Many Models, Little Adoption—What Accounts for Low Uptake of Machine Learning Models for Atrial Fibrillation Prediction and Detection?
Abstract
1. Introduction
1.1. Background
1.2. Scope and Key Questions
- (KQ1) In adult patients without a known history of stroke or AF or cardiovascular comorbidities, what are the performance statistics, data features and processing steps, and limitations of ML models in predicting incidence of AF?
- (KQ2) In adult patients with a previous history of stroke, what are the performance statistics, data features and processing steps, and limitations of ML models for AF detection?
- A PICOTS (population, interventions, comparators, outcomes, timing, and setting) table with details on the key questions is shown below (Table 1).
2. Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Data Collection
3. Results
3.1. AI for Primary Stroke Prevention: Prediction of Atrial Fibrillation in the General Population
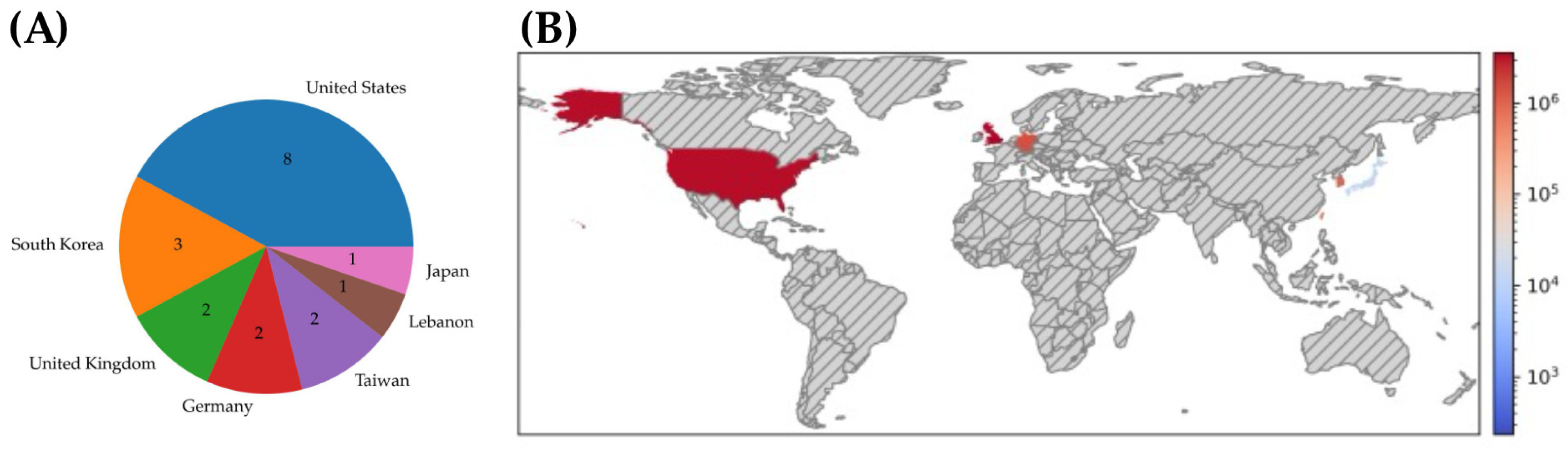
3.1.1. Search Results and Study Characteristics
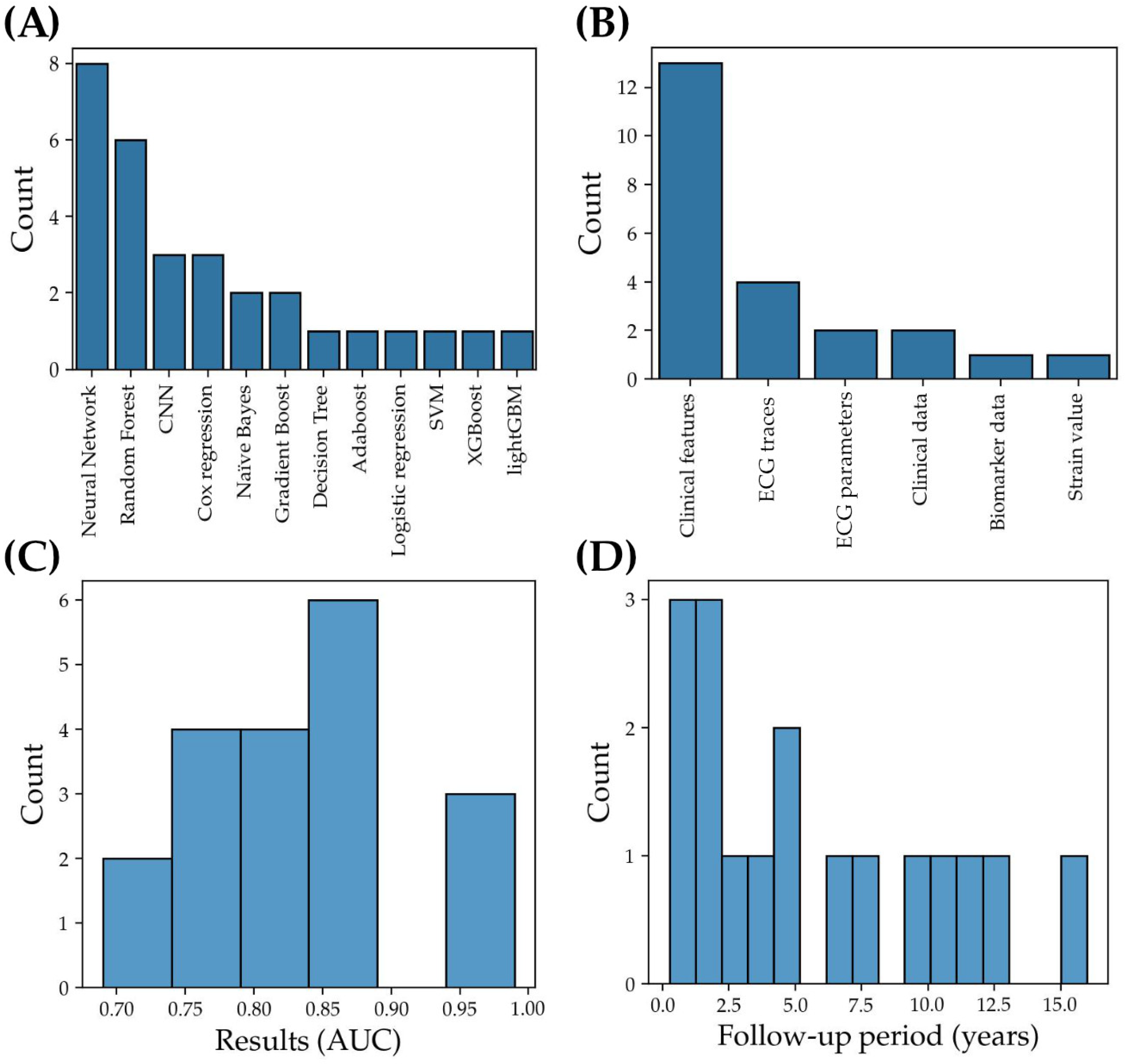
3.1.2. Machine Learning Models: Characteristics and Performance Metrics
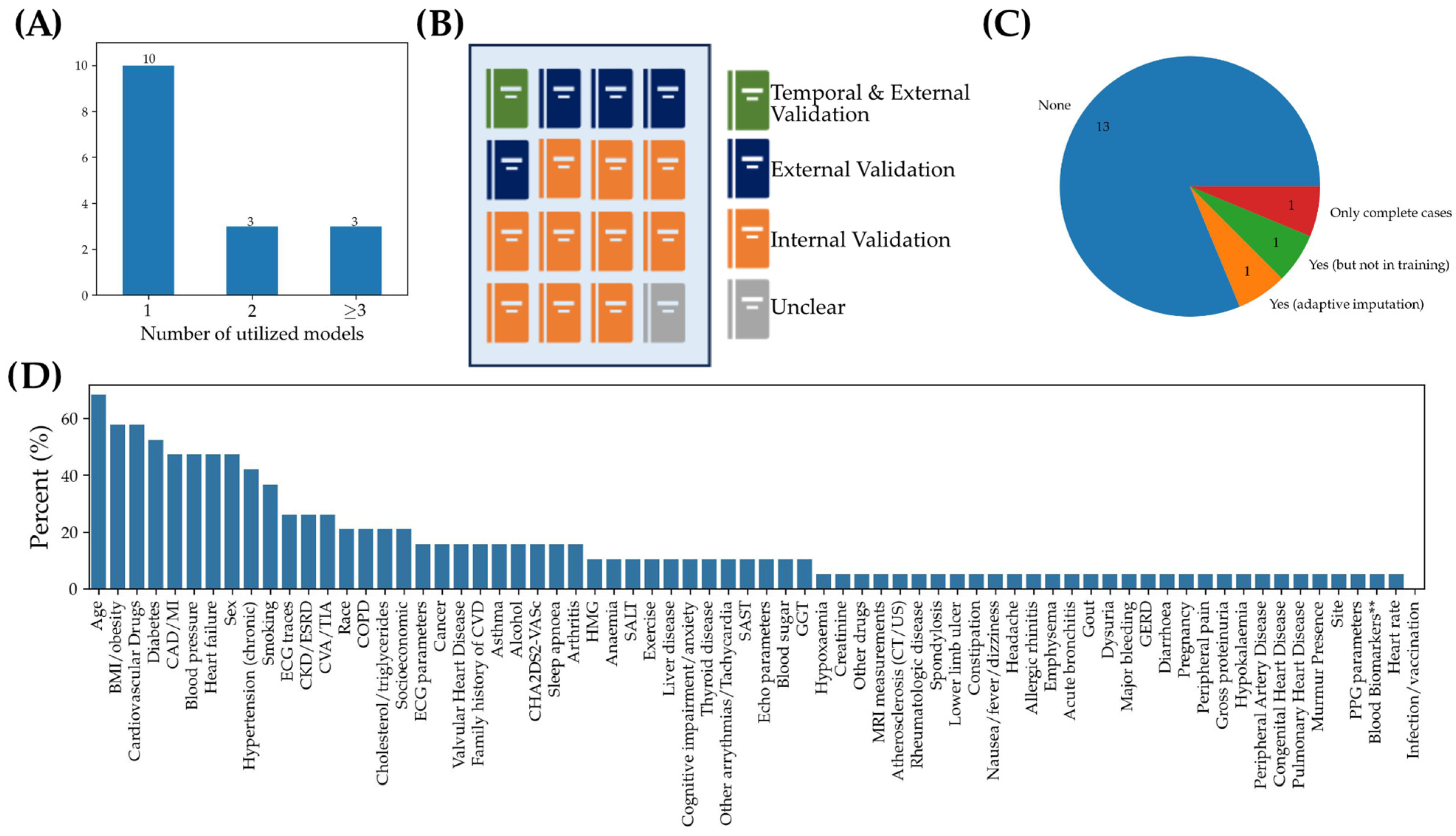
3.1.3. Analysis of Limiting Factors and Best Practices: From Data Pre-Processing to Model Validation
