Polymorphisms of the HTR2C Gene as Predictors of Metabolic Disturbances During Clozapine Therapy: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Inclusion Criteria
2.2. Exclusion Criteria
2.3. Search Strategy
2.4. Quality Assessment
2.5. Data Extraction and Study Grouping for Synthesis
2.6. Data Synthesis
3. Results
3.1. Weight Gain
3.2. Metabolic Syndrome
3.3. Other Metabolic Disturbances
4. Meta-Analysis
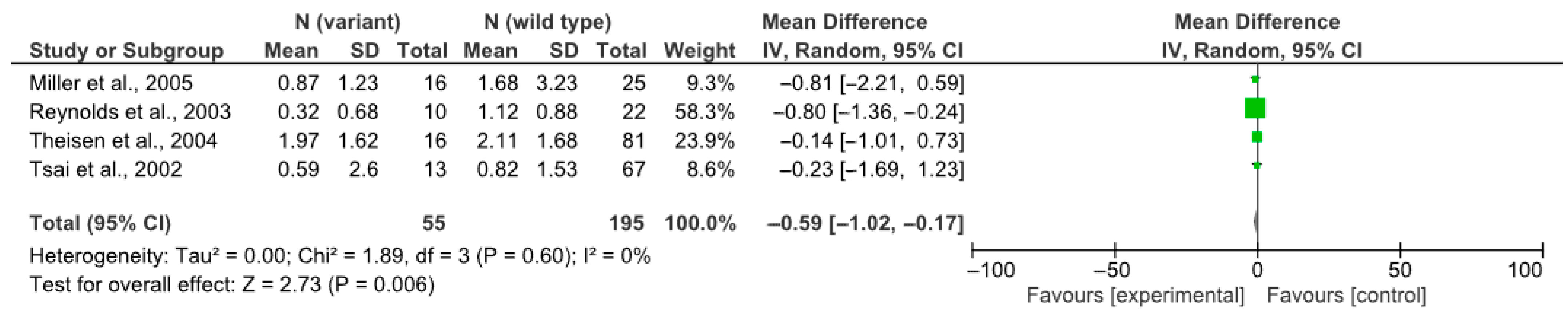
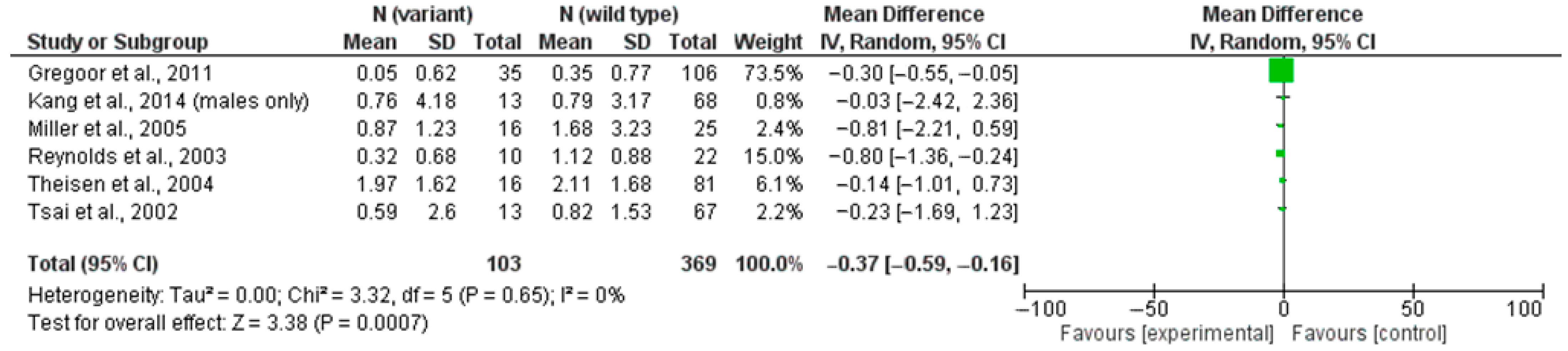
4.1. rs3813929 (–759C/T) and Weight Gain
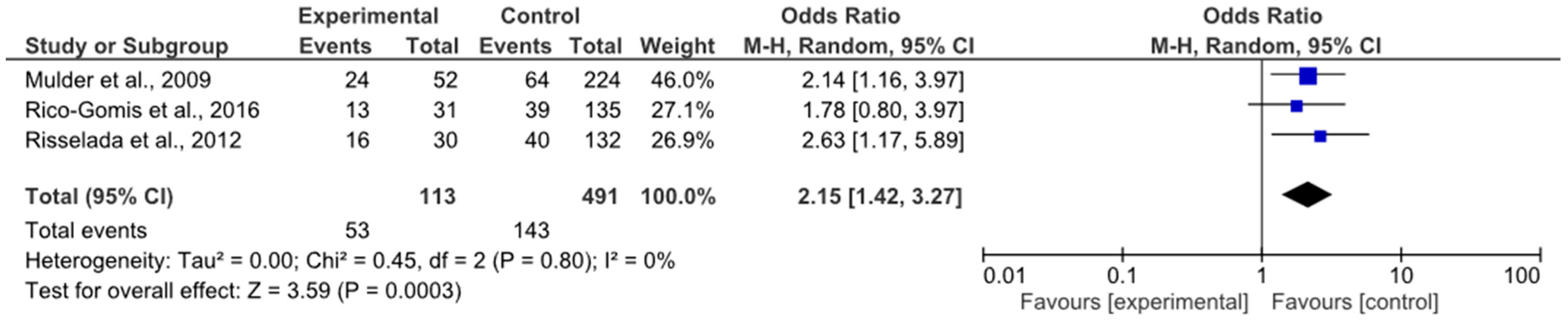
4.2. rs1414334 (C>G) and Metabolic Syndrome (MetS)
5. Discussion
5.1. Certainty of Evidence
5.2. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandey, A.; Kalita, K. Treatment-resistant schizophrenia: How far have we traveled? Front. Psychiatry 2022, 30, 994425. [Google Scholar] [CrossRef] [PubMed]
- Hurley, K.; O’brien, S.; Halleran, C.; Byrne, D.; Foley, E.; Cunningham, J.; Hoctor, F.; Sahm, L.J. Metabolic Syndrome in Adults Receiving Clozapine; The Need for Pharmacist Support. Pharmacy 2023, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation 2002, 106, 3143–3421. [Google Scholar] [CrossRef]
- Alberti, K.; Zimmet, P.; Shaw, J. Metabolic syndrome—A new world-wide definition. A Consensus Statement from the International Diabetes Federation. Diabet. Med. 2006, 23, 469–480. [Google Scholar]
- Lind, P.A.; Parker, R.K.; Northwood, K.; Siskind, D.J.; Medland, S.E. Clozapine Efficacy and Adverse Drug Reactions Among a Nationwide Study of 1021 Australians Prescribed Clozapine: The ClozaGene Study. Schizophr. Bull. 2025, 51, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Citrome, L.; McEvoy, J.P.; Saklad, S.R. Guide to the Management of Clozapine-Related Tolerability and Safety Concerns. Clin. Schizophr. Relat. Psychoses 2016, 10, 163–177. [Google Scholar] [CrossRef]
- Nemani, K.L.; Greene, M.C.; Ulloa, M.; Vincenzi, B.; Copeland, P.M.; Al-Khadari, S.; Henderson, D.C. Cardiovascular Risk and Mortality: Results of a 21-Year Naturalistic Study in Patients with Schizophrenia and Schizoaffective Disorder. Clin. Schizophr. Relat. Psychoses 2019, 12, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, W.; Li, H.; Ye, X.; Qin, J.-J. Changes of metabolic syndrome status alter the risks of cardiovascular diseases. Sci. Rep. 2025, 15, 5448. [Google Scholar] [CrossRef]
- Guembe, M.J.; Fernandez-Lazaro, C.I.; Sayon-Orea, C.; Toledo, E.; Moreno-Iribas, C.; for the RIVANA Study Investigators ; Cosials, J.B.; Reyero, J.B.; Martínez, J.D.; Diego, P.G.; et al. Risk for cardiovascular disease associated with metabolic syndrome and its components: A 13-year prospective study in the RIVANA cohort. Cardiovasc. Diabetol. 2020, 19, 195. [Google Scholar] [CrossRef]
- Peritogiannis, V.; Ninou, A.; Samakouri, M. Mortality in Schizophrenia-Spectrum Disorders: Recent Advances in Understanding and Management. Healthcare 2022, 10, 2366. [Google Scholar] [CrossRef]
