Publicly Available Datasets for Artificial Intelligence in Neurosurgery: A Systematic Review
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Screening
2.3. Data Extraction and Analysis
2.4. Quality Assessment
3. Results
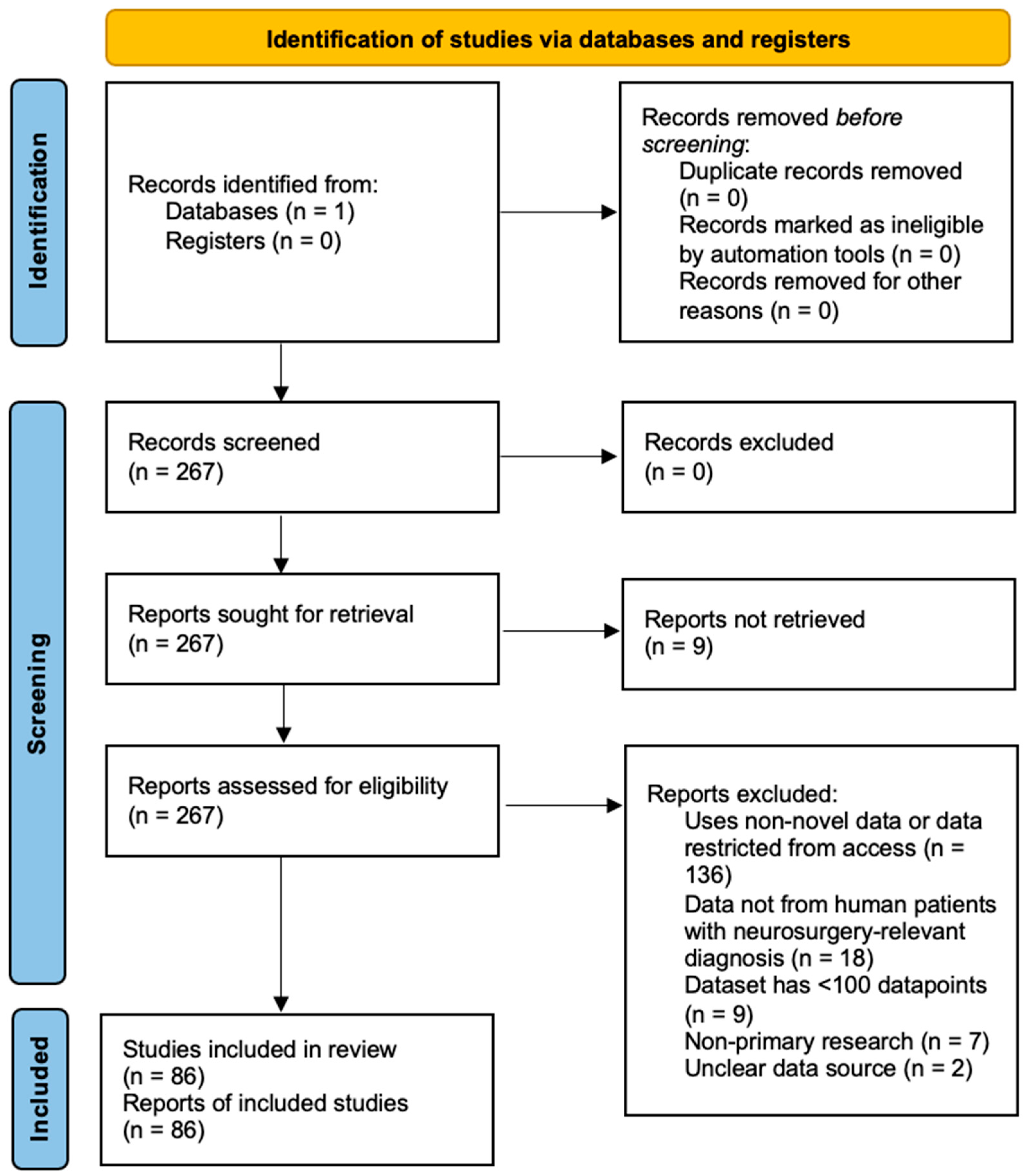
3.1. Study Selection
3.2. Study Characteristics
3.2.1. Analysis of Dataset Characteristics
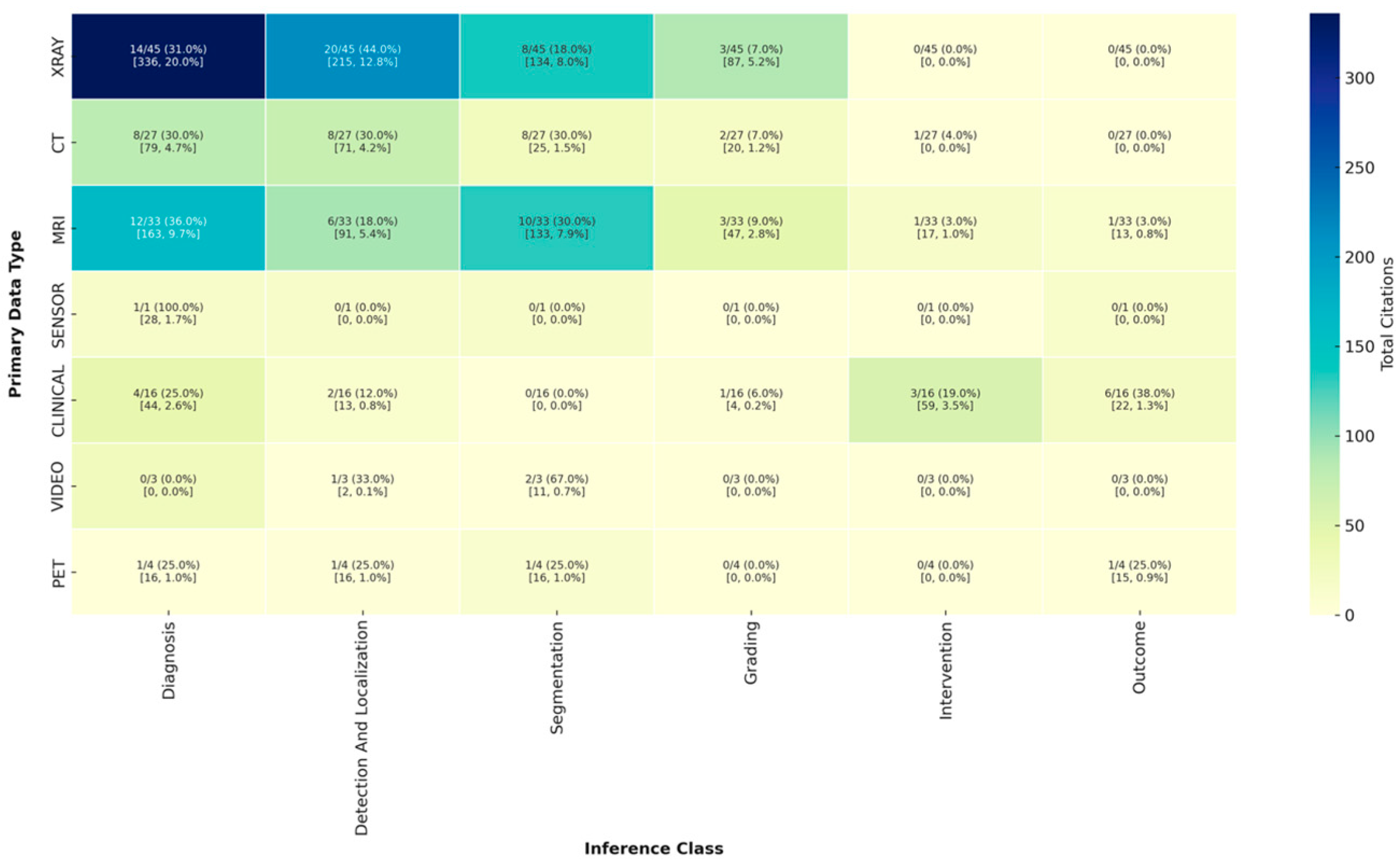
3.2.2. Analysis of Inference Type by Data Modality
3.2.3. Analysis of Model Validation and Public Application
3.2.4. Analysis of Performance Metrics
3.3. Risk of Bias in Included Studies
3.4. Reporting Completeness
3.5. Key Findings from High-Impact Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ou-Yang, D.; Burger, E.L.; Kleck, C.J. Pre-Operative Planning in Complex Deformities and Use of Patient-Specific UNiDTM Instrumentation. Glob. Spine J. 2022, 12 (Suppl. S2), 40S–44S. [Google Scholar] [CrossRef] [PubMed]
- Anisuzzaman, D.M.; Malins, J.G.; Friedman, P.A.; Attia, Z.I. Fine-Tuning Large Language Models for Specialized Use Cases. Mayo Clin. Proc. Digit. Health 2025, 3, 100184. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, T.; Mattos, D.I.; Bosch, J.; Olsson, H.H. Data Labeling: An Empirical Investigation into Industrial Challenges and Mitigation Strategies. In Product-Focused Software Process Improvement; Morisio, M., Torchiano, M., Jedlitschka, A., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 202–216. [Google Scholar] [CrossRef]
- Roh, Y.; Heo, G.; Whang, S.E. A Survey on Data Collection for Machine Learning: A Big Data—AI Integration Perspective. IEEE Trans. Knowl. Data Eng. 2021, 33, 1328–1347. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, K.; Ning, Z.; Wang, K.; Dong, Y.; Liu, X.; Liu, S.; Wang, J.; Zhu, C.; Yu, Q.; et al. Deep learning of lumbar spine X-ray for osteopenia and osteoporosis screening: A multicenter retrospective cohort study. Bone 2020, 140, 115561. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Chen, H.H.; Horng-Shing Lu, H.; Hondar Wu, H.T.; Chang, M.C.; Chou, P.H. Can a Deep-learning Model for the Automated Detection of Vertebral Fractures Approach the Performance Level of Human Subspecialists? Clin. Orthop. Relat. Res. 2021, 479, 1598–1612. [Google Scholar] [CrossRef] [PubMed]
