Machine Learning-Based Identification of Risk Factors for ICU Mortality in 8902 Critically Ill Patients with Pandemic Viral Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Design
2.2. Study Population
2.3. Definitions
2.4. Study Variables
2.5. Missing Data Management
2.6. Analysis Plan and Statistical Analysis
3. Results
3.1. Whole Population
3.2. Factors Associated with Crude ICU Mortality According to General Linear Model (GLM)
3.3. Linear Model (GLM) Validation
3.4. Development of the GLM Model with Correction of Class Imbalance
3.5. Factors Associated with ICU Mortality According to No-Linear Model (Random Forest)
3.6. Non-Linear Model (RFc) Validation
3.7. Patient Classification by Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention (CDC). COVID-19 Case Surveillance Public Use Data. 2021. Available online: https://data.cdc.gov (accessed on 24 April 2025).
- Viboud, C.; Simonsen, L. Global mortality of 2009 pandemic influenza A H1N1. Lancet Infect. Dis. 2012, 12, 651–653. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, L.; Spreeuwenberg, P.; Lustig, R.; Taylor, R.J.; Fleming, D.M.; Kroneman, M.; Van Kerkhove, M.D.; Mounts, A.W.; Paget, W.J.; the GLaMOR Collaborating Teams. Global Mortality Estimates for the 2009 Influenza Pandemic from the GLaMOR Project: A Modeling Study. PLoS Med. 2013, 10, e1001558. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). WHO Coronavirus (COVID-19) Dashboard—Deaths. 2023. Available online: https://data.who.int/dashboards/covid19/deaths (accessed on 24 April 2025).
- Mathieu, E.; Ritchie, H.; Rodés-Guirao, L.; Appel, C.; Gavrilov, D.; Giattino, C.; Hasell, J.; Macdonald, B.; Dattani, S.; Beltekian, D.; et al. COVID-19 Pandemic. Our World in Data. 2020. Available online: https://ourworldindata.org/coronavirus (accessed on 24 April 2025).
- Olesińska, W.; Biernatek, M.; Lachowicz-Wiśniewska, S.; Piątek, J. Systematic review of the impact of COVID-19 on healthcare systems and society—The role of diagnostics and nutrition in pandemic response. J. Clin. Med. 2025, 14, 2482. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Iuliano, A.D.; Reed, C.; Meltzer, M.I.; Shay, D.K.; Cheng, P.Y.; Bandaranayake, D.; Breiman, R.F.; Brooks, W.A.; Buchy, P.; et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: A modelling study. Lancet Infect. Dis. 2012, 12, 687–695. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Z.; Pei, Y.; Huang, Y.; Chen, C.; Zhou, T. Clinical Characteristics and Prognostic Factors of Severe COVID-19. Infect. Drug Resist. 2025, 18, 3117–3125. [Google Scholar] [CrossRef]
- Rezahosseini, O.; Roed, C.; Sejdic, A.; Eiberg, M.F.; Nielsen, L.; Boel, J.; Johannesen, C.K.; van Wijhe, M.; Franck, K.T.; Ostrowski, S.R.; et al. Risk of intensive care unit admission and mortality in patients hospitalized due to influenza A or B and SARS-CoV-2 variants Omicron or Delta. Immun. Inflamm. Dis. 2024, 12, e1269. [Google Scholar] [CrossRef]
- Rodríguez, A.H.; Ruiz-Botella, M.; Martín-Loeches, I.; Jiménez Herrera, M.; Solé-Violán, J.; Gómez, J.; Bodí, M.; Trefler, S.; Papiol, E.; Díaz, E.; et al. Deploying unsupervised clustering analysis to derive clinical phenotypes and risk factors associated with mortality risk in 2022 critically ill patients with COVID-19 in Spain. Crit. Care 2021, 25, 63. [Google Scholar] [CrossRef]
- Beam, A.L.; Kohane, I.S. Big data and machine learning in health care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef]
- Christodoulou, E.; Ma, J.; Collins, G.S.; Steyerberg, E.W.; Verbakel, J.Y.; Van Calster, B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 2019, 110, 12–22. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Ghassemi, M.M.; Nemati, S.; Niehaus, K.E.; Clifton, D.A.; Clifford, G.D. Machine Learning and Decision Support in Critical Care. Proc. IEEE 2016, 104, 444–466. [Google Scholar] [CrossRef]
- Rodríguez, A.H.; Avilés-Jurado, F.X.; Díaz, E.; Schuetz, P.; Trefler, S.I.; Solé-Violán, J.; Cordero, L.; Vidaur, L.; Estella, Á.; Socias, L.; et al. Procalcitonin (PCT) Levels for Ruling-Out Bacterial Coinfection in ICU Patients with Influenza: A CHAID Decision-Tree Analysis. J. Infect. 2016, 72, 143–151. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef] [PubMed]
- Cuschieri, S. The STROBE Guidelines. Saudi J. Anaesth. 2019, 13 (Suppl. 1), S31–S34. [Google Scholar] [CrossRef] [PubMed]
- Uyeki, T.M.; Bernstein, H.H.; Bradley, J.S.; Englund, J.A.; File, T.M.; Fry, A.M.; Gravenstein, S.; Hayden, F.G.; A Harper, S.; Hirshon, J.M.; et al. Clinical Practice Guidelines by the Infectious Diseases Society of America: 2018 Update on Diagnosis, Treatment, Chemoprophylaxis, and Institutional Outbreak Management of Seasonal Influenza. Clin. Infect. Dis. 2019, 68, 895–902. [Google Scholar] [CrossRef]
- World Health Organization. Clinical Management of Severe Acute Respiratory Infection (SARI) When COVID-19 Disease is Suspected: Interim Guidance, 13 March 2020. World Health Organization. 2020. Available online: https://apps.who.int/iris/handle/10665/331446 (accessed on 24 April 2025).