- Sosin, D.; Khasanova, A.; Moshevitin, S.; Mosolov, S. Pharmacogenetic predictors of clozapine metabolic disturbances. Curr. Ther. Ment. Disord. 2024, 4, 30–40. [Google Scholar]
- Akshatha, C.; Kumar, C.; Mangalwedhe, S. Insulin resistance in first episode antipsychotic naïve psychosis patients—A cross sectional study. Indian. J. Psychiatry 2022, 64, S545. [Google Scholar]
- Lee, J.; Xue, X.; Au, E.; McIntyre, W.B.; Asgariroozbehani, R.; Tseng, G.C.; Papoulias, M.; Panganiban, K.; Agarwal, S.M.; Mccullumsmith, R.; et al. Central insulin dysregulation in antipsychotic-naïve first-episode psychosis: In silico exploration of gene expression signatures. Psychiatry Res. 2024, 331, 115636. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; He, J.; Cui, Z.; Wang, R.; Bao, K.; Huang, Y.; Wang, R.; Liu, T. Central 5-HTR2C in the Control of Metabolic Homeostasis. Front. Endocrinol. 2021, 12, 694204. [Google Scholar] [CrossRef]
- Khasanova, A.K. Pharmacogenetic factors of clozapine-induced metabolic syndrome. Pers. Psychiatry Neurol. 2023, 3, 38–47. [Google Scholar] [CrossRef]
- Little, J.; Higgins, J.; Ioannidis, J.; Moher, D.; Gagnon, F.; von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. Strengthening the Reporting of Genetic Association Studies (STREGA)—An extension of the STROBE statement. Genet. Epidemiol. 2009, 33, 581–598. [Google Scholar] [CrossRef]
- De Luca, V.; Müller, D.J.; Hwang, R.; Lieberman, J.A.; Volavka, J.; Meltzer, H.Y.; Kennedy, J.L. HTR2C haplotypes and antipsychotics-induced weight gain: X-linked multimarker analysis. Hum. Psychopharmacol. Clin. Exp. 2007, 22, 463–467. [Google Scholar] [CrossRef]
- Moola, S.; Munn, Z.; Sears, K.; Sfetcu, R.; Currie, M.; Lisy, K.; Tufanaru, C.; Qureshi, R.; Mattis, P.; Mu, P. Conducting systematic reviews of association (etiology): The Joanna Briggs Institute’s approach. Int. J. Evid. Based Heal. 2015, 13, 163–169. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thomas, J. Cochrane Handbook for Systematic Reviews of Interventions Version 6.2, 2021. Cochrane. 2021. Available online: https://training.cochrane.org/handbook/current (accessed on 20 March 2021).
- Grade Working Group. Available online: https://www.gradeworkinggroup.org/ (accessed on 20 March 2025).
- Rietschel, M.; Naber, D.; Fimmers, R.; Möller, H.-J.; Propping, P.; Nöthen, M.M. Efficacy and side-effects of clozapine not associated with variation in the 5-HT2C receptor. NeuroReport 1997, 8, 1999–2003. [Google Scholar] [CrossRef]
- Hong, C.-J.; Lin, C.-H.; Yu, Y.W.-Y.; Yang, K.-H.; Tsai, S.-J. Genetic Variants of the Serotonin System and Weight Change during Clozapine Treatment. Pharmacogenetics 2001, 11, 265–268. [Google Scholar] [CrossRef]
- Basile, V.S.; Masellis, M.; De Luca, V.; Meltzer, H.Y.; Kennedy, J.L. 759C/T genetic variation of 5HT(2C) receptor and clozapine-induced weight gain. Lancet 2002, 360, 1790-1. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, G.P.; Zhang, Z.-J.; Zhang, X.-B. Association of antipsychotic drug-induced weight gain with a 5-HT2C receptor gene polymorphism. Lancet 2002, 359, 2086–2087. [Google Scholar] [CrossRef]
