A Rare ABCB5 Variant in a Familial Case of Intrahepatic Cholestasis of Pregnancy: A Potential Novel Genetic Contributor
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Presentation and Control Group
2.2. Whole-Exome Sequencing (WES)
2.3. Variant Filtering and Prioritizing
2.4. Protein Structure Impact and Cross-Species Conservation Analysis
2.5. Confirmation Analyses
3. Results
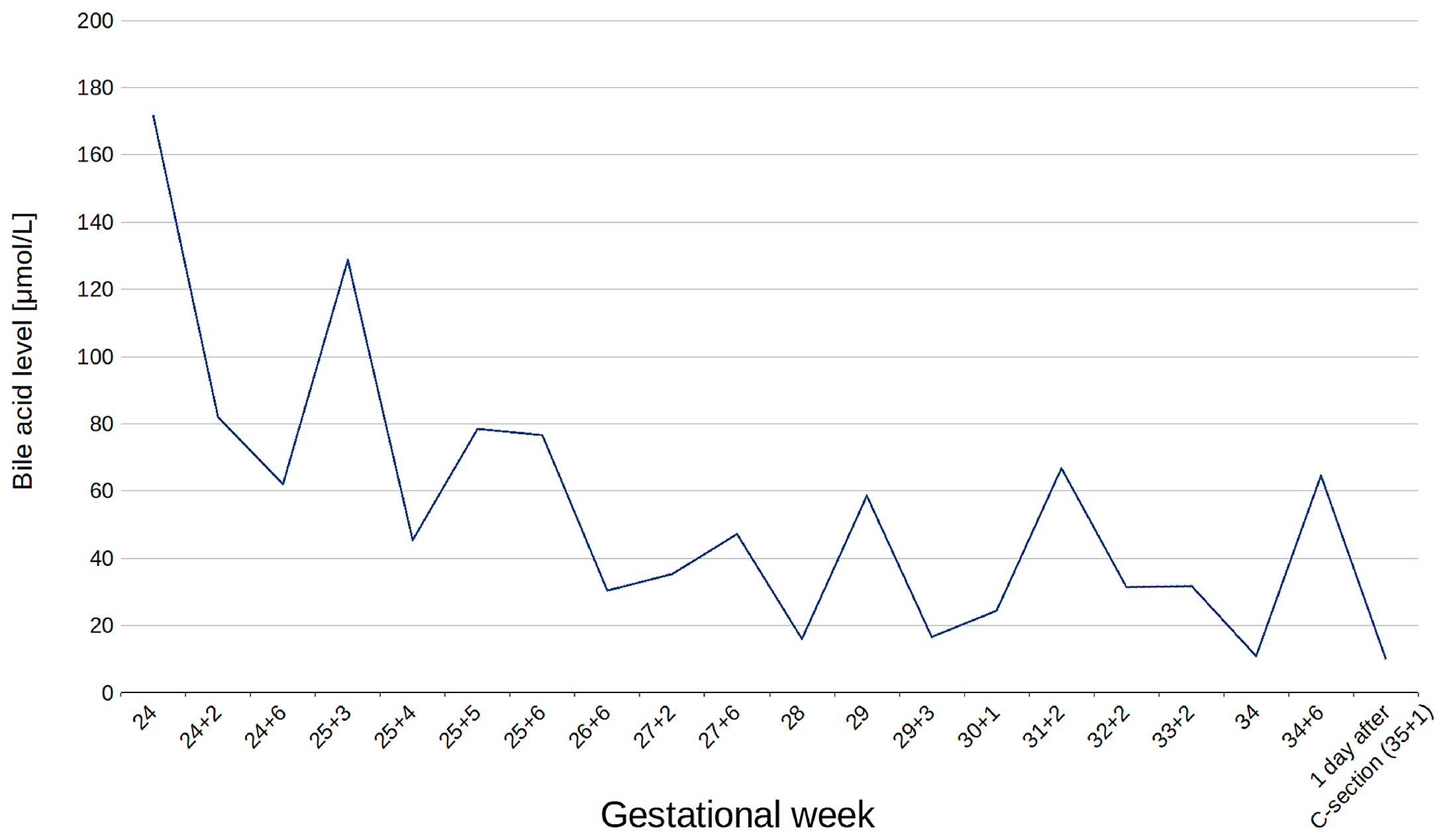
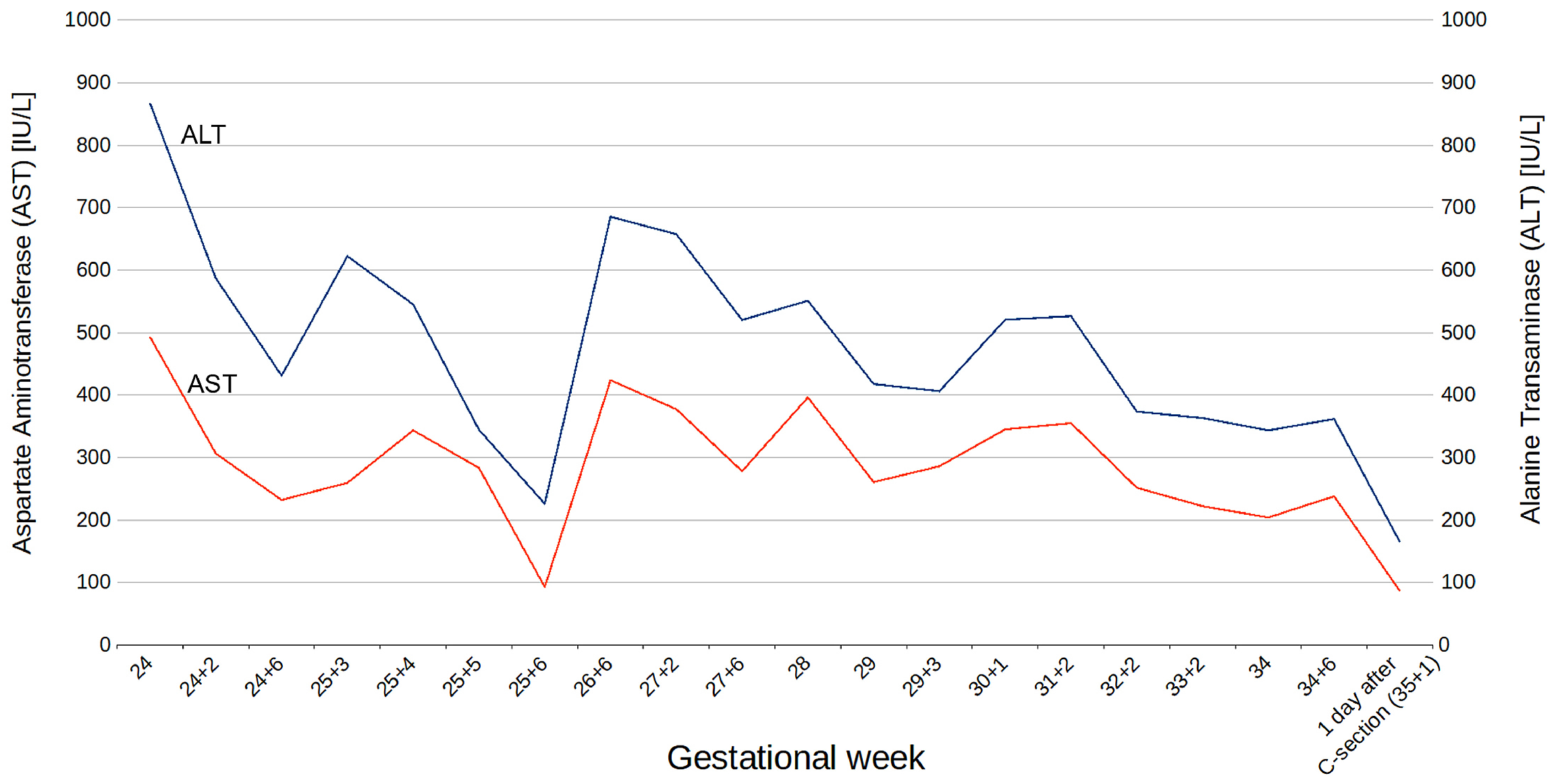
3.1. Clinical Findings
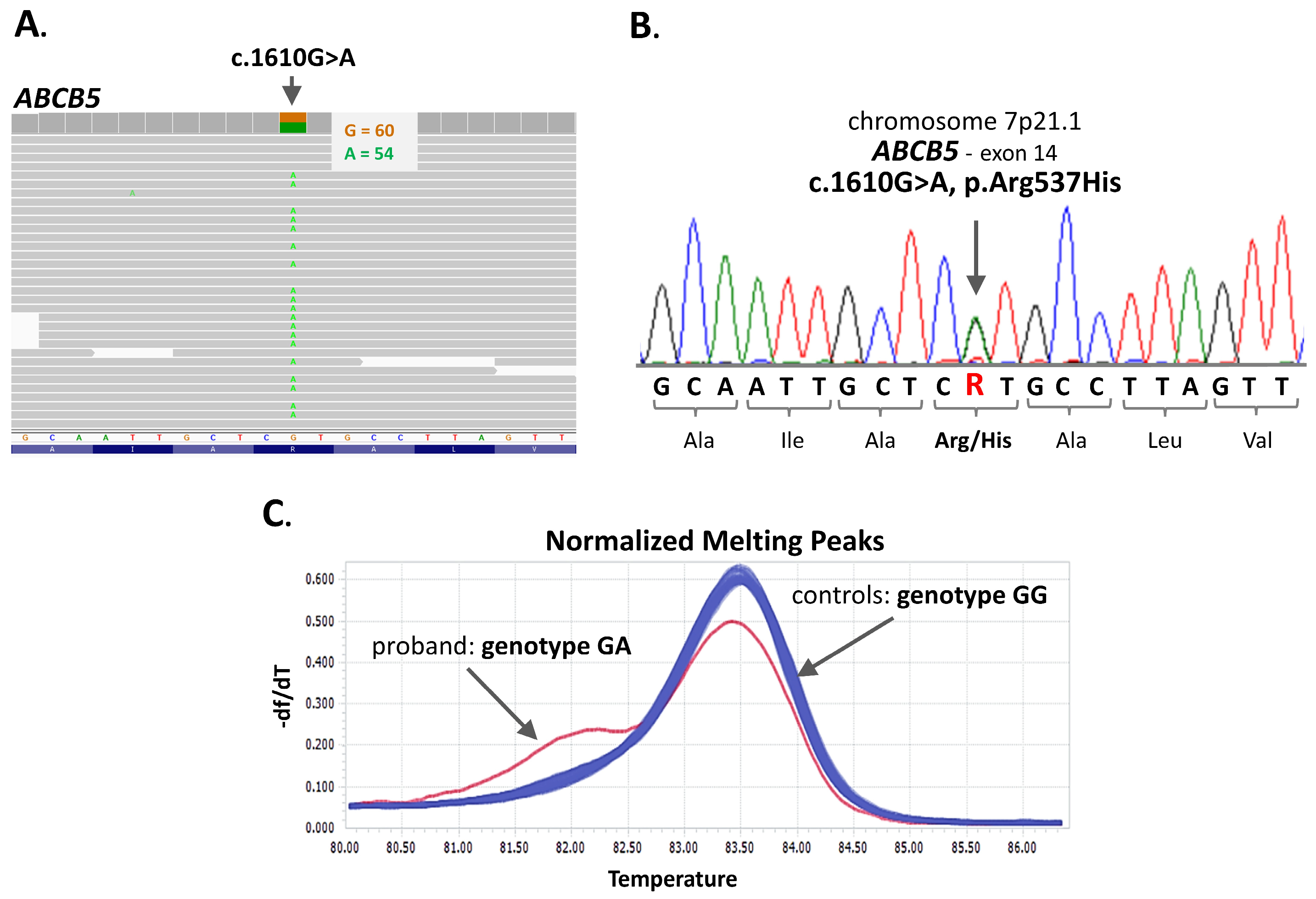
3.2. Molecular Analyses
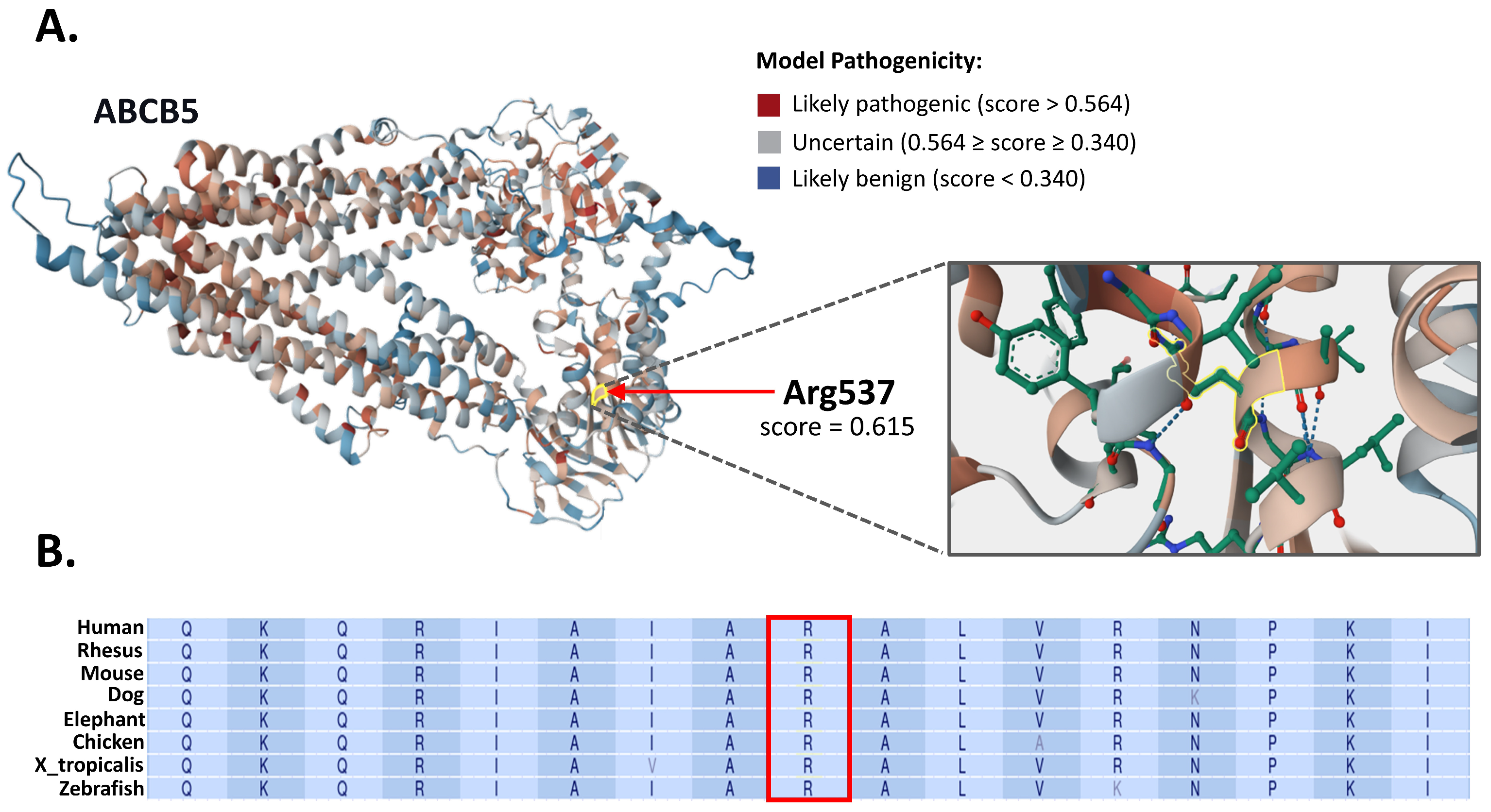
3.3. In Silico Structural and Evolutionary Conservation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADA | Adaptive Boosting |
| CADD | Combined Annotation-Dependent Depletion |
| GWAS | Genome-Wide Association Study |
| HRM | High-Resolution Melting curve |