- LewandrowskI, K.-U.; Muraleedharan, N.; Eddy, S.A.; Sobti, V.; Reece, B.D.; León, J.F.R.; Shah, S. Feasibility of Deep Learning Algorithms for Reporting in Routine Spine Magnetic Resonance Imaging. Int. J. Spine Surg. 2020, 14 (Suppl. S3), S86–S97. [Google Scholar] [CrossRef] [PubMed]
- Cepeda, S.; Esteban-Sinovas, O.; Singh, V.; Moiyadi, A.; Zemmoura, I.; Del Bene, M.; Barbotti, A.; DiMeco, F.; West, T.R.; Nahed, B.V.; et al. Prognostic Modeling of Overall Survival in Glioblastoma Using Radiomic Features Derived from Intraoperative Ultrasound: A Multi-Institutional Study. Cancers 2025, 17, 280. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Cho, W.; Choi, D.; Sim, G.; Jeong, S.Y.; Baik, S.H.; Bae, Y.J.; Choi, B.S.; Kim, J.H.; Yoo, S.; et al. Prediction of treatment response after stereotactic radiosurgery of brain metastasis using deep learning and radiomics on longitudinal MRI data. Sci. Rep. 2024, 14, 11085. [Google Scholar] [CrossRef] [PubMed]
- Park, J.J.; Doiphode, N.; Zhang, X.; Pan, L.; Blue, R.; Shi, J.; Buch, V.P. Developing the surgeon-machine interface: Using a novel instance-segmentation framework for intraoperative landmark labelling. Front. Surg. 2023, 10, 1259756. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, D.; Poonamallee, L.; Joshi, S.; Bahel, V. Automated intracranial hemorrhage detection in traumatic brain injury using 3D CNN. J. Neurosci. Rural Pract. 2023, 14, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, F.; Xu, J.; Zhao, Q.; Huang, C.; Yu, Y.; Yuan, H. Automated detection and classification of acute vertebral body fractures using a convolutional neural network on computed tomography. Front. Endocrinol. 2023, 14, 1132725. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, L.; You, S.; Wang, Z.; Zhu, S.; Chen, C.; Ma, X.; Yang, L.; Zhang, S.; Yang, Q. Automatic Detection and Classification of Modic Changes in MRI Images Using Deep Learning: Intelligent Assisted Diagnosis System. Orthop. Surg. 2024, 16, 196–206. [Google Scholar] [CrossRef] [PubMed]