- Tsai, S.-J.; Hong, C.-J.; Yu, Y.W.-Y.; Lin, C.-H. −759C/T genetic variation of 5HT(2C) receptor and clozapine-induced weight gain. Lancet 2002, 360, 1790. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, G.P.; Zhang, Z.; Zhang, X. Polymorphism of the Promoter Region of the Serotonin 5-HT 2C Receptor Gene and Clozapine-Induced Weight Gain. Am. J. Psychiatry 2003, 160, 677–679. [Google Scholar] [CrossRef]
- Theisen, F.M.; Hinney, A.; Brömel, T.; Heinzel-Gutenbrunner, M.; Martin, M.; Krieg, J.-C.; Remschmidt, H.; Hebebrand, J. Lack of association between the –759C/T polymorphism of the 5-HT2C receptor gene and clozapine-induced weight gain among German schizophrenic individuals. Psychiatr. Genet. 2004, 14, 139–142. [Google Scholar] [CrossRef]
- Miller, D.D.; Ellingrod, V.L.; Holman, T.L.; Buckley, P.F.; Arndt, S. Clozapine-induced weight gain associated with the 5HT2C receptor –759C/T polymorphism. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2005, 133B, 97–100. [Google Scholar] [CrossRef]
- Mulder, H.; Franke, B.; Beek van der, A.A.v.d.-P.; Arends, J.; Wilmink, F.W.; Egberts, A.C.G.; Scheffer, H. The association between HTR2C polymorphisms and obesity in psychiatric patients using antipsychotics: A cross-sectional study. Pharmacogenom. J. 2007, 7, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Popp, J.; Leucht, S.; Heres, S.; Steimer, W. DRD4 48bp VNTR but not 5-HT2C Cys23Ser receptor polymorphism is related to anti-psychotic-induced weight gain. Pharmacogenomics J. 2009, 9, 71–77. [Google Scholar] [CrossRef][Green Version]
- Gunes, A.; Melkersson, K.I.; Scordo, M.G.; Dahl, M.-L. Association between HTR2C and HTR2A polymorphisms and metabolic abnormalities in patients treated with olanzapine or clozapine. J. Clin. Psychopharmacol. 2009, 29, 65–68. [Google Scholar] [CrossRef]
- Opgen-Rhein, C.; Brandl, E.J.; Müller, D.J.; Neuhaus, A.H.; Tiwari, A.K.; Sander, T.; Dettling, M. genes with antipsychotic-induced weight gain in a German sample. Pharmacogenomics 2010, 11, 773–780. [Google Scholar] [CrossRef]
- Gregoor, J.G.; Mulder, H.; Cohen, D.; van Megen, H.J.; Egberts, T.C.; Heerdink, E.R.; van der Weide, J. Combined HTR2C-LEP genotype as a determinant of obesity in patients using anti-psychotic medication. J. Clin. Psychopharmacol. 2010, 30, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Gregoor, J.; Weide, J.; Loovers, H.; Van Megen, H.J.; Egberts, T.C.; Heerdink, E.R. Polymorphisms of the LEP, LEPR and HTR2C gene: Obesity and BMI change in patients using antipsychotic medication in a naturalistic setting. Pharmacogenomics 2011, 12, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Lee, J.-I.; Han, H.R.; Soh, M.; Hong, J.P. Polymorphisms of the leptin and HTR2C genes and clozapine-induced weight change and baseline BMI in patients with chronic schizophrenia. Psychiatr. Genet. 2014, 24, 249–256. [Google Scholar] [CrossRef]