- Martin-Loeches, I.; Schultz, M.J.; Vincent, J.L.; Alvarez-Lerma, F.; Bos, L.D.; Solé-Violán, J.; Torres, A.; Rodriguez, A. Increased Incidence of Co-Infection in Critically Ill Patients with Influenza. Intensive Care Med. 2017, 43, 48–58. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Clinical Guidance for Hospitalized and Non-Hospitalized Patients when SARS-CoV-2 and Influenza Viruses Are Co-Circulating; Public Health Guidance; CDC: Atlanta, GA, USA, 2024. Available online: https://www.cdc.gov/flu/hcp/clinical-guidance/testing-guidance-for-clinicians.html (accessed on 25 April 2025).
- Kellum, J.A.; Lameire, N.; Aspelin, P.; MacLeod, A.M.; Barsoum, R.S.; Metha, R.L.; Burdmann, E.A.; Murray, P.T.; Goldstein, S.L.; Kidney Disease: Improving Global Outcomes (KDIGO) Acute Kidney Injury Work Group; et al. KDIGO Clinical Practice Guideline for Acute Kidney Injury. Kidney Int. Suppl. 2012, 2, 1–138. [Google Scholar] [CrossRef]
- Knaus, W.A.; Draper, E.A.; Wagner, D.P.; Zimmerman, J.E. APACHE II: A Severity of Disease Classification System. Crit. Care Med. 1985, 13, 818–829. [Google Scholar] [CrossRef]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonça, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The SOFA (Sepsis-Related Organ Failure Assessment) Score to Describe Organ Dysfunction/Failure. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Menardi, G.; Torelli, N. Training and Assessing Classification Rules with Imbalanced Data. Data Min. Knowl. Discov. 2014, 28, 92–122. [Google Scholar] [CrossRef]
- Lunardon, N.; Menardi, G.; Torelli, N. ROSE: A Package for Binary Imbalanced Learning. R J. 2014, 6, 79–89. [Google Scholar] [CrossRef]
- Huang, H.; Cai, S.; Li, Y.; Li, Y.; Fan, Y.; Li, L.; Lei, C.; Tang, X.; Hu, F.; Li, F.; et al. Prognostic Factors for COVID-19 Pneumonia Progression to Severe Symptoms Based on Earlier Clinical Features: A Retrospective Analysis. Front. Med. 2020, 7, 557453. [Google Scholar] [CrossRef]