| ICP | Intrahepatic Cholestasis of Pregnancy |
| IUFD | Intrauterine Fetal Death |
| NBS1 | Nucleotide-Binding Site 1 |
| pLDDT | Predicted Local Distance Difference Test |
| TBA | Total Bile Acids |
| TSBA | Total Serum Bile Acid |
| WES | Whole-Exome Sequencing |
References
- Geenes, V.; Williamson, C. Intrahepatic cholestasis of pregnancy. World J. Gastroenterol. 2009, 15, 2049–2066. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P.H.; Levine, A.P.; Cebola, I.; Chan, M.M.Y.; Amin, A.S.; Aich, A.; Mozere, M.; Maude, H.; Mitchell, A.L.; Zhang, J.; et al. GWAS meta-analysis of intrahepatic cholestasis of pregnancy implicates multiple hepatic genes and regulatory elements. Nat. Commun. 2022, 13, 4840. [Google Scholar] [CrossRef]
- Ovadia, C.; Seed, P.T.; Sklavounos, A.; Geenes, V.; Di Ilio, C.; Chambers, J.; Kohari, K.; Bacq, Y.; Bozkurt, N.; Brun-Furrer, R.; et al. Association of adverse perinatal outcomes of intrahepatic cholestasis of pregnancy with biochemical markers: Results of aggregate and individual patient data meta-analyses. Lancet 2019, 393, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Ovadia, C.; Syngelaki, A.; Souretis, K.; Martineau, M.; Girling, J.; Vasavan, T.; Fan, H.M.; Seed, P.T.; Chambers, J.; et al. Re-evaluating diagnostic thresholds for intrahepatic cholestasis of pregnancy: Case-control and cohort study. BJOG 2021, 128, 1635–1644. [Google Scholar] [CrossRef] [PubMed]
- Guszczynska-Losy, M.; Wirstlein, P.K.; Wender-Ozegowska, E.; Kedzia, M. Evaluation of predictive value of biochemical markers for adverse obstetrics outcomes in pregnancies complicated by cholestasis. Ginekol. Pol. 2020, 91, 269–276. [Google Scholar] [CrossRef]