- Klemettilä, J.-P.; Kampman, O.; Seppälä, N.; Viikki, M.; Hämäläinen, M.; Moilanen, E.; Mononen, N.; Lehtimäki, T.; Leinonen, E. leptin and adiponectin genes and serum marker analyses in clozapine treated long-term patients with schizophrenia. Eur. Psychiatry 2015, 30, 296–302. [Google Scholar] [CrossRef]
- Vasudev, K.; Choi, Y.-H.; Norman, R.; Kim, R.B.; Schwarz, U.I. Genetic Determinants of Clozapine-Induced Metabolic Side Effects. Can. J. Psychiatry 2017, 62, 138–149. [Google Scholar] [CrossRef]
- Puangpetch, A.; Srisawasdi, P.; Unaharassamee, W.; Jiratjintana, N.; Vanavanan, S.; Punprasit, S.; Nakorn, C.N.; Sukasem, C.; Kroll, M.H. Association between polymorphisms of LEP, LEPR, DRD2, HTR2A and HTR2C genes and risperidone-or clozapine-induced hyperglycemia. Pharmgenom. Pers. Med. 2019, 12, 155–166. [Google Scholar]
- Mulder, H.; Franke, B.; Beek van der, A.A.v.d.-P.; Arends, J.; Wilmink, F.W.; Scheffer, H.; Egberts, A.C. The association between HTR2C gene polymorphisms and the metabolic syndrome in patients with schizophrenia. J. Clin. Psychopharmacol. 2007, 27, 338–343. [Google Scholar] [CrossRef]
- Yevtushenko, O.O.; Cooper, S.J.; O’Neill, R.; Doherty, J.K.; Woodside, J.V.; Reynolds, G.P. smoking and drug treatment on metabolic disturbances in patients with schizophrenia. Br. J. Psychiatry 2008, 192, 424–428. [Google Scholar] [CrossRef]
- Mulder, H.; Cohen, D.; Scheffer, H.; Gispen-de Wied, C.; Arends, J.; Wilmink, F.W.; Franke, B.; Egberts, A.C.G. HTR2C gene polymorphisms and the metabolic syndrome in patients with schiz-ophrenia: A replication study. J. Clin. Psychopharmacol. 2009, 29, 16–20. [Google Scholar] [CrossRef]
- Risselada, A.J.; Vehof, J.; Bruggeman, R.; Wilffert, B.; Cohen, D.; Al Hadithy, A.F.; Arends, J.; Mulder, H. Association between HTR2C gene polymorphisms and the metabolic syndrome in patients using antipsychotics: A replication study. Pharmacogenom. J. 2012, 12, 62–67. [Google Scholar] [CrossRef]
- Bai, Y.M.; Chen, T.-T.; Liou, Y.-J.; Hong, C.-J.; Tsai, S.-J. Association between HTR2C polymorphisms and metabolic syndrome in patients with schizophrenia treated with atypical antipsychotics. Schizophr. Res. 2011, 125, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Lee, J.I.; Chang, A.K.; Joo, Y.H.; Kim, C.Y.; Kim, S.Y. Genetic polymorphisms in the HTR2C and peroxisome proliferator-activated receptors are not associated with metabolic syndrome in patients with schizophrenia taking clozapine. Psychiatry Investig. 2011, 8, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Rico-Gomis, J.M.; Palazón-Bru, A.; Triano-García, I.; Mahecha-García, L.F.; García-Monsalve, A.; Navarro-Ruiz, A.; Villagordo-Peñalver, B.; Jiménez-Abril, J.; Martínez-Hortelano, A.; Gil-Guillén, V.F. Association between the HTR2C rs1414334 C/G gene polymorphism and the development of the metabolic syndrome in patients treated with atypical antipsychotics. PeerJ 2016, 4, e2163. [Google Scholar] [CrossRef]
