.; Muren, L.P. Prediction of Rectum and Bladder Morbidity Following Radiotherapy of Prostate Cancer Based on Motion-Inclusive Dose Distributions. Radiother. Oncol. 2013, 107, 147–152. [Google Scholar] [CrossRef]
- Valle, L.F.; Ruan, D.; Dang, A.; Levin-Epstein, R.G.; Patel, A.P.; Weidhaas, J.B.; Nickols, N.G.; Lee, P.P.; Low, D.A.; Qi, X.S.; et al. Development and Validation of a Comprehensive Multivariate Dosimetric Model for Predicting Late Genitourinary Toxicity Following Prostate Cancer Stereotactic Body Radiotherapy. Front. Oncol. 2020, 10, 786. [Google Scholar] [CrossRef]
- Van Den Bosch, L.; Van Der Schaaf, A.; Van Der Laan, H.P.; Hoebers, F.J.P.; Wijers, O.B.; Van Den Hoek, J.G.M.; Moons, K.G.M.; Reitsma, J.B.; Steenbakkers, R.J.H.M.; Schuit, E.; et al. Comprehensive Toxicity Risk Profiling in Radiation Therapy for Head and Neck Cancer: A New Concept for Individually Optimised Treatment. Radiother. Oncol. 2021, 157, 147–154. [Google Scholar] [CrossRef]
- El Naqa, I.; Kerns, S.L.; Coates, J.; Luo, Y.; Speers, C.; West, C.M.L.; Rosenstein, B.S.; Ten Haken, R.K. Radiogenomics and Radiotherapy Response Modeling. Phys. Med. Biol. 2017, 62, R179–R206. [Google Scholar] [CrossRef]
- Patrick, H.M.; Kildea, J. Technical Note: Rtdsm—An Open-source Software for Radiotherapy Dose-surface Map Generation and Analysis. Med. Phys. 2022, 49, 7327–7335. [Google Scholar] [CrossRef]
- Mathotaarachchi, S.; Wang, S.; Shin, M.; Pascoal, T.A.; Benedet, A.L.; Kang, M.S.; Beaudry, T.; Fonov, V.S.; Gauthier, S.; Labbe, A.; et al. VoxelStats: A MATLAB Package for Multi-Modal Voxel-Wise Brain Image Analysis. Front. Neuroinformatics 2016, 10, 20. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- Crippa, A.; Orsini, N. Multivariate Dose-Response Meta-Analysis: The Dosresmeta R Package. J. Stat. Softw. 2016, 72, 1–15. [Google Scholar] [CrossRef]
- Van Herk, M.; De Jaeger, K.; De Munck, J.; Hoogeman, M.; Meinders, J.; Ploeger, L.; Zijp, L. A Delineation System for N Modalities—Software Aspects. In The Use of Computers in Radiation Therapy; Schlegel, W., Bortfeld, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2000; pp. 73–75. ISBN 978-3-540-67176-3. [Google Scholar]
- Green, A.; Vasquez Osorio, E.; Aznar, M.C.; McWilliam, A.; Van Herk, M. Image Based Data Mining Using Per-Voxel Cox Regression. Front. Oncol. 2020, 10, 1178. [Google Scholar] [CrossRef]

| One-Sided n1 (Δ = 2 Gy) | One-Sided n1 (Δ = 4 Gy) | Two-Sided n1 (Δ = 2 Gy) | Two-Sided n1 (Δ = 4 Gy) | n2 (Assume n2 Fixed for the Calculation of n1) |
|---|---|---|---|---|
| 734 | 41 | 1744 | 47 | 100 |
| 220 | 36 | 266 | 41 | 200 |
| 178 | 35 | 207 | 39 | 300 |
| 163 | 34 | 187 | 39 | 400 |
| 155 | 34 | 176 | 38 | 500 |
| 150 | 34 | 170 | 38 | 600 |
| 147 | 34 | 166 | 38 | 700 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puri, T. Voxel-Based Dose–Toxicity Modeling for Predicting Post-Radiotherapy Toxicity: A Critical Perspective. J. Clin. Med. 2025, 14, 7248. https://doi.org/10.3390/jcm14207248
Puri T. Voxel-Based Dose–Toxicity Modeling for Predicting Post-Radiotherapy Toxicity: A Critical Perspective. Journal of Clinical Medicine. 2025; 14(20):7248. https://doi.org/10.3390/jcm14207248
Chicago/Turabian StylePuri, Tanuj. 2025. "Voxel-Based Dose–Toxicity Modeling for Predicting Post-Radiotherapy Toxicity: A Critical Perspective" Journal of Clinical Medicine 14, no. 20: 7248. https://doi.org/10.3390/jcm14207248
APA StylePuri, T. (2025). Voxel-Based Dose–Toxicity Modeling for Predicting Post-Radiotherapy Toxicity: A Critical Perspective. Journal of Clinical Medicine, 14(20), 7248. https://doi.org/10.3390/jcm14207248






