Diagnostic Utility of Intratumoral Susceptibility Signals in Adult Diffuse Gliomas: Tumor Grade Prediction and Correlation with Molecular Markers Within the WHO CNS5 (2021) Classification
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Imaging Acquisition
2.3. Histopathological and Molecular Study
2.4. Data Collection
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Cohort
3.2. MRI Interpretation
3.3. Correlation Between Semiquantitative ITSS Grading and Histological and Molecular Findings
3.4. Prediction of Tumor Histological Grade
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area under the curve |
| CDKN2A/B | Cyclin-dependent kinase inhibitors 2A and 2B |
| DSC | Dynamic susceptibility contrast |
| HGG | High-grade glioma |
| IDH | Isocitrate dehydrogenase |
| ITSS | Intratumoral susceptibility signals |
| LGG | Low-grade glioma |
| rCVB | Relative cerebral blood volume |
| ROC | Receiver operating characteristic |
| SWAN | Susceptibility-weighted angiography |
| SWI | Susceptibility-weighted imaging |
| WHO CNS5 | Fifth edition of the World Health Organization Classification of Tumors of the Central Nervous System |
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; A Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro Oncol. 2022, 24 (Suppl. S5), v1–v95. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.; Ahrendsen, J.T. What’s new in neuropathology 2024: CNS WHO 5th edition updates. J. Pathol. Transl. Med. 2024, 58, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-C.; Zhang, H.; Tan, Y.; Qin, J.-B.; Wu, X.-F.; Wang, L.; Zhang, L. Combined value of susceptibility-weighted and perfusion-weighted imaging in assessing WHO grade for brain astrocytomas. J. Magn. Reson. Imaging 2014, 39, 1569–1574. [Google Scholar] [CrossRef]
- Yang, X.; Xing, Z.; She, D.; Lin, Y.; Zhang, H.; Su, Y.; Cao, D. Grading of IDH-mutant astrocytoma using diffusion, susceptibility and perfusion-weighted imaging. BMC Med. Imaging 2022, 2, 105. [Google Scholar] [CrossRef]
- Yang, X.; Hu, C.; Xing, Z.; Lin, Y.; Su, Y.; Wang, X.; Cao, D. Prediction of Ki-67 labeling index, ATRX mutation, and MGMT promoter methylation status in IDH-mutant astrocytoma by morphological MRI, SWI, DWI, and DSC-PWI. Eur. Radiol. 2023, 33, 1003–1014. [Google Scholar] [CrossRef]