- Puangpetch, A.; Unaharassamee, W.; Jiratjintana, N.; Koomdee, N.; Sukasem, C. Genetic polymorphisms of HTR2C, LEP and LEPR on metabolic syndromes in patients treated with atypical antipsychotic drugs. J. Pharm. Pharmacol. 2018, 70, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Reporting Checklist for Genetic Association Study. Based on the STREGA Guidelines. Available online: https://www.equator-network.org/reporting-guidelines/strobe-strega/ (accessed on 20 March 2025).

| Authors | Study Design | Sample | Population | Therapy | Polymor- Phisms | Results |
|---|---|---|---|---|---|---|
| Rietschel et al., 1997 [21] | Cross-sectional (AP ≥ 28 days) | 152 SZ, SZA, PD | Europeans | MONO CLOZ | rs6318 (Cys23Ser) | No association |
| Hong et al., 2001 [22] | Prospective (4 months) | 93 TR SZ | Europeans | MONO CLOZ | rs6318 (Cys23Ser) | No association |
| Basile et al., 2002 [23] | Prospective (6 weeks) | 80 SZ | Europeans and African Americans | MONO CLOZ | rs3813929 (–759C/T) | Hemizygous T allele men gainedmore weight thanhemizygous C allele men |
| Reynolds et al., 2002 [24] | Prospective (10 weeks) | 123 FE SZ | Chinese | CLOZ-4, AP | rs3813929 (–759C/T) | T-allele carriers gained less weight |
| Tsai et al., 2002 [25] | Prospective (4 months) | 80 SZ, SZA | Chinese | MONO CLOZ | rs3813929 (–759C/T) | No association |
| Reynolds et al., 2003 [26] | Prospective (6 weeks) | 32 SZ | Chinese | MONO CLOZ | rs3813929 (–759C/T) | Men with T allele gained less weight |
| Theisen et al., 2004 [27] | Prospective (12 weeks) | 97 SZ | Europeans | MONO CLOZ | rs3813929 (–759C/T) | No association |
| Miller et al., 2005 [28] | Prospective (6 months) | 41 TR SZ | Caucasian, African American, Hispanic | MONO CLOZ | rs3813929 (–759C/T) | T-allele carriers gained less weight |
| Mulder et al., 2007 [29] | Cross-sectional (AP ≥ 3 months) | 127 SZ, SZA, PD | Europeans | CLOZ-44, AP PHC | rs3813929 (–759C/T), rs3813928 (–997G/A), rs518147 (–697G/C), rs1414334 (C>G), HTR2C c.1–142948(GT)n 13R | Carriers of the combined genotype rs1414334 C, rs518147 C, (GT)n 13, and rs3813929 C, and carriers of s1414334 C are related to an increased risk of obesity |
| De Luca et al., 2007 [17] | Prospective (6–14 weeks) | 139 SZ | various ethnicities, including African Americans | CLOZ-92, AP | rs3813929 (–759C/T), rs6318 (Ser23Cys), (GT)12–18/(CT)4–5 | The Long-C-Ser haplotype confers a protective effect against weight gain |
| Popp et al., 2009 [30] | Prospective (4 weeks) | 102 SZ, SZA, PD | Europeans | CLOZ-16, AP, AD, SD, NT, and PHC | rs6318 (Cys23Ser) | No association |
| Gunes et al., 2009 [31] | Cross-sectional (AP ≥ 6 months months) | 46 SZ, SZA | white origin | CLOZ-18, OL | rs3813929 (–759C/T), rs518147 (–697G/C), rs6318 (Cys23Ser) | Haplotype (759C, 697G, 23Cys) is associated with an increased risk of obesity |
| Opgen-Rhein et al., 2010 [32] | Retrospective case–control (AP ≥ 6 weeks) | 128 SZ | Europeans, Turks | CLOZ-24, AP, AD, NT, and PHC | rs498207 (–1165A/G), rs3813928 (–997G/A), rs3813929 (–759C/T), rs6318 (Cys23Ser) | Weight gain was higher in hemizygous males carrying the rs498207 A allele and in females with the rs498207 AA genotype, as well as in carriers of rs3813928 and rs3813929. The AGC haplotype (rs498207 A, rs3813928 G, rs3813929 C) is associated with increased weight gain |