- Abu-Hayyeh, S.; Martinez-Becerra, P.; Sheikh Abdul Kadir, S.H.; Selden, C.; Romero, M.R.; Rees, M.; Marschall, H.U.; Marin, J.J.; Williamson, C. Inhibition of Na+-taurocholate Co-transporting polypeptide-mediated bile acid transport by cholestatic sulfated progesterone metabolites. J. Biol. Chem. 2010, 285, 16504–16512. [Google Scholar] [CrossRef]
- Abu-Hayyeh, S.; Papacleovoulou, G.; Lövgren-Sandblom, A.; Tahir, M.; Oduwole, O.; Jamaludin, N.A.; Ravat, S.; Nikolova, V.; Chambers, J.; Selden, C.; et al. Intrahepatic cholestasis of pregnancy levels of sulfated progesterone metabolites inhibit farnesoid X receptor resulting in a cholestatic phenotype. Hepatology 2013, 57, 716–726. [Google Scholar] [CrossRef]
- Brites, D.; Rodrigues, C.M.; van-Zeller, H.; Brito, A.; Silva, R. Relevance of serum bile acid profile in the diagnosis of intrahepatic cholestasis of pregnancy in an high incidence area: Portugal. Eur. J. Obstet. Gynecol. Reprod. Biol. 1998, 80, 31–38. [Google Scholar] [CrossRef]
- Dixon, P.H.; Weerasekera, N.; Linton, K.J.; Donaldson, O.; Chambers, J.; Egginton, E.; Weaver, J.; Nelson-Piercy, C.; de Swiet, M.; Warnes, G.; et al. Heterozygous MDR3 missense mutation associated with intrahepatic cholestasis of pregnancy: Evidence for a defect in protein trafficking. Hum. Mol. Genet. 2000, 9, 1209–1217. [Google Scholar] [CrossRef]
- Stättermayer, A.F.; Halilbasic, E.; Wrba, F.; Ferenci, P.; Trauner, M. Variants in ABCB4 (MDR3) across the spectrum of cholestatic liver diseases in adults. J. Hepatol. 2020, 73, 651–663. [Google Scholar] [CrossRef]
- Pillarisetty, L.S.; Sharma, A. Pregnancy Intrahepatic Cholestasis. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK551503/ (accessed on 10 June 2025).