- Li, X.; Zhu, Y.; Kang, H.; Zhang, Y.; Liang, H.; Wang, S.; Zhang, W. Glioma grading by microvascular permeability parameters derived from dynamic contrast-enhanced MRI and intratumoral susceptibility signal on susceptibility weighted imaging Head & neck imaging. Cancer Imaging 2015, 15, 4. [Google Scholar]
- Haller, S.; Haacke, E.M.; Thurnher, M.M.; Barkhof, F. Susceptibility-weighted imaging: Technical essentials and clinical neurologic applications. Radiology 2021, 299, 3–26. [Google Scholar] [CrossRef]
- Su, C.Q.; Lu, S.S.; Han, Q.Y.; Zhou, M.D.; Hong, X.N. Intergrating conventional MRI, texture analysis of dynamic contrast-enhanced MRI, and susceptibility weighted imaging for glioma grading. Acta Radiol. 2019, 60, 777–787. [Google Scholar] [CrossRef]
- Kong, L.W.; Chen, J.; Zhao, H.; Yao, K.; Fang, S.Y.; Wang, Z.; Wang, Y.Y.; Li, S.W. Intratumoral Susceptibility Signals Reflect Biomarker Status in Gliomas. Sci. Rep. 2019, 9, 17080. [Google Scholar] [CrossRef]
- Di Ieva, A.; Lam, T.; Alcaide-Leon, P.; Bharatha, A.; Montanera, W.; Cusimano, M.D. Magnetic resonance susceptibility weighted imaging in neurosurgery: Current applications and future perspectives. J. Neurosurg. 2015, 123, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.C.; Chen, Y.L.; Shui, W.P.; Chung, H.W.; Hsu, S.S.; Lai, P.H. Intra-tumoral susceptibility signal: A post-processing technique for objective grading of astrocytoma with susceptibility-weighted imaging. Quant. Imaging Med. Surg. 2022, 12, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, H.; Zhang, W.; Zheng, J. Contribution of susceptibility- and diffusion-weighted magnetic resonance imaging for grading gliomas. Exp. Ther. Med. 2018, 15, 5113–5118. [Google Scholar] [CrossRef] [PubMed]
- Aydin, O.; Buyukkaya, R.; Hakyemez, B. Susceptibility imaging in glial tumor grading; using 3 tesla magnetic resonance (MR) system and 32 channel head coil. Polish J. Radiol. 2017, 82, 179–187. [Google Scholar] [CrossRef]
- Yang, X.; Lin, Y.; Xing, Z.; She, D.; Su, Y.; Cao, D. Predicting 1p/19q codeletion status using diffusion-, susceptibility-, perfusion-weighted, and conventional MRI in IDH-mutant lower-grade gliomas. Acta Radiol. 2021, 62, 1657–1665. [Google Scholar] [CrossRef]