3.2. AI for Secondary Stroke Prevention: Detection of Atrial Fibrillation in Stroke Cohorts
3.2.1. Search Results and Study Characteristics
3.2.2. Machine Learning Models: Characteristics and Performance Metrics
3.2.3. Analysis of Limiting Factors and Best Practices: From Data Pre-Processing to Model Validation
4. Discussion
4.1. Recommendations for Clinical Implementation of ML Models
4.2. Further Considerations for ML Models in Clinical Practice
4.3. Limitations of the Study
4.4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolf, P.A.; Abbott, R.D.; Kannel, W.B. Atrial fibrillation as an independent risk factor for stroke: The Framingham Study. Stroke 1991, 22, 983–988. [Google Scholar] [CrossRef]
- Penado, S.; Cano, M.; Acha, O.; Hernández, J.L.; Riancho, J.A. Atrial fibrillation as a risk factor for stroke recurrence. Am. J. Med. 2003, 114, 206–210. [Google Scholar] [CrossRef]
- Seiffge, D.J.; Werring, D.J.; Paciaroni, M.; Dawson, J.; Warach, S.; Milling, T.J.; Engelter, S.T.; Fischer, U.; Norrving, B. Timing of anticoagulation after recent ischaemic stroke in patients with atrial fibrillation. Lancet Neurol. 2019, 18, 117–126. [Google Scholar] [CrossRef]