- Zhu, Z.; Cai, T.; Fan, L.; Lou, K.; Hua, X.; Huang, Z.; Gao, G. Clinical Value of Immune-Inflammatory Parameters to Assess the Severity of Coronavirus Disease 2019. Int. J. Infect. Dis. 2020, 95, 332–339. [Google Scholar] [CrossRef]
- Gong, J.; Ou, J.; Qiu, X.; Jie, Y.; Chen, Y.; Yuan, L.; Cao, J.; Tan, M.; Xu, W.; Zheng, F.; et al. A tool to early predict Severe Coronavirus Disease 2019 (COVID-19): A Multicenter Study Using the Risk Nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis. 2020, 71, 833–840. [Google Scholar] [CrossRef]
- Aloisio, E.; Chibireva, M.; Serafini, L.; Pasqualetti, S.; Falvella, F.S.; Dolci, A.; Panteghini, M. A Comprehensive Appraisal of Labor-atory Biochemistry Tests as Major Predictors of COVID-19 Severity. Arch. Pathol. Lab. Med. 2020, 144, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Song, N.C.; Zheng, Z.K.; Li, J.S.; Li, S.K. Laboratory Findings and a Combined Multifactorial Approach to Predict Death in Critically Ill Patients with COVID-19: A Retrospective Study. Epidemiol. Infect. 2020, 148, e129. [Google Scholar] [CrossRef] [PubMed]
- Reina Reina, A.; Barrera, J.M.; Valdivieso, B.; Gas, M.E.; Maté, A.; Trujillo, J.C. Machine Learning Model from a Spanish Cohort for Prediction of SARS-COV-2 Mortality Risk and Critical Patients. Sci. Rep. 2022, 12, 5723. [Google Scholar] [CrossRef] [PubMed]
- Pourhomayoun, M.; Shakibi, M. Predicting Mortality Risk in Patients with COVID-19 Using Machine Learning to Help Medical Decision-Making. Smart Health 2021, 20, 100178. [Google Scholar] [CrossRef]
- Alballa, N.; Al-Turaiki, I. Machine Learning Approaches in COVID-19 Diagnosis, Mortality, and Severity Risk Prediction: A Review. Inform. Med. Unlocked 2021, 24, 100622. [Google Scholar] [CrossRef]