- Park, M.J.; Kim, H.S.; Jahng, G.H.; Ryu, C.W.; Park, S.M.; Kim, S.Y. Semiquantitative assessment of intratumoral susceptibility signals using non-contrast-enhanced high-field high-resolution susceptibility-weighted imaging in patients with gliomas: Comparison with MR perfusion imaging. AJNR Am. J. Neuroradiol. 2009, 30, 1402–1408. [Google Scholar] [CrossRef]
- Sreenivasan, S.; Madhugiri, V.; Sasidharan, G.; Kumar, R.V.R. Measuring glioma volumes: A comparison of linear measurement based formulae with the manual image segmentation technique. J. Cancer Res. Ther. 2016, 12, 161–168. [Google Scholar]
- Aiudi, D.; Iacoangeli, A.; Dobran, M.; Polonara, G.; Chiapponi, M.; Mattioli, A.; Gladi, M.; Iacoangeli, M. The Prognostic Role of Volumetric MRI Evaluation in the Surgical Treatment of Glioblastoma. J. Clin. Med. 2024, 13, 849. [Google Scholar] [CrossRef]
- Fahrendorf, D.; Hesselmann, V.; Schwindt, W.; Wölfer, J.; Jeibmann, A.; Kooijman, H.; Kugel, H.; Heindel, W.; Bink, A. Variations of ITSS-Morphology and their Relationship to Location and Tumor Volume in Patients with Glioblastoma. J. Neuroimaging 2015, 25, 1015–1022. [Google Scholar] [CrossRef]
- Fahrendorf, D.; Schwindt, W.; Wölfer, J.; Jeibmann, A.; Kooijman, H.; Kugel, H.; Grauer, O.; Heindel, W.; Hesselmann, V.; Bink, A. Benefits of contrast-enhanced SWI in patients with glioblastoma multiforme. Eur. Radiol. 2013, 23, 2868–2879. [Google Scholar] [CrossRef]
- Saini, J.; Gupta, P.K.; Sahoo, P.; Singh, A.; Patir, R.; Ahlawat, S.; Beniwal, M.; Thennarasu, K.; Santosh, V.; Gupta, R.K. Differentiation of grade II/III and grade IV glioma by combining “T1 contrast-enhanced brain perfusion imaging” and susceptibility-weighted quantitative imaging. Neuroradiology 2018, 60, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Esperante, D.; Galicia, K.D.; Rivas-Cuervo, K.G.; Cacho-Díaz, B.; Trejo-Becerril, C.; Taja-Chayeb, L.; Aboud, O.; Carlos-Escalante, J.A.; Wegman-Ostrosky, T. TP53 oncogenic variants as prognostic factors in individuals with glioblastoma: A systematic review and meta-analysis. Front. Neurol. 2024, 15, 1490246. [Google Scholar] [CrossRef] [PubMed]