- Turro, E.; Astle, W.J.; Megy, K.; Gräf, S.; Greene, D.; Shamardina, O.; Allen, H.L.; Sanchis-Juan, A.; Frontini, M.; Thys, C.; et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 2020, 583, 96–102. [Google Scholar] [CrossRef]
- Meier, Y.; Zodan, T.; Lang, C.; Zimmermann, R.; Kullak-Ublick, G.A.; Meier, P.J.; Stieger, B.; Pauli-Magnus, C. Increased susceptibility for intrahepatic cholestasis of pregnancy and contraceptive-induced cholestasis in carriers of the 1331T>C polymorphism in the bile salt export pump. World J. Gastroenterol. 2008, 14, 38–45. [Google Scholar] [CrossRef]
- Pauli-Magnus, C.; Meier, P.J.; Stieger, B. Genetic determinants of drug-induced cholestasis and intrahepatic cholestasis of pregnancy. Semin. Liver Dis. 2010, 30, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xiong, L.; Cai, J.; Fu, J.; Liu, H.; Ye, Y.; Yang, L.; Xing, S.; Yang, X. Intrahepatic cholestasis of pregnancy: Insights into pathogenesis and advances in omics studies. Hepatol. Int. 2024, 18, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lai, H.; Zeng, X.; Xin, S.; Nie, L.; Liang, Z.; Wu, M.; Chen, Y.; Zheng, J.; Zou, Y. Whole-exome sequencing reveals ANO8 as a genetic risk factor for intrahepatic cholestasis of pregnancy. BMC Pregnancy Childbirth 2020, 20, 544. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lai, H.; Xin, S.; Li, Z.; Zeng, X.; Nie, L.; Liang, Z.; Wu, M.; Zheng, J.; Zou, Y. Whole-exome sequencing identifies novel mutations in ABC transporter genes associated with intrahepatic cholestasis of pregnancy disease: A case-control study. BMC Pregnancy Childbirth 2021, 21, 110. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, J.; Xin, S.; Zeng, Y.; Wu, X.; Zeng, X.; Lai, H.; Zou, Y. Whole-exome sequencing expands the roles of novel mutations of organic anion transporting polypeptide, ATP-binding cassette transporter, and receptor genes in intrahepatic cholestasis of pregnancy. Front. Genet. 2022, 13, 941027. [Google Scholar] [CrossRef]
- Girling, J.; Knight, C.L.; Chappell, L.; Royal College of Obstetricians and Gynaecologists. Intrahepatic cholestasis of pregnancy: Green-top Guideline No. 43 June 2022. BJOG 2022, 129, e95–e114. [Google Scholar] [CrossRef]
- Grochowalski, Ł.; Jarczak, J.; Urbanowicz, M.; Słomka, M.; Szargut, M.; Borówka, P.; Sobalska-Kwapis, M.; Marciniak, B.; Ossowski, A.; Lorkiewicz, W.; et al. Y-Chromosome Genetic Analysis of Modern Polish Population. Front. Genet. 2020, 11, 567309. [Google Scholar] [CrossRef]
- Biedziak, B.; Dąbrowska, J.; Szponar-Żurowska, A.; Bukowska-Olech, E.; Jamsheer, A.; Mojs, E.; Mulle, J.; Płoski, R.; Mostowska, A. Identification of a new familial case of 3q29 deletion syndrome associated with cleft lip and palate via whole-exome sequencing. Am. J. Med. Genet. Part A 2023, 191, 205–219. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Stockner, T.; Gradisch, R.; Schmitt, L. The role of the degenerate nucleotide binding site in type I ABC exporters. FEBS Lett. 2020, 594, 3815–3838. [Google Scholar] [CrossRef]
- Gerard, L.; Gillet, J.P. The uniqueness of ABCB5 as a full transporter ABCB5FL and a half-transporter-like ABCB5β. Cancer Drug Resist. 2024, 7, 29. [Google Scholar] [CrossRef]
- Dixon, P.H.; Williamson, C. The pathophysiology of intrahepatic cholestasis of pregnancy. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Mikucka-Niczyporuk, A.; Pierzynski, P.; Lemancewicz, A.; Kosinski, P.; Charkiewicz, K.; Knas, M.; Kacerovsky, M.; Blachnio-Zabielska, A.; Laudanski, P. Role of sphingolipids in the pathogenesis of intrahepatic cholestasis. Prostaglandins Other Lipid Mediat. 2020, 147, 106399. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, J.; Lee, P.M.Y.; Zhang, J.; Li, F.; Ren, T. Familial clustering of intrahepatic cholestasis of pregnancy: A nationwide population-based study in Denmark. Hepatology 2023, 78, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Turunen, K.; Helander, K.; Mattila, K.J.; Sumanen, M. Intrahepatic cholestasis of pregnancy is common among patients’ first-degree relatives. Acta Obstet. Gynecol. Scand. 2013, 92, 1108–1110. [Google Scholar] [CrossRef]