| Gregoor et al., 2010 [33] | Cross-sectional (AP ≥ 3 months) | 200 SZ, SZA, PD | Europeans | CLOZ-67, AP, PHC | rs3813929 (–759C/T), rs1414334 (c.551-3008 C>G) | The combination of the absence of the –759T allele and the presence of the LEP –2548G allele is associated with an increased risk of obesity |
| Gregoor et al., 2011 [34] | Prospective (per year) | 141 PSD, AFD, PD | Europeans | CLOZ-68, AP, PHC | rs3813929 (–759C/T) | The HTR2C –759T allele exhibited a trend toward protection against weight gain |
| Kang et al., 2014 [35] | Retrospective (AP ≥ 1 year) | 113 SZ | Koreans, Chinese | CLOZ MONO-68, AP, NT, PHC | rs3813928 (–759C/T) | The HTR2C –759T allele exhibited a trend toward protection against weight gain in men |
| Klemettilä et al., 2015 [36] | Cross-sectional (AP ≥ 1 year) | 190 SZ | Europeans | CLOZ MONO-121, CLOZ + AP-69 | rs1414334 (c.551-3008 C>G) | No association |
| Vasudev et al., 2017 [37] | Cross-sectional (AP ≥ 6 months) | 60 SZ, SZA, DD | overwhelming majority Europeans, First Nations | CLOZ-60, CLOZ MONO-18, CLOZ + AP/AD/NT | rs3813929 (–759C/T), rs3813928 (–697G/A), rs1414334 (c.551-3008 C>G) | The HTR2C rs1414334 G allele is associated with weight loss in men |
| Puangpetch et al., 2019 [38] | Cross-sectional (AP ≥ 1 year) | 180 SZ | Thais | CLOZ-50, AP | rs518147 (–697 G/C), rs12836771 (A/G)—tag-SNP for rs3813929 (–759C/T) | No association |
| Authors | Study Design | Sample | Population | Therapy | Subject of Study | Polymorphisms | Results |
|---|---|---|---|---|---|---|---|
| Mulder et al., 2007 [39] | Cross-sectional (AP ≥ 3 months) | 112 SZ, SZA, PD | White | CLOZ-41, AP, and PHC | MetS (NCEP: ATP III) | rs3813929 (–759C/T), rs3813928 (–997G/A), rs518147 (–697G/C), rs1414334 (C>G), HTR2C c.1–142948(GT)n 13R | rs1414334 C, rs518147 C, (GT)n 13R are associated with an increased risk of MetS; strong positive association of the combined haplotype (especially in men) (13 repeat + rs518147 C + rs1414334 C) with MetS |
| Yevtushenko et al., 2008 [40] | Cross-sectional (AP long) | 134 SZ, SZA | Europeans | CLOZ MONO-21, AP | MetS (IDF) | rs3813929 (–759C/T) | The combination of the HTR2C C allele and the LEP G allele is positively associated with MetS |
| Mulder et al., 2009 [41] | Cross-sectional replication (AP ≥ 3 months), pooled analysis | 164 in replication sample, 276 in pooled analysis | Asian, African, Mediterranean, Hindustan origins | CLOZ-37 (replication sample), AP, PHC | MetS (NCEP: ATP III) | rs3813929 (–759C/T), rs518147 (–697G/C), rs1414334 (c.551-3008 C>G), HTR2C c.1–142948(GT)n 13R | The rs1414334 C allele showed a trend toward positive association with MetS in the replication cohort. In the combined analysis, both rs1414334 C and the HTR2C c.1–142948(GT)n 13R variant are associated with an increased risk of MetS |
| Risselada et al., 2012 [42] | Cross-sectional replication (AP long) | 186 SZ, SZA, PD | Overwhelmingly Europeans | CLOZ-31, AP, PHC | MetS (NCEP: ATP III) | rs3813929 (–759C/T), rs1414334 (c.551-3008 C>G) | The rs1414334 C allele is associated with an increased risk of MetS |