- MohammadiNasrabadi, A.; Moammer, G.; Quateen, A.; Bhanot, K.; McPhee, J. Landet: An efficient physics-informed deep learning approach for automatic detection of anatomical landmarks and measurement of spinopelvic alignment. J. Orthop. Surg. Res. 2024, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Liu, G.; Zhang, H.; Wang, L.; Zhu, S.; Bao, M.D.L.H.; Wang, Y.; Chen, C.; Wang, Z.; Liu, X.; et al. Evaluating CNN Architectures for the Automated Detection and Grading of Modic Changes in MRI: A Comparative Study. Orthop. Surg. 2024, 17, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Cha, J.H.; Lee, S.; Han, L.; Park, W.; Ahn, J.S.; Park, S.-C. Deep-Learning-Based Cerebral Artery Semantic Segmentation in Neurosurgical Operating Microscope Vision Using Indocyanine Green Fluorescence Videoangiography. Front. Neurorobotics 2022, 15, 735177. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Mou, L.; Yan, Q.; Ma, S.; Yue, X.; Zhou, S.; Lin, Z.; Zhang, J.; Liu, J.; Zhao, Y. Automated Segmentation of Trigeminal Nerve and Cerebrovasculature in MR-Angiography Images by Deep Learning. Front. Neurosci. 2021, 15, 744967. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.; Soto-Rey, I.; Kramer, F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Res. Notes 2022, 15, 210. [Google Scholar] [CrossRef] [PubMed]
| Variable | Analysis |
|---|---|
| Neurosurgical subspeciality: | |
| Spine | 72/86 (83.7%) |
| Tumor | 3/86 (3.5%) |
| Vascular | 4/86 (4.7%) |
| Trauma | 6/86 (7.0%) |
| Other | 1/86 (1.2%) |
| Data type(s) included: | |
| X-ray | 32/86 (37.2%) |
| CT | 18/86 (20.9%) |
| MRI | 25/86 (29.1%) |
| PET | 2/86 (2.3%) |
| Clinical | 14/86 (16.3%) |
| Sensor | 1/86 (1.2%) |
| Video | 2/86 (2.3%) |
| Label type: | |
| Diagnosis | 23/86 (26.7%) |
| Segmentation | 31/86 (36.0%) |
| Detection/localization | 18/86 (20.9%) |
| Intervention | 5/86 (5.8%) |
| Grading | 18/86 (20.9%) |
| Image | 2/86 (2.3%) |
| Outcome | 2/86 (2.3%) |
| Number of Patients | Mean: 756.4, SD: 925.8, Median: 100, Range: 1–4963 |
| Multi-institutional data | 23/86 (26.7%) |
| Number of institutions | Mean: 3.47, SD: 18.5, Median: 1, Range: 1–167 |