- Turakhia, M.P.; Shafrin, J.; Bognar, K.; Trocio, J.; Abdulsattar, Y.; Wiederkehr, D.; Goldman, D.P. Estimated prevalence of undiagnosed atrial fibrillation in the United States. PLoS ONE 2018, 13, e0195088. [Google Scholar] [CrossRef] [PubMed]
- Healey, J.S.; Connolly, S.J.; Gold, M.R.; Israel, C.W.; Van Gelder, I.C.; Capucci, A.; Lau, C.P.; Fain, E.; Yang, S.; Bailleul, C.; et al. Subclinical Atrial Fibrillation and the Risk of Stroke. N. Engl. J. Med. 2012, 366, 120–129. [Google Scholar] [CrossRef]
- Haowen, J.; Shyn Yi, T.; Jeremy King, W.; Jiaqi, L.; Tian Ming, T.; Vern Hsen, T.; Colin, Y. A meta-analysis of extended ECG monitoring in detection of atrial fibrillation in patients with cryptogenic stroke. Open Heart 2022, 9, e002081. [Google Scholar] [CrossRef]
- Etgen, T.; Hochreiter, M.; Mundel, M.; Freudenberger, T. Insertable Cardiac Event Recorder in Detection of Atrial Fibrillation After Cryptogenic Stroke. Stroke 2013, 44, 2007–2009. [Google Scholar] [CrossRef] [PubMed]
- Tayal, A.H.; Tian, M.; Kelly, K.M.; Jones, S.C.; Wright, D.G.; Singh, D.; Jarouse, J.; Brillman, J.; Murali, S.; Gupta, R. Atrial fibrillation detected by mobile cardiac outpatient telemetry in cryptogenic TIA or stroke. Neurology 2008, 71, 1696–1701. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, P.D.; Koehler, J.L.; Mehra, R. Comparison of continuous versus intermittent monitoring of atrial arrhythmias. Heart Rhythm 2006, 3, 1445–1452. [Google Scholar] [CrossRef]