- Cornelius, E. COVID-19 Mortality Prediction Using Machine Learning-Integrated Random Forest Algorithm Under Varying Patient Frailty. Master’s Thesis, Illinois State University, Normal, IL, USA, 2021. [Google Scholar] [CrossRef]
- Hernández-Cárdenas, C.M.; Choreño-Parra, J.A.; Torruco-Sotelo, C.; Jurado, F.; Serna-Secundino, H.; Aguilar, C.; García-Olazarán, J.G.; Hernández-García, D.; Choreño-Parra, E.M.; Zúñiga, J.; et al. Clinical Risk Factors for Mortality Among Critically Ill Mexican Patients with COVID-19. Front. Med. 2021, 8, 699607. [Google Scholar] [CrossRef]
- Banoei, M.M.; Dinparastisaleh, R.; Zadeh, A.V.; Mirsaeidi, M. Machine-Learning-Based COVID-19 Mortality Prediction Model and Identification of Patients at Low and High Risk of Dying. Crit. Care 2021, 25, 328. [Google Scholar] [CrossRef]
- Wang, M.; Li, W.; Wang, H.; Song, P. Development and Validation of Machine Learning-Based Models for Predicting Healthcare-Associated Bacterial/Fungal Infections Among COVID-19 Inpatients: A Retrospective Cohort Study. Antimicrob. Resist. Infect. Control 2024, 13, 42. [Google Scholar] [CrossRef]
- Karasneh, R.A.; Khassawneh, B.Y.; Al-Azzam, S.; Al-Mistarehi, A.H.; Lattyak, W.J.; Aldiab, M.; Kabbaha, S.; Hasan, S.S.; Conway, B.R.; Aldeyab, M.A. Risk Factors Associ-ated with Mortality in COVID-19 Hospitalized Patients: Data from the Middle East. Int. J. Clin. Pract. 2022, 2022, 9617319. [Google Scholar] [CrossRef]
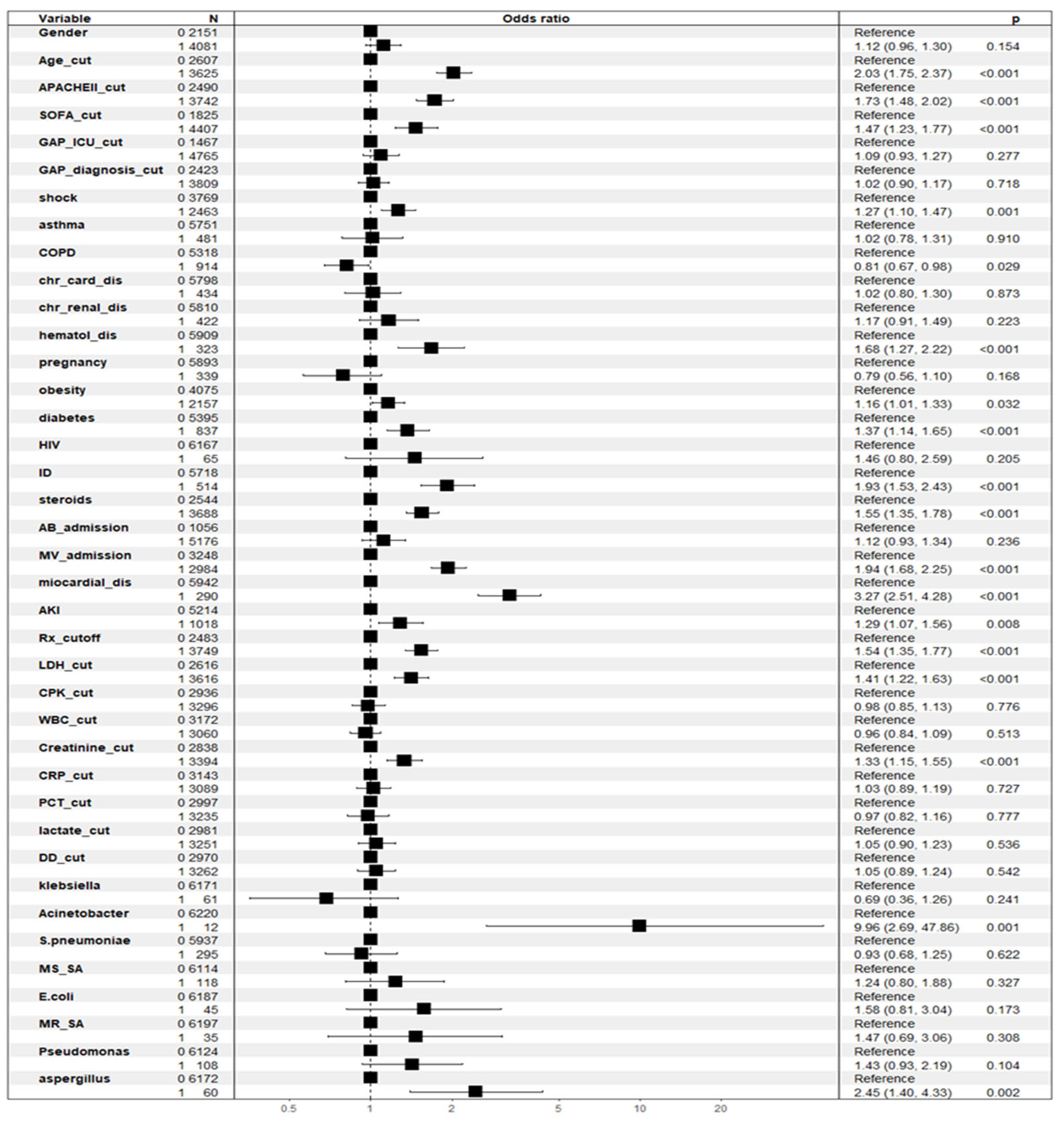

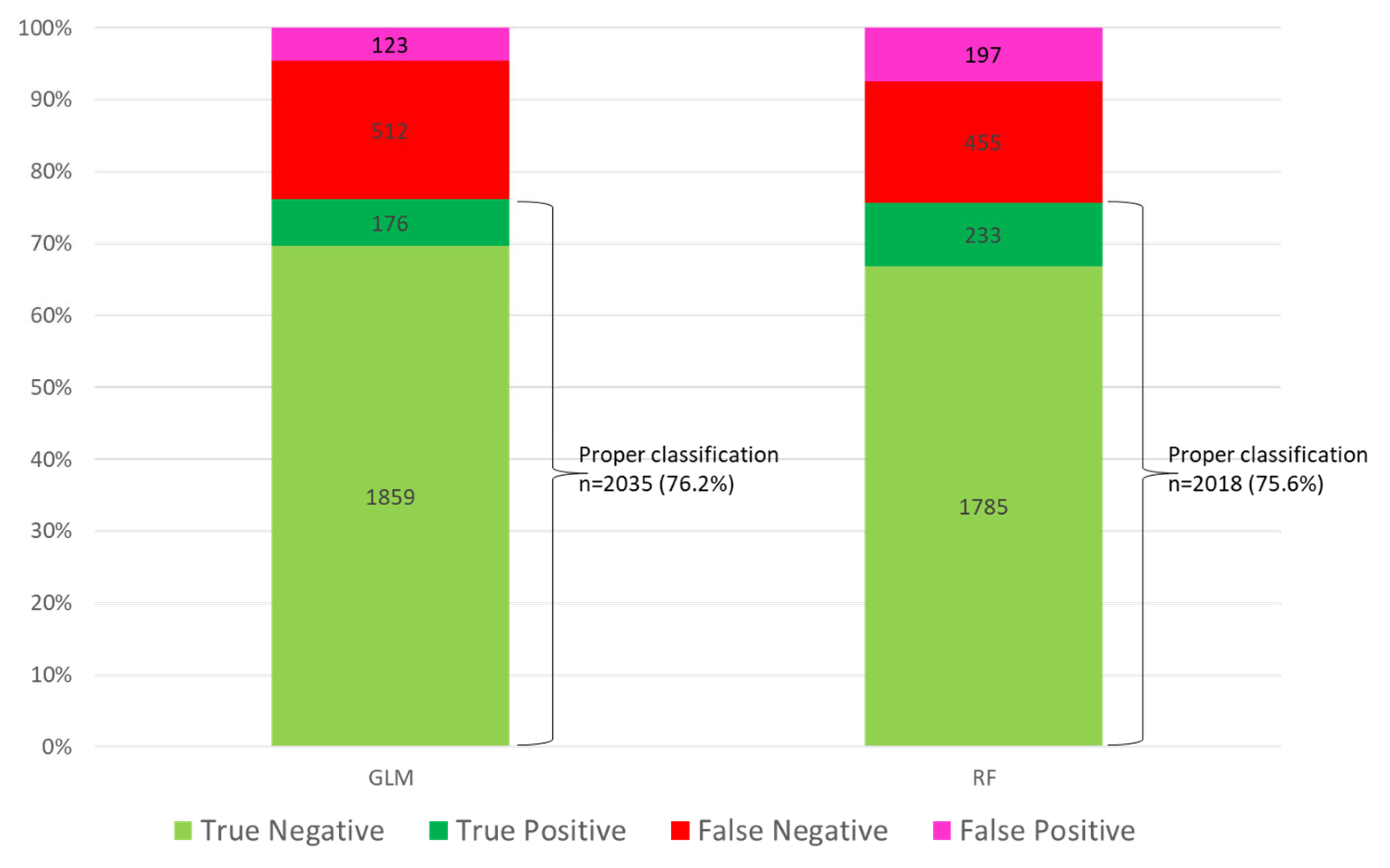
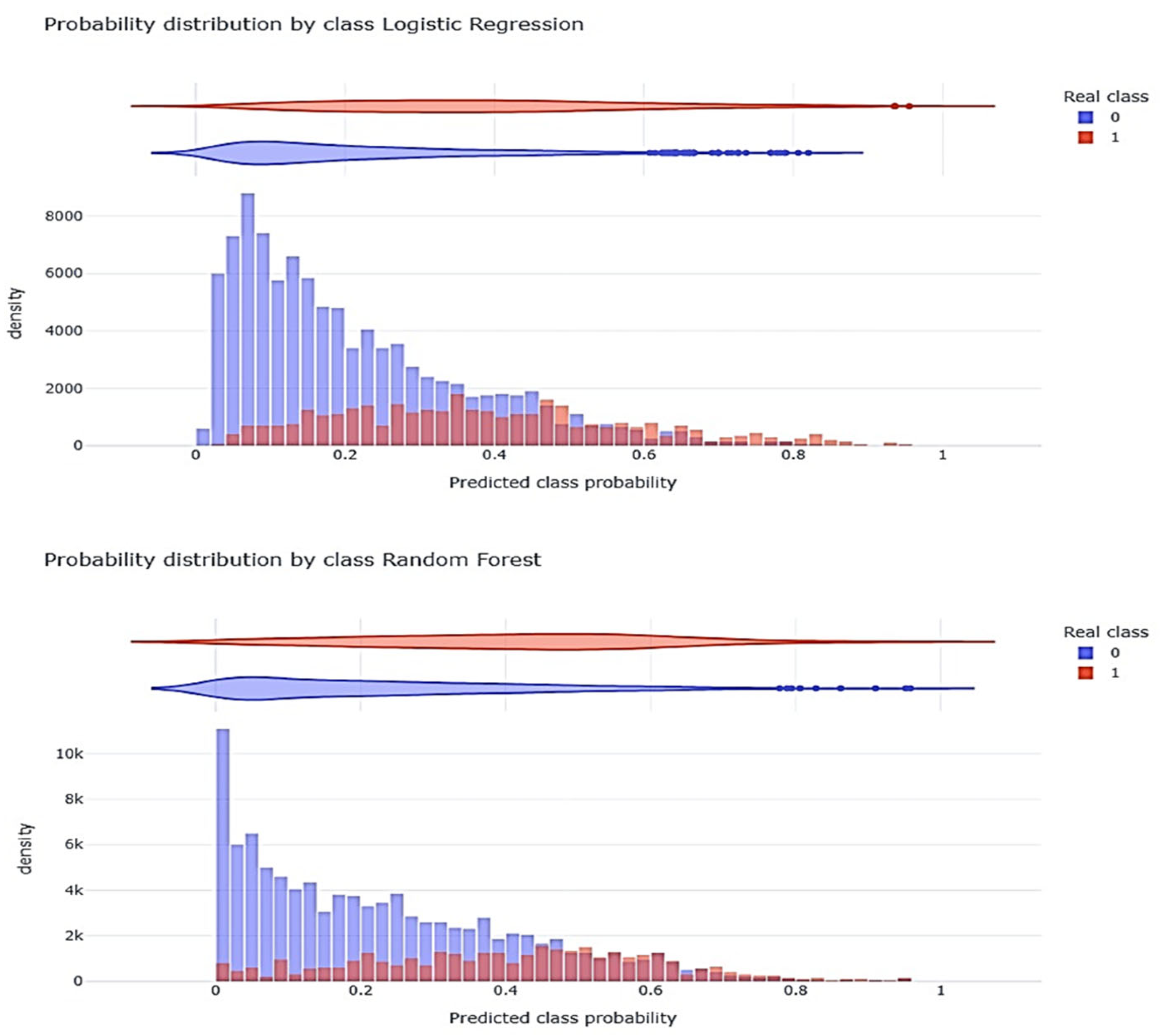
| Variable | Whole Population (n = 8902) | Survival (n = 6608) | Non-Survival (n = 2294) | p-Value |
|---|---|---|---|---|
| General | ||||
| Age, median (Q1–Q3) years | 60 (49–70) | 58 (48–68) | 67 (57–74) | <0.001 |
| Age cut-off > 58 years, n (%) | 5177(58.1) | 3473 (52.6) | 1704 (74.3) | <0.001 |
| Male sex, n (%) | 5855 (65.8) | 4248 (64,3) | 1607 (70.1) | <0.001 |
| APACHE II, median (Q1–Q3) | 14 (10–19) | 13 (10–17) | 17 (13–22) | <0.001 |