| Includes baseline model | 84/86 (97.7%) |
| Inference class of baseline model: | |
| Diagnosis | 36/84 (42.9%) |
| Detection/localization | 35/84 (41.7%) |
| Segmentation | 29/84 (34.5%) |
| Intervention | 4/84 (4.8%) |
| Measurement | 7/84 (8.3%) |
| Grading | 9/84 (10.7%) |
| Outcome | 8/84 (9.5%) |
| Generative | 1/84 (1.2%) |
| Model type: | |
| CNN | 52/84 (61.9%) |
| Segmentation | 26/84 (31.0%) |
| Linear | 10/84 (11.9%) |
| Non-transformer NLP | 3/84 (3.6%) |
| Transformer | 3/84 (3.6%) |
| GAN | 1/84 (1.2%) |
| Model externally validated | 19/84 (22.6%) |
| Model/code publicly available | 17/84 (20.2%) |
| Application publicly available | 6/84 (7.1%) |
| Number of citations | Mean: 12.0, SD: 17.9, Median: 6.5, Range: 0–127 |
| Model Used | External Validation | Public Code | Public Application |
|---|---|---|---|
| Linear | 0/10 (0%) | 1/10 (10.0%) | 1/10 (10.0%) |
| CNN | 12/52 (23.1%) | 11/52 (21.2%) | 4/52 (7.7%) |
| Segmentation | 6/26 (23.1%) | 5/26 (19.2%) | 2/26 (7.7%) |
| GAN | 0/1 (0.0%) | 0/1 (0.0%) | 0/1 (0.0%) |
| Non-transformer NLP | 1/3 (33.3%) | 0/3 (0.0%) | 1/3 (33.3%) |
| Transformer | 1/3 (33.3%) | 1/3 (33.3%) | 0/3 (0.0%) |
| Model Used | External Validation | Public Code | Public Application |
|---|---|---|---|
| X-ray | 10/31 (32.3%) | 7/31 (22.6%) | 3/31 (9.7%) |
| CT | 5/17 (29.4%) | 4/17 (23.5%) | 3/17 (17.6%) |
| MRI | 6/24 (25.0%) | 6/24 (25.0%) | 1/24 (4.2%) |
| PET | 0/2 (0.0%) | 1/2 (50.0%) | 0/2 (0.0%) |
| Clinical | 0/13 (0.0%) | 2/13 (15.4%) | 1/13 (7.7%) |
| Sensor | 0/1 (0.0%) | 0/1 (0.0%) | 0/1 (0.0%) |
| Video | 0/2 (0.0%) | 0/2 (0.0%) | 0/2 (0.0%) |
| Metric | Detection and Localization | Diagnosis | Generative | Grading | Intervention | Measurement | Outcome | Segmentation |
|---|---|---|---|---|---|---|---|---|
| Accuracy | 12/26 (46.2%) | 22/31 (71.0%) | 0/1 (0.0%) | 2/6 (33.3%) | 3/4 (75.0%) | 3/6 (50.0%) | 5/5 (100.0%) | 12/25 (48.0%) |
| AUC/AUROC | 5/26 (19.2%) | 9/31 (29.0%) | 0/1 (0.0%) | 2/6 (33.3%) | 2/4 (50.0%) | 0/6 (0.0%) | 2/5 (40.0%) | 2/25 (8.0%) |
| Dice | 5/26 (19.2%) | 3/31 (9.7%) | 0/1 (0.0%) | 0/6 (0.0%) | 0/4 (0.0%) | 2/6 (33.3%) | 0/5 (0.0%) | 15/25 (60.0%) |