- Yuile, A.; Satgunaseelan, L.; Wei, J.Q.; Rodriguez, M.; Back, M.; Pavlakis, N.; Hudson, A.; Kastelan, M.; Wheeler, H.R.; Lee, A. CDKN2A/B Homozygous Deletions in Astrocytomas: A Literature Review. Curr. Issues Mol. Biol. 2023, 45, 5276–5292. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Bonekamp, D.; Nowosielski, M.; Kratz, A.; Sill, M.; Burth, S.; Wick, A.; Eidel, O.; Schlemmer, H.P.; Radbruch, A.; et al. Radiogenomics of Glioblastoma: Machine Learning—Based Classification of Molecular Characteristics by Using. Radiology 2016, 281, 907–918. [Google Scholar] [CrossRef]
- Lu, V.M.; O’connor, K.P.; Shah, A.H.; Eichberg, D.G.; Luther, E.M.; Komotar, R.J.; Ivan, M.E. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: A systematic review of the contemporary literature. J. Neurooncol. 2020, 148, 221–229. [Google Scholar] [CrossRef]
- Wach, J.; Basaran, A.E.; Arlt, F.; Vychopen, M.; Seidel, C.; Barrantes-Freer, A.; Müller, W.; Gaunitz, F.; Güresir, E. CDKN2A/B deletions are strongly associated with meningioma progression: A meta-analysis of individual patient data. Acta Neuropathol. Commun. 2023, 11, 189. [Google Scholar] [CrossRef]
- Tong, S.; Wu, J.; Song, Y.; Fu, W.; Yuan, Y.; Zhong, P.; Wang, B. IDH1-mutant metabolite D-2-hydroxyglutarate inhibits proliferation and sensitizes glioma to temozolomide via down-regulating ITGB4/PI3K/AKT. Cell Death Discov. 2024, 10, 317. [Google Scholar] [CrossRef]
- Lin, Y.; Xing, Z.; She, D.; Yang, X.; Zheng, Y.; Xiao, Z.; Wang, X.; Cao, D. IDH mutant and 1p/19q co-deleted oligodendrogliomas: Tumor grade stratification using diffusion-, susceptibility-, and perfusion-weighted MRI. Neuroradiology 2017, 59, 555–562. [Google Scholar] [CrossRef]
- Chan, A.K.-Y.; Shi, Z.-F.; Li, K.K.-W.; Wang, W.-W.; Chen, H.; Chung, N.Y.-F.; Chan, D.T.-M.; Poon, W.-S.; Loong, H.H.-F.; Liu, X.-Z.; et al. Combinations of Single-Gene Biomarkers Can Precisely Stratify 1,028 Adult Gliomas for Prognostication. Front. Oncol. 2022, 12, 839302. [Google Scholar] [CrossRef]