| APACHE II cut-off > 13, n (%) | 5309 (59.6) | 3536 (53.5) | 1773 (77.3) | <0.001 |
| SOFA score, median (Q1–Q3) | 5 (3–7) | 4(3–7) | 6(4–9) | <0.001 |
| SOFA cut-off > 4, n (%) | 6274 (70.5) | 4299 (65.1) | 1975 (86.1) | <0.001 |
| GAP UCI, median (Q1–Q3) | 1 (1–3) | 1 (1–3) | 2 (0–4) | <0.001 |
| GAP UCI cut-off > 1 day, n (%) | 6804 (76.4) | 5085 (77.0) | 1719 (74.9) | 0.053 |
| GAP diagnosis, median (Q1–Q3) | 4 (1–7) | 3 (1–7) | 4 (1–7) | 0.012 |
| GAP diagnosis cut-off > 3 days, n (%) | 5413 (60.8) | 3943 (59.7) | 1470 (64.1) | <0.001 |
| > 2 fields with infiltrations in chest X-ray, n (%) | 5343 (60.0) | 3775 (57.1) | 1568 (68.4) | <0.001 |
| Antiviral vaccine, n (%) | 1333 (14.9) | 885 (13.4) | 448 (19.5) | <0.001 |
| Shock at ICU admission, n (%) | 3549 (39.9) | 2286 (34.6) | 1263 (55.1) | <0.001 |
| Laboratory | ||||
| White blood cells count, median (Q1–Q3) × 103 | 8.6 (5.7–12.5) | 8.5 (5.7–12.1) | 9.0 (5.8–13.7) | <0.001 |
| White blood cells count cut-off < 8.5 × 103, n (%) | 4405 (49.5) | 3351 (50.7) | 1054 (45.9) | <0.001 |
| Lactate dehydrogenase, median (Q1–Q3) U/L | 542 (403–687) | 524 (378–665) | 590 (458–749) | <0.001 |
| Lactate dehydrogenase cut-off > 500 U/L, n (%) | 5157 (57.9) | 3593 (54.4) | 1564 (68.2) | <0.001 |
| C-reactive protein, median (Q1–Q3) mg/dL | 19.6 (9.8–34.7) | 19.0(9.5–34.4) | 21.1 (10.4–35.4) | 0.001 |
| C-reactive protein cut-off >20 mg/dL, n (%) | 4387 (49.3) | 3184 (48.2) | 1203 (52.4) | <0.001 |
| Procalcitonin, median (Q1–Q3) ng/mL | 0.88 (0.20–5.67) | 0.83 (0.20–5.08) | 1.04 (0.23–8.20) | <0.001 |
| Procalcitonin cut-off >0.80 ng/mL, n (%) | 4606 (51.7) | 3350 (50.7) | 1256 (54.8) | 0.001 |
| Lactate, median (Q1–Q3) mmol/L | 2.0 (1.4–3.3) | 2.0 (1.3–3.2) | 2.2 (1.4–3.8) | <0.001 |
| Lactate cut-off > 2 mmol/L, n (%) | 4660 (52.3) | 3369 (51.0) | 1291 (56.3) | <0.001 |