| F1 Score | 6/26 (23.1%) | 10/31 (32.3%) | 0/1 (0.0%) | 2/6 (33.3%) | 1/4 (25.0%) | 0/6 (0.0%) | 2/5 (40.0%) | 3/25 (12.0%) |
| Gwet k | 0/26 (0.0%) | 1/31 (3.2%) | 0/1 (0.0%) | 1/6 (16.7%) | 0/4 (0.0%) | 0/6 (0.0%) | 0/5 (0.0%) | 0/25 (0.0%) |
| ICC | 3/26 (11.5%) | 1/31 (3.2%) | 1/1 (100.0%) | 0/6 (0.0%) | 0/4 (0.0%) | 2/6 (33.3%) | 0/5 (0.0%) | 1/25 (4.0%) |
| IOU | 0/26 (0.0%) | 0/31 (0.0%) | 0/1 (0.0%) | 0/6 (0.0%) | 1/4 (25.0%) | 0/6 (0.0%) | 0/5 (0.0%) | 3/25 (12.0%) |
| MAE | 3/26 (11.5%) | 0/31 (0.0%) | 0/1 (0.0%) | 0/6 (0.0%) | 0/4 (0.0%) | 2/6 (33.3%) | 0/5 (0.0%) | 0/25 (0.0%) |
| NPV | 0/26 (0.0%) | 4/31 (12.9%) | 0/1 (0.0%) | 0/6 (0.0%) | 1/4 (25.0%) | 0/6 (0.0%) | 0/5 (0.0%) | 1/25 (4.0%) |
| PR AUC | 0/26 (0.0%) | 0/31 (0.0%) | 0/1 (0.0%) | 0/6 (0.0%) | 0/4 (0.0%) | 0/6 (0.0%) | 0/5 (0.0%) | 1/25 (4.0%) |
| PPV | 6/26 (23.1%) | 15/31 (48.4%) | 0/1 (0.0%) | 2/6 (33.3%) | 3/4 (75.0%) | 1/6 (16.7%) | 0/5 (0.0%) | 5/25 (20.0%) |
| Sensitivity | 16/26 (61.5%) | 27/31 (87.1%) | 0/1 (0.0%) | 6/6 (100.0%) | 3/4 (75.0%) | 2/6 (33.3%) | 4/5 (80.0%) | 14/25 (56.0%) |
| Specificity | 11/26 (42.3%) | 18/31 (58.1%) | 0/1 (0.0%) | 5/6 (83.3%) | 2/4 (50.0%) | 2/6 (33.3%) | 3/5 (60.0%) | 10/25 (40.0%) |
| Metric | Detection and Localization | Diagnosis | Generative | Grading | Intervention | Measurement | Outcome | Segmentation |
|---|---|---|---|---|---|---|---|---|
| Accuracy | 136/732 (18.6%) | 320/2109 (15.2%) | 0/0 | 100/480 (20.8%) | 42/214 (19.6%) | 44/287 (15.3%) | 19/58 (32.8%) | 178/1020 (17.5%) |
| AUC/AUROC | 45/732 (6.1%) | 236/2109 (11.2%) | 0/0 | 20/480 (4.2%) | 46/214 (21.5%) | 0/287 (0.0%) | 7/58 (12.1%) | 18/1020 (1.8%) |
| Dice | 69/732 (9.4%) | 33/2109 (1.6%) | 0/0 | 0/480 (0.0%) | 0/214 (0.0%) | 69/287 (24.0%) | 0/58 (0.0%) | 206/1020 (20.2%) |
| F1 Score | 48/732 (6.6%) | 109/2109 (5.2%) | 0/0 | 26/480 (5.4%) | 13/214 (6.1%) | 0/287 (0.0%) | 13/58 (22.4%) | 37/1020 (3.6%) |
| Gwet k | 0/732 (0.0%) | 26/2109 (1.2%) | 0/0 | 26/480 (5.4%) | 0/214 (0.0%) | 0/287 (0.0%) | 0/58 (0.0%) | 0/1020 (0.0%) |
| ICC | 13/732 (1.8%) | 0/2109 (0.0%) | 0/0 | 0/480 (0.0%) | 0/214 (0.0%) | 1/287 (0.3%) | 0/58 (0.0%) | 0/1020 (0.0%) |