- Familiari, P.; Lapolla, P.; Picotti, V.; Palmieri, M.; Pesce, A.; Carosi, G.; Relucenti, M.; Nottola, S.; Gianno, F.; Minasi, S.; et al. Role of 1p/19q Codeletion in Diffuse Low-grade Glioma Tumour Prognosis. Anticancer Res. 2023, 43, 2659–2670. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Gupta, R.K.; Patir, R.; Vaishya, S.; Ahlawat, S.; Singh, A. Quantitative vs. semiquantitative assessment of intratumoral susceptibility signals in patients with different grades of glioma. J. Magn. Reson. Imaging 2020, 51, 225–233. [Google Scholar] [CrossRef]


| Characteristics | ITSS Grade | ||||
|---|---|---|---|---|---|
| Grade 0 (n = 12) | Grade 1 (n = 25) | Grade 2 (n = 16) | Grade 3 (n = 46) | ||
| Imaging features | |||||
| rCBV 1 | 1.1 (0.8–1.2) | 1.6 (1–1.9) | 2.4 (1.2–2.5) | 5.3 (4.1–6.1) | |
| Volume in mL 1 | 3.1 (1.3–3.2) | 15.9 (2–27.3) | 19.4 (4.2–20.4) | 43.6 (16.7–62.2) | |
| Tumor type | |||||
| Oligodendroglioma 2 | 1 (8.3%) | 11 (44%) | 2 (12.5%) | 4 (8.7%) | |
| Astrocytoma 2 | 11 (91.7%) | 13 (52%) | 10 (62.5%) | 4 (8.7%) | |
| Glioblastoma 2 | 0 (0%) | 1 (4%) | 4 (25%) | 38 (82.6%) | |
| Glioma grade | |||||
| Low grade | Grade 1 2 | 6 (50%) | 3 (12%) | 0 (0%) | 0 (0%) |
| Grade 2 2 | 3 (25%) | 19 (76%) | 0 (0%) | 0 (0%) | |
| High grade | Grade 3 2 | 3 (25%) | 1 (4%) | 11 (68.8%) | 6 (13%) |
| Grade 4 2 | 0 (0%) | 2 (8%) | 5 (31.2%) | 40 (87%) | |
| Morphological features | |||||
| Mitosis/10 HPF 1,3 | 0 (0–4) | 2 (0–3) | 7 (4–12) | 14 (6–20) | |
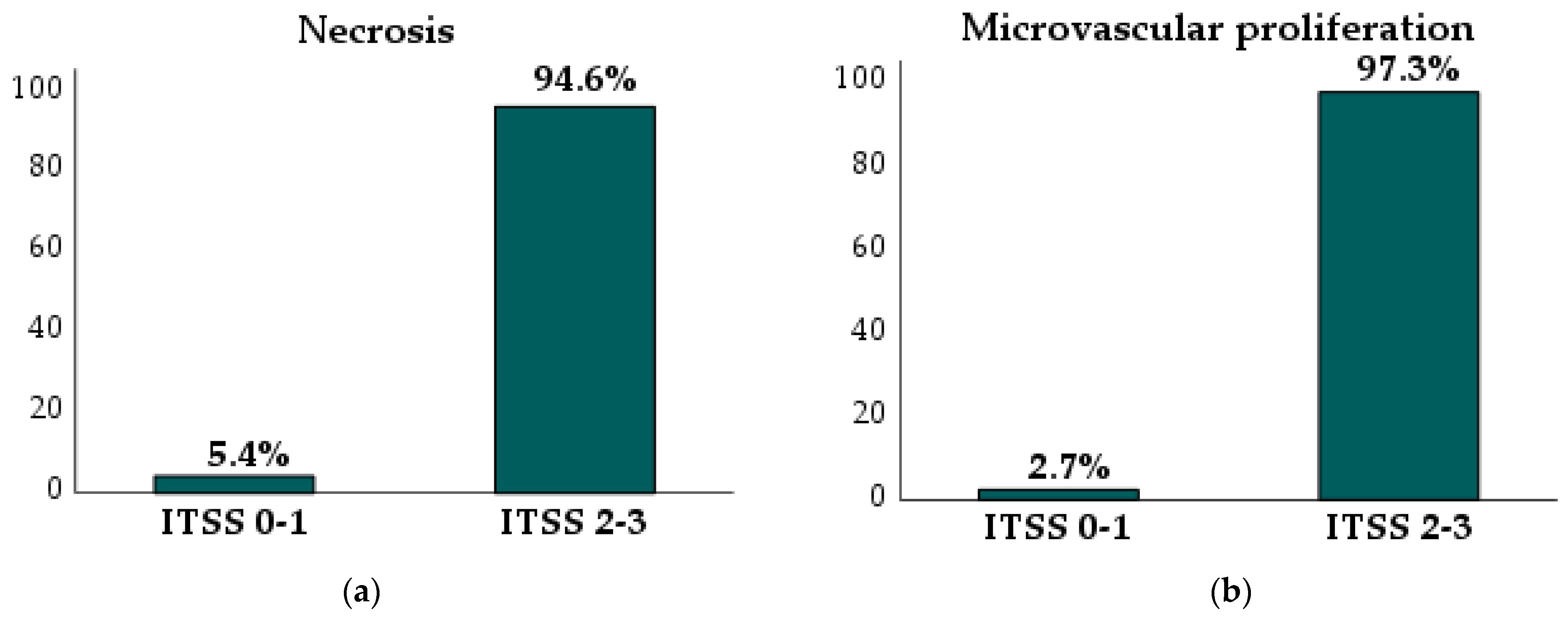
| Microvascular proliferation 2 | 0 (0%) | 1 (4%) | 3 (18.8%) | 38 (82.6%) | |
| Necrosis 2 | 1 (8.3%) | 1 (4%) | 4 (25%) | 42 (91.3%) | |
| Immunohistochemistry | |||||
| Ki-67 index 1 | 3.75 (1–5) | 7.24 (3–8) | 27.00 (10–40) | 31.17 (20–40) | |
| p53 mutation 2 | 6 (50%) | 13 (52%) | 10 (62%) | 14 (30.4%) | |
| Molecular analysis | |||||
| IDH mutation 2 | 7 (58.3%) | 17 (68%) | 8 (50%) | 6 (13.3%) | |
| 1p/19q co-deletion 2 | 1 (8.3%) | 10 (40%) | 2 (12.5%) | 4 (8.7%) | |
| CDKN2A/B deletion | 1 (11.1%) | 3 (15%) | 6 (40%) | 22 (49.9%) | |
| Homozygous deletion 2 | 1 (11.1%) | 2 (10%) | 5 (33.3%) | 21 (46.7%) | |
| Heterozygous deletion 2 | 0 (0%) | 1 (5%) | 1 (6.7%) | 1 (2.2%) | |
| Variable | Coefficient (β) | OR 1 (95%CI) | p Value |
|---|---|---|---|
| ITSS | −3.388 | 0.034 (0.007–0.158) | p < 0.001 |
| rCBV | 0.87 | 2.387 (1.365–4.172) | p = 0.002 |