| Creatinine, median (Q1–Q3) mg/dL | 0.89 (0.7–1.2) | 0.85 (0.68–1.12) | 1.01 (0.75–1.50) | <0.001 |
| Creatinine cut-off >0.85 mg/dL, n (%) | 4841 (54.4) | 3330 (50.4) | 1511 (65.9) | <0.001 |
| D-dimer, median (Q1–Q3) ng/mL | 3071 (971–6604) | 2716 (900–6000) | 4180 (1200–8680) | <0.001 |
| D-dimer cut-off > 2700 ng/mL, n (%) | 4663 (52.4) | 3314 (50.2) | 1349 (58.8) | <0.001 |
| creatine phosphokinase, median (Q1–Q3) U/L | 216 (100–420) | 210 (97–414) | 234 (111–442) | 0.001 |
| Creatine phosphokinase cut-off > 200 U/L, n (%) | 4707 (52.9) | 3433 (52.0) | 1274 (55.5) | 0.003 |
| Comorbidities | ||||
| Diabetes mellitus, n (%) | 1196 (13.4) | 756 (11.4) | 440 (19.2) | <0.001 |
| Asthma, n (%) | 698 (7.7) | 556 (8.4) | 142 (6.2) | 0.001 |
| COPD, n (%) | 1281 (14.4) | 936 (14.2) | 345 (15.0) | 0.32 |
| Chronic heart disease, n (%) | 623 (7.0) | 418 (6.3) | 205 (8.9) | <0.001 |
| Chronic liver disease, n (%) | 595 (6.7) | 357 (5.4) | 238 (10.4) | <0.001 |
| Pregnancy, n (%) | 480 (5.4) | 399 (6.0) | 81 (3.5) | <0.001 |
| Obesity, n (%) | 3046 (34.2) | 2256 (34.1) | 790 (34.4) | 0.81 |
| Human immunodeficiency virus, n (%) | 144 (1.6) | 107 (1.6) | 37 (1.6) | 1.00 |
| Hematologic disease, n (%) | 436 (4.8) | 237 (3.6) | 199 (8.7) | <0.001 |
| Immunosuppression, n (%) | 711 (8.0) | 401 (6.0) | 310 (13.5) | <0.001 |
| Treatment | ||||
| Steroids, n (%) | 5275 (59.2) | 3746 (56.7) | 1529 (66.7) | <0.001 |
| Antibiotics (AB) at ICU admission, n (%) | 7410 (83.2) | 5428 (82.1) | 1982 (86.4) | <0.001 |
| Appropriate empiric AB treatment, n (%) | 951 ((10.7) | 671 (10.2) | 280 (12.2) | 0.007 |
| High flow nasal cannula at admission, n (%) | 1438 (16.1) | 1138 (17.2) | 300 (13.1) | <0.001 |
| Invasive mechanical ventilation, n (%) | 4252 (47.8) | 2751 (41.6) | 1501 (65.4) | <0.001 |
| Most common aetiology of coinfection | ||||
| Coinfection, n (%) | 1211 (100) | 810 (12.3) | 401 (17.5) | <0.001 |