- Perez, M.V.; Mahaffey, K.W.; Hedlin, H.; Rumsfeld, J.S.; Garcia, A.; Ferris, T.; Balasubramanian, V.; Russo, A.M.; Rajmane, A.; Cheung, L.; et al. Large-Scale Assessment of a Smartwatch to Identify Atrial Fibrillation. N. Engl. J. Med. 2019, 381, 1909–1917. [Google Scholar] [CrossRef]
- Lubitz, S.A.; Faranesh, A.Z.; Selvaggi, C.; Atlas, S.J.; McManus, D.D.; Singer, D.E.; Pagoto, S.; McConnell, M.V.; Pantelopoulos, A.; Foulkes, A.S. Detection of Atrial Fibrillation in a Large Population Using Wearable Devices: The Fitbit Heart Study. Circulation 2022, 146, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Muehlematter, U.J.; Daniore, P.; Vokinger, K.N. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–2020): A comparative analysis. Lancet Digit. Health 2021, 3, e195–e203. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Mansour, S.; Zgheib, A.; Safatly, L.; Hajj, A.E.; Baydoun, M.; Ghaziri, H.; Aridi, H.; Ismaeel, H. Using Artificial Intelligence to Uncover Association of Left Atrial Strain with The Framingham Risk Score for Atrial Fibrillation Development. J. Am. Coll. Cardiol. 2020, 75, 455. [Google Scholar] [CrossRef