| Constant | −0.242 | 0.785 | p = 0.695 |
| Variable | Coefficient (β) | OR 1 (95%CI) | p Value |
|---|---|---|---|
| ITSS | −4.605 | 0.010 (0.001–0.074) | p < 0.001 |
| Tumor volume | 0.081 | 1.085 (1.024–1.149) | p = 0.006 |
| Constant | 0.677 | 1.968 | p = 0.142 |
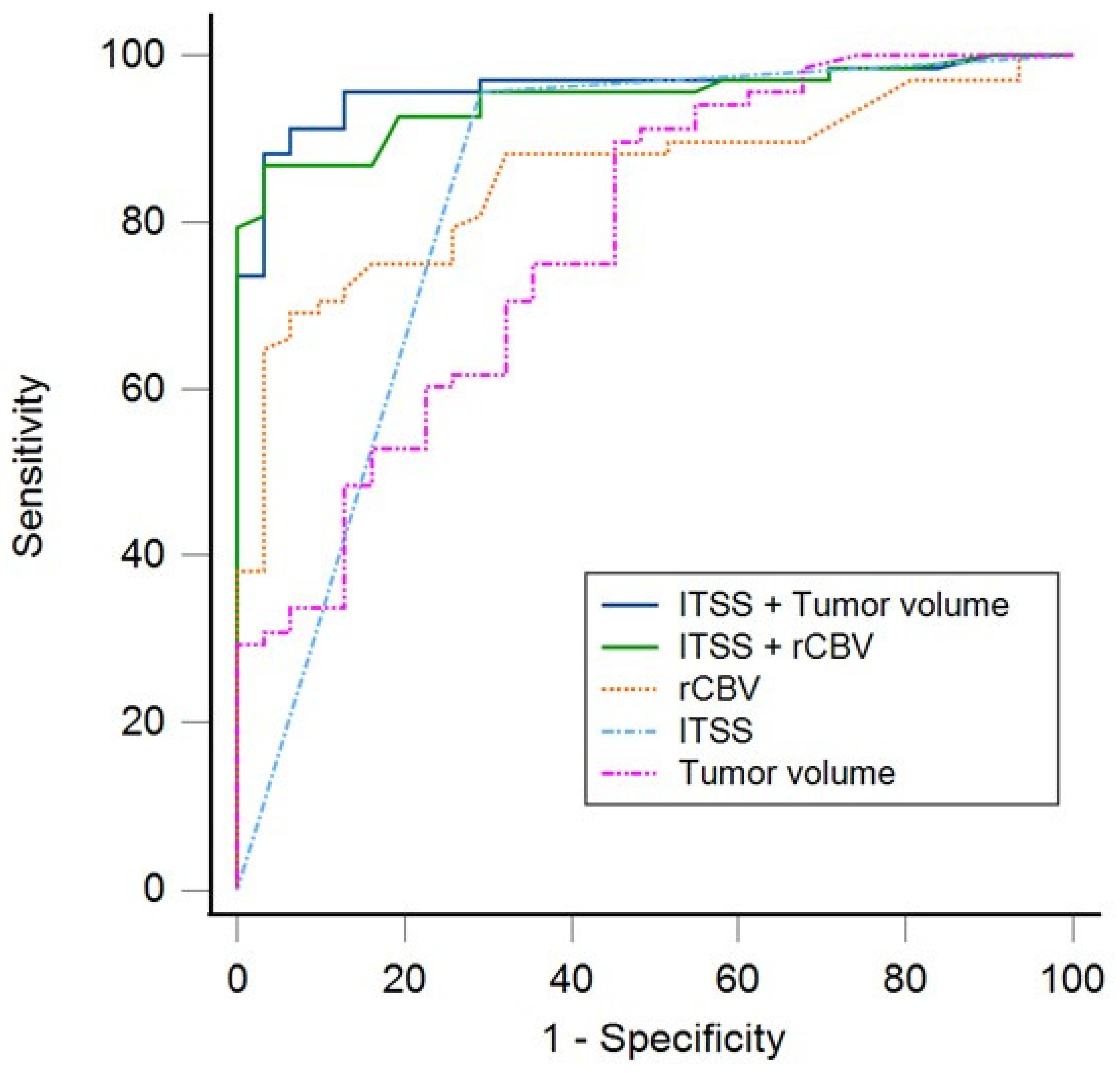
| Model | Cutoff | SE | Sp | ACC | PPV | NPV | AUC (95%CI) |
|---|---|---|---|---|---|---|---|
| ITSS + rCBV | 0.64 | 95.2% | 71.4% | 80.8% | 71.4% | 95.2% | 0.94 (0.89–0.99) |
| ITSS + volume | 0.68 | 95.6% | 74.1% | 87.8% | 71% | 95.8% | 0.96 (0.92–0.99) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tudela Martínez, J.I.; Vázquez Sáez, V.; Carbonell, G.; Rodrigo Lara, H.; Guzmán-Aroca, F.; Berna Mestre, J.d.D. Diagnostic Utility of Intratumoral Susceptibility Signals in Adult Diffuse Gliomas: Tumor Grade Prediction and Correlation with Molecular Markers Within the WHO CNS5 (2021) Classification. J. Clin. Med. 2025, 14, 4004. https://doi.org/10.3390/jcm14114004
Tudela Martínez JI, Vázquez Sáez V, Carbonell G, Rodrigo Lara H, Guzmán-Aroca F, Berna Mestre JdD. Diagnostic Utility of Intratumoral Susceptibility Signals in Adult Diffuse Gliomas: Tumor Grade Prediction and Correlation with Molecular Markers Within the WHO CNS5 (2021) Classification. Journal of Clinical Medicine. 2025; 14(11):4004. https://doi.org/10.3390/jcm14114004
Chicago/Turabian StyleTudela Martínez, José Ignacio, Victoria Vázquez Sáez, Guillermo Carbonell, Héctor Rodrigo Lara, Florentina Guzmán-Aroca, and Juan de Dios Berna Mestre. 2025. "Diagnostic Utility of Intratumoral Susceptibility Signals in Adult Diffuse Gliomas: Tumor Grade Prediction and Correlation with Molecular Markers Within the WHO CNS5 (2021) Classification" Journal of Clinical Medicine 14, no. 11: 4004. https://doi.org/10.3390/jcm14114004
APA StyleTudela Martínez, J. I., Vázquez Sáez, V., Carbonell, G., Rodrigo Lara, H., Guzmán-Aroca, F., & Berna Mestre, J. d. D. (2025). Diagnostic Utility of Intratumoral Susceptibility Signals in Adult Diffuse Gliomas: Tumor Grade Prediction and Correlation with Molecular Markers Within the WHO CNS5 (2021) Classification. Journal of Clinical Medicine, 14(11), 4004. https://doi.org/10.3390/jcm14114004