| Methicillin-sensitive S. aureus (MSSA), n (%) | 172 (14.2) | 111 (13.7) | 61 (15.2) | 0.47 |
| Pseudomonas aeruginosa, n (%) | 143 (11.8) | 82 (10.1) | 61 (15.2) | 0.01 |
| Klebsiella spp. N (%) | 85 (7.0) | 60 (7.4) | 25 (6.2)) | 0.45 |
| Aspergillus spp., n (%) | 78 (6.5) | 33 (4.0) | 45 (11.2) | <0.001 |
| E. coli, n (%) | 69 (5.7) | 43 (5.3) | 26 (6.3) | 0.40 |
| Methicillin-resistant S. aureus (MRSA). n (%) | 56 (4.6) | 33 (4.0) | 23 (5.7) | 0.19 |
| Acinetobacter spp., n (%) | 17 (1.4) | 4 (0.5) | 13 (3.2) | <0.001 |
| Outcomes | ||||
| ICU LOS, median (Q1–Q3) days | 13 (6–23) | 12 (6–23) | 14 (7–24) | 0.03 |
| Acute kidney injury, n (%) | 1435 (16.1) | 855 (12.9) | 580 (25.3) | <0.001 |
| GLM Model | Random Forest Model | |||
|---|---|---|---|---|
| Variable | OR | 95%CI | Decreased Accuracy | Decreased Gini |
| Age ≥ 58 years | 2.03 | 1.74–2.36 | 34.9% | 79.2% |
| APACHE II ≥ 13 points | 1.72 | 1.48–2.02 | 19.1% | 88.1% |
| SOFA ≥ 4 points | 1.47 | 1.23–1.76 | 26.0% | 65.1% |
| Shock | 1.27 | 1.09–1.47 | 16.4% | 77.4% |
| Hematologic disease | 1.67 | 1.26–2.22 | 19.5% | 39.4% |
| Obesity | 1.16 | 1.01–1.32 | ----- | 92.4% |
| Diabetes | 1.37 | 1.14–1.65 | 16.5% | 60.6% |
| Immunosuppression | 1.92 | 1.53–2.42 | 18.9% | 53.0% |
| Steroids | 1.54 | 1.34–1.77 | 12.7% | 81.6% |
| Mechanical ventilation | 1.94 | 1.67–2.25 | 33.0% | 88.1% |
| Myocardial dysfunction | 3.27 | 2.53–4.28 | 47.2% | 63.6% |
| Acute kidney injury | 1.29 | 1.07–1.55 | ---- | ----- |
| >2 fields with infiltrations in chest X-ray | 1.54 | 1.34–1.77 | 16.8% | 81.3% |
| LDH ≥ 500 U/L | 1.41 | 1.22–1.63 | 11.5% | 79.7% |
| Creatinine ≥ 0.85 mg/dL | 1.33 | 1.14–1.55 | 13.3% | 73.8% |
| Acinetobacter spp. | 9.95 | 2.61–47.8 | ---- | ---- |
| Aspergillus spp. | 2.45 | 1.39–4.33 | 11.2% | ---- |
| Procalcitonin ≥2 ng/mL | ---- | ---- | 23.0% | 68.1% |
| D-dimer ≥ 2700 ng/mL | ---- | ---- | 21.7% | 75.9% |