| Bai et al., 2011 [43] | Case–control study (AP ≥ 3 months) | 456 SZ | Chinese | CLOZ MONO-171, AP | MetS (IDF Asia) | rs521018, rs498177, rs2192371, rs5988072, rs12833104, rs6318 (Cys23Ser) | The rs498177 CC genotype is associated with an increased risk of MetS in women, and the rs521018–rs498177 haplotype is more frequent among individuals with MetS |
| Kang et al., 2011 [44] | Cross-sectional (AP ≥ 1 year) | 146 SZ | Korean | CLOZ-146, CLOZ + AP, CLOZ + NT | MetS (NCEP ATP IIIA Asia) | rs3813928 (–759C/T), rs518147 (–697G/C) | No association |
| Vasudev et al., 2017 [37] | Cross-sectional (AP ≥ 6 months) | 60 SZ, SZA, DD | Overwhelmingly Europeans, First Nations | CLOZ-60, CLOZ MONO-18, CLOZ + AP/AD/NT | MetS (NCEP: ATP III) | rs3813929 (–759C/T), rs3813928 (–697G/A), rs1414334 (c.551-3008 C>G) | No association |
| Rico-Gomis et al., 2016 [45] | Cross-sectional (AP ≥ 3 months) | 166 SZ, SZA, SZF, PD, BD | Europeans | CLOZ, AP | MetS (IDF) | rs1414334 (c.551-3008 C>G) | No association |
| Puangpetch et al., 2018 [46] | Cross-sectional (AP long) | 113 SZ | Thais | CLOZ-47, AP, PHC | MetS (IDF) | rs518147 (–697G/C), rs12836771 (A/G)—tag-SNP for rs3813929 (–759C/T) | The rs518147 CC and rs12836771 GG genotypes are associated with an increased risk of MetS |
| Authors | Study Design | Sample | Population | Therapy | Subject of Study | Polymorphisms | Results |
|---|---|---|---|---|---|---|---|
| Mulder et al., 2007 [39] | Cross-sectional (AP ≥ 3 months) | 112 SZ, SZA, PD | White | CLOZ—41, AP, and PHC | WC, TG, HDL, BP | rs3813929 (–759C/T), rs3813928 (–997G/A), rs518147 (–697G/C), rs1414334 (C>G), HTR2C:c.1–142948(GT)n 13R | The HTR2C:c.1—142948(GT)n 13R, rs518147 (–697) C, rs1414334 C alleles are associated with higher WC |
| Yevtushenko et al., 2008 [40] | Cross-sectional (AP ≥ 3 months) | 134 SZ, SZA | Europeans | CLOZ MONO—21, AP | WC, TG, HDL, BP, GC | rs3813929 (–759C/T) | The combination of the HTR2C (–759C/T) CC genotype and the LEP (72548A/G) G allele is positively associated with higher WC |
| Mulder et al., 2009 [41] | Cross-sectional (AP long) | 164 in replication sample, 276 in pooled analysis | Asian, African, Mediterranean, and Hindustan origins | CLOZ—37 (replication sample), AP, PHC | WC, TG, HDL, BP | rs3813929 (–759C/T), rs518147 (–697G/C), rs1414334 (c.551-3008 C>G), HTR2C:c.1–142948(GT)n 13R | rs1414334 C allele has a positive association with hypertriglyceridemia and higher WC |
| Gunes et al., 2009 [31] | Cross-sectional replication (AP ≥ 3 months), pooled analysis | 46 SZ, SZA | white origin | CLOZ—18, OL | WG, insulin, C-peptide, TG, cholesterol, HOMA-IR | rs3813929 (–759C/T), rs518147 (–697G/C), rs6318 (Cys23Ser) | No association |
| Risselada et al., 2010 [42] | Cross-sectional (AP ≥ 6 months) | 186 SZ, SZA, PD | Overwhelmingly Europeans | CLOZ-31, AP, PHC | WC, TG, HDL, BP, GC | rs3813929 (–759C/T), rs1414334 (c.551-3008 C>G) | The rs1414334 C allele is positively associated with increased TG. There was a trend toward lower HDL cholesterol associated with the rs1414334 allele |