- Davit-Spraul, A.; Gonzales, E.; Baussan, C.; Jacquemin, E. Progressive familial intrahepatic cholestasis. Orphanet J. Rare Dis. 2009, 4, 1. [Google Scholar] [CrossRef]
- Pasmant, E.; Goussard, P.; Baranes, L.; Laurendeau, I.; Quentin, S.; Ponsot, P.; Consigny, Y.; Farges, O.; Condat, B.; Vidaud, D.; et al. First description of ABCB4 gene deletions in familial low phospholipid-associated cholelithiasis and oral contraceptives-induced cholestasis. Eur. J. Hum. Genet. 2012, 20, 277–282. [Google Scholar] [CrossRef]
- Burdick, K.J.; Cogan, J.D.; Rives, L.C.; Robertson, A.K.; Koziura, M.E.; Brokamp, E.; Duncan, L.; Hannig, V.; Pfotenhauer, J.; Vanzo, R.; et al. Limitations of exome sequencing in detecting rare and undiagnosed diseases. Am. J. Med. Genet. Part A 2020, 182, 1400–1406. [Google Scholar] [CrossRef]
- Corominas, J.; Smeekens, S.P.; Nelen, M.R.; Yntema, H.G.; Kamsteeg, E.J.; Pfundt, R.; Gilissen, C. Clinical exome sequencing-Mistakes and caveats. Hum. Mutat. 2022, 43, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Ewans, L.J.; Minoche, A.E.; Schofield, D.; Shrestha, R.; Puttick, C.; Zhu, Y.; Drew, A.; Gayevskiy, V.; Elakis, G.; Walsh, C.; et al. Whole exome and genome sequencing in mendelian disorders: A diagnostic and health economic analysis. Eur. J. Hum. Genet. 2022, 30, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Anaya, A.M.; Gerard, L.; Albert, M.; Gaussin, J.F.; Boonen, M.; Gillet, J.P. The β Isoform of Human ATP-Binding Cassette B5 Transporter, ABCB5β, Localizes to the Endoplasmic Reticulum. Int. J. Mol. Sci. 2023, 24, 15847. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.G.; Szakács, G.; Annereau, J.P.; Rouzaud, F.; Liang, X.J.; Valencia, J.C.; Nagineni, C.N.; Hooks, J.J.; Hearing, V.J.; Gottesman, M.M. Principal expression of two mRNA isoforms (ABCB 5alpha and ABCB 5beta ) of the ATP-binding cassette transporter gene ABCB 5 in melanoma cells and melanocytes. Pigment. Cell Res. 2005, 18, 102–112. [Google Scholar] [CrossRef]
- Moitra, K.; Scally, M.; McGee, K.; Lancaster, G.; Gold, B.; Dean, M. Molecular evolutionary analysis of ABCB5: The ancestral gene is a full transporter with potentially deleterious single nucleotide polymorphisms. PLoS ONE 2011, 6, e16318. [Google Scholar] [CrossRef]
- Saeed, M.E.M.; Boulos, J.C.; Machel, K.; Andabili, N.; Marouni, T.; Roth, W.; Efferth, T. Expression of the Stem Cell Marker ABCB5 in Normal and Tumor Tissues. In Vivo 2022, 36, 1651–1666. [Google Scholar] [CrossRef]
- Frank, N.Y.; Margaryan, A.; Huang, Y.; Schatton, T.; Waaga-Gasser, A.M.; Gasser, M.; Sayegh, M.H.; Sadee, W.; Frank, M.H. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005, 65, 4320–4333. [Google Scholar] [CrossRef]
- Frank, N.Y.; Frank, M.H. ABCB5 gene amplification in human leukemia cells. Leuk. Res. 2009, 33, 1303–1305. [Google Scholar] [CrossRef]
- Grimm, M.; Krimmel, M.; Polligkeit, J.; Alexander, D.; Munz, A.; Kluba, S.; Keutel, C.; Hoffmann, J.; Reinert, S.; Hoefert, S. ABCB5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eur. J. Cancer 2012, 48, 3186–3197. [Google Scholar] [CrossRef]
- Guo, Q.; Grimmig, T.; Gonzalez, G.; Giobbie-Hurder, A.; Berg, G.; Carr, N.; Wilson, B.J.; Banerjee, P.; Ma, J.; Gold, J.S.; et al. ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer. J. Biol. Chem. 2018, 293, 11166–11178. [Google Scholar] [CrossRef]