| Lactate ≥ 2 mmol/L | ---- | ---- | 18.1% | 79.5% |
| COPD | ---- | ---- | 17.4% | 61.3% |
| CPK ≥ 200 U/L | ---- | ---- | 13.1% | 90.6% |
| GAP-Diagnosis ≥ 3 days | ---- | ---- | ---- | 96.9% |
| WBC count < 8.5 × 103 | ---- | ---- | ---- | 93.3% |
| Male | ---- | ---- | ---- | 81.3% |
| GAP-ICU < 1 day | ---- | ---- | ---- | 77.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papiol, E.; Ferrer, R.; Ruiz-Rodríguez, J.C.; Díaz, E.; Zaragoza, R.; Borges-Sa, M.; Berrueta, J.; Gómez, J.; Bodí, M.; Sancho, S.; et al. Machine Learning-Based Identification of Risk Factors for ICU Mortality in 8902 Critically Ill Patients with Pandemic Viral Infection. J. Clin. Med. 2025, 14, 5383. https://doi.org/10.3390/jcm14155383
Papiol E, Ferrer R, Ruiz-Rodríguez JC, Díaz E, Zaragoza R, Borges-Sa M, Berrueta J, Gómez J, Bodí M, Sancho S, et al. Machine Learning-Based Identification of Risk Factors for ICU Mortality in 8902 Critically Ill Patients with Pandemic Viral Infection. Journal of Clinical Medicine. 2025; 14(15):5383. https://doi.org/10.3390/jcm14155383
Chicago/Turabian StylePapiol, Elisabeth, Ricard Ferrer, Juan C. Ruiz-Rodríguez, Emili Díaz, Rafael Zaragoza, Marcio Borges-Sa, Julen Berrueta, Josep Gómez, María Bodí, Susana Sancho, and et al. 2025. "Machine Learning-Based Identification of Risk Factors for ICU Mortality in 8902 Critically Ill Patients with Pandemic Viral Infection" Journal of Clinical Medicine 14, no. 15: 5383. https://doi.org/10.3390/jcm14155383
APA StylePapiol, E., Ferrer, R., Ruiz-Rodríguez, J. C., Díaz, E., Zaragoza, R., Borges-Sa, M., Berrueta, J., Gómez, J., Bodí, M., Sancho, S., Suberviola, B., Trefler, S., & Rodríguez, A. (2025). Machine Learning-Based Identification of Risk Factors for ICU Mortality in 8902 Critically Ill Patients with Pandemic Viral Infection. Journal of Clinical Medicine, 14(15), 5383. https://doi.org/10.3390/jcm14155383