| Bai et al., 2011 [43] | Cross-sectional replication (AP long) | 456 SZ | Chinese | CLOZ MONO-171, AP | TG, HDL, BP, GC | rs521018, rs498177, rs2192371, rs5988072, rs12833104, rs6318 (Cys23Ser) | No association |
| Kang et al., 2011 [44] | Case–control study (AP ≥ 3 months) | 146 SZ | Korean | CLOZ-146, CLOZ + AP, CLOZ + NT | WC, TG, HDL, BP, GC | rs3813928 (–759C/T), rs518147 (–697G/C) | No association |
| Klemettilä et al., 2015 [36] | Cross-sectional (AP ≥ 1 year) | 190 SZ | Europeans | CLOZ MONO-121, CLOZ + AP-69 | WG, leptin, adiponectin, adipsin, IL-6, IL-1Ra, HOMA-IR, TG, HDL | rs1414334 (c.551-3008 C>G) | The GG genotype is positively associated with higher levels of TG and lower levels of HDL |
| Vasudev et al., 2017 [37] | Cross-sectional (AP ≥ 1 year) | 60 SZ, SZA, DD | Overwhelmingly Europeans, First Nations | CLOZ-60, CLOZ MONO-18, CLOZ + AP/AD/NT | WG, WC, TG, HDL, BP, GC | rs3813929 (–759C/T), rs3813928 (–697G/A), rs1414334 (c.551-3008 C>G) | No association |
| Rico-Gomis et al., 2016 [45] | Cross-sectional (AP ≥ 6 months) | 166 SZ, SZA, SZF, PD, BD | Europeans | CLOZ, AP | WC, TG, HDL, BP, GC | rs1414334 (c.551-3008 C>G) | No association |
| Puangpetch et al., 2018 [46] | Cross-sectional (AP ≥ 3 months) | 113 SZ | Thais | CLOZ-47, AP, PHC | WC, TG, HDL, BP, GC | rs518147 (–697G/C), rs12836771 (A/G)—tag-SNP for rs3813929 (–759C/T) | No association |
| Puangpetch et al., 2019 [38] | Cross-sectional (AP long) | 180 SZ | Thais | CLOZ-50, AP | GC, BMI, WC, HOMA-IR, leptin, adiponectin, prolactin, lipid profile | rs518147 (–697 G/C), rs12836771 (A/G)—tag-SNP for rs3813929 (–759C/T) | No association |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khasanova, A.K.; Sosin, D.N.; Mosolov, S.N.; Mirzaev, K.B.; Sychev, D.A. Polymorphisms of the HTR2C Gene as Predictors of Metabolic Disturbances During Clozapine Therapy: A Systematic Review and Meta-Analysis. J. Clin. Med. 2025, 14, 3861. https://doi.org/10.3390/jcm14113861
Khasanova AK, Sosin DN, Mosolov SN, Mirzaev KB, Sychev DA. Polymorphisms of the HTR2C Gene as Predictors of Metabolic Disturbances During Clozapine Therapy: A Systematic Review and Meta-Analysis. Journal of Clinical Medicine. 2025; 14(11):3861. https://doi.org/10.3390/jcm14113861
Chicago/Turabian StyleKhasanova, Aiperi K., Dmitriy N. Sosin, Sergey N. Mosolov, Karin B. Mirzaev, and Dmitriy A. Sychev. 2025. "Polymorphisms of the HTR2C Gene as Predictors of Metabolic Disturbances During Clozapine Therapy: A Systematic Review and Meta-Analysis" Journal of Clinical Medicine 14, no. 11: 3861. https://doi.org/10.3390/jcm14113861
APA StyleKhasanova, A. K., Sosin, D. N., Mosolov, S. N., Mirzaev, K. B., & Sychev, D. A. (2025). Polymorphisms of the HTR2C Gene as Predictors of Metabolic Disturbances During Clozapine Therapy: A Systematic Review and Meta-Analysis. Journal of Clinical Medicine, 14(11), 3861. https://doi.org/10.3390/jcm14113861