- Kugimiya, N.; Nishimoto, A.; Hosoyama, T.; Ueno, K.; Enoki, T.; Li, T.S.; Hamano, K. The c-MYC-ABCB5 axis plays a pivotal role in 5-fluorouracil resistance in human colon cancer cells. J. Cell. Mol. Med. 2015, 19, 1569–1581. [Google Scholar] [CrossRef]
- Leung, I.C.; Chong, C.C.; Cheung, T.T.; Yeung, P.C.; Ng, K.K.; Lai, P.B.; Chan, S.L.; Chan, A.W.; Tang, P.M.; Cheung, S.T. Genetic variation in ABCB5 associates with risk of hepatocellular carcinoma. J. Cell. Mol. Med. 2020, 24, 10705–10713. [Google Scholar] [CrossRef]
- Duvivier, L.; Gillet, J.P. Deciphering the roles of ABCB5 in normal and cancer cells. Trends Cancer 2022, 8, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Ksander, B.R.; Kolovou, P.E.; Wilson, B.J.; Saab, K.R.; Guo, Q.; Ma, J.; McGuire, S.P.; Gregory, M.S.; Vincent, W.J.; Perez, V.L.; et al. ABCB5 is a limbal stem cell gene required for corneal development and repair. Nature 2014, 511, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, L.; Heitsch, L.; Carrera, C.; Farias, F.H.G.; Del Aguila, J.L.; Dhar, R.; Budde, J.; Bergmann, K.; Bradley, J.; Harari, O.; et al. Multi-ancestry GWAS reveals excitotoxicity associated with outcome after ischaemic stroke. Brain 2022, 145, 2394–2406. [Google Scholar] [CrossRef] [PubMed]
- Glessner, J.T.; Bradfield, J.P.; Wang, K.; Takahashi, N.; Zhang, H.; Sleiman, P.M.; Mentch, F.D.; Kim, C.E.; Hou, C.; Thomas, K.A.; et al. A genome-wide study reveals copy number variants exclusive to childhood obesity cases. Am. J. Hum. Genet. 2010, 87, 661–666. [Google Scholar] [CrossRef]
- Lee, H.A.; Ahn, E.H.; Kim, J.H.; Kim, J.O.; Ryu, C.S.; Lee, J.Y.; Cho, S.H.; Lee, W.S.; Kim, N.K. Association study of frameshift and splice variant polymorphisms with risk of idiopathic recurrent pregnancy loss. Mol. Med. Rep. 2018, 18, 2417–2426. [Google Scholar] [CrossRef]
- Wadén, K.; Karlöf, E.; Narayanan, S.; Lengquist, M.; Hansson, G.K.; Hedin, U.; Roy, J.; Matic, L. Clinical risk scores for stroke correlate with molecular signatures of vulnerability in symptomatic carotid patients. iScience 2022, 25, 104219. [Google Scholar] [CrossRef]
- Sawicka-Gutaj, N.; Gruszczyński, D.; Guzik, P.; Mostowska, A.; Walkowiak, J. Ethics of Human Studies in the Light of the Declaration of Helsinki—A Mini-Review. J. Med. Sci. 2022, 91, e700. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kędzia, M.; Wender-Ożegowska, E.; Dąbrowska, J.; Jagodziński, P.P.; Mostowska, A. A Rare ABCB5 Variant in a Familial Case of Intrahepatic Cholestasis of Pregnancy: A Potential Novel Genetic Contributor. J. Clin. Med. 2025, 14, 5618. https://doi.org/10.3390/jcm14165618
Kędzia M, Wender-Ożegowska E, Dąbrowska J, Jagodziński PP, Mostowska A. A Rare ABCB5 Variant in a Familial Case of Intrahepatic Cholestasis of Pregnancy: A Potential Novel Genetic Contributor. Journal of Clinical Medicine. 2025; 14(16):5618. https://doi.org/10.3390/jcm14165618
Chicago/Turabian StyleKędzia, Małgorzata, Ewa Wender-Ożegowska, Justyna Dąbrowska, Paweł P. Jagodziński, and Adrianna Mostowska. 2025. "A Rare ABCB5 Variant in a Familial Case of Intrahepatic Cholestasis of Pregnancy: A Potential Novel Genetic Contributor" Journal of Clinical Medicine 14, no. 16: 5618. https://doi.org/10.3390/jcm14165618
APA StyleKędzia, M., Wender-Ożegowska, E., Dąbrowska, J., Jagodziński, P. P., & Mostowska, A. (2025). A Rare ABCB5 Variant in a Familial Case of Intrahepatic Cholestasis of Pregnancy: A Potential Novel Genetic Contributor. Journal of Clinical Medicine, 14(16), 5618. https://doi.org/10.3390/jcm14165618