| IOU | 0/732 (0.0%) | 0/2109 (0.0%) | 0/0 | 0/480 (0.0%) | 0/214 (0.0%) | 0/287 (0.0%) | 0/58 (0.0%) | 22/1020 (2.2%) |
| MAE | 13/732 (1.8%) | 0/2109 (0.0%) | 0/0 | 0/480 (0.0%) | 0/214 (0.0%) | 1/287 (0.3%) | 0/58 (0.0%) | 0/1020 (0.0%) |
| NPV | 0/732 (0.0%) | 151/2109 (7.2%) | 0/0 | 0/480 (0.0%) | 0/214 (0.0%) | 0/287 (0.0%) | 0/58 (0.0%) | 0/1020 (0.0%) |
| PR AUC | 0/732 (0.0%) | 0/2109 (0.0%) | 0/0 | 0/480 (0.0%) | 0/214 (0.0%) | 0/287 (0.0%) | 0/58 (0.0%) | 43/1020 (4.2%) |
| PPV | 21/732 (2.9%) | 275/2109 (13.0%) | 0/0 | 26/480 (5.4%) | 42/214 (19.6%) | 0/287 (0.0%) | 0/58 (0.0%) | 26/1020 (2.5%) |
| Sensitivity | 204/732 (27.9%) | 532/2109 (25.2%) | 0/0 | 141/480 (29.4%) | 42/214 (19.6%) | 86/287 (30.0%) | 13/58 (22.4%) | 260/1020 (25.5%) |
| Specificity | 183/732 (25.0%) | 427/2109 (20.2%) | 0/0 | 141/480 (29.4%) | 29/214 (13.6%) | 86/287 (30.0%) | 6/58 (10.3%) | 230/1020 (22.5%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, B.; Kim, B.; Schonfeld, E.; Nageeb, G.; Pant, A.; Sjoholm, A.; Medikonda, R.; Hani, U.; Veeravagu, A. Publicly Available Datasets for Artificial Intelligence in Neurosurgery: A Systematic Review. J. Clin. Med. 2025, 14, 5674. https://doi.org/10.3390/jcm14165674
Chan B, Kim B, Schonfeld E, Nageeb G, Pant A, Sjoholm A, Medikonda R, Hani U, Veeravagu A. Publicly Available Datasets for Artificial Intelligence in Neurosurgery: A Systematic Review. Journal of Clinical Medicine. 2025; 14(16):5674. https://doi.org/10.3390/jcm14165674
Chicago/Turabian StyleChan, Bianca, Brandon Kim, Ethan Schonfeld, George Nageeb, Aaradhya Pant, Adam Sjoholm, Ravi Medikonda, Ummey Hani, and Anand Veeravagu. 2025. "Publicly Available Datasets for Artificial Intelligence in Neurosurgery: A Systematic Review" Journal of Clinical Medicine 14, no. 16: 5674. https://doi.org/10.3390/jcm14165674
APA StyleChan, B., Kim, B., Schonfeld, E., Nageeb, G., Pant, A., Sjoholm, A., Medikonda, R., Hani, U., & Veeravagu, A. (2025). Publicly Available Datasets for Artificial Intelligence in Neurosurgery: A Systematic Review. Journal of Clinical Medicine, 14(16), 5674. https://doi.org/10.3390/jcm14165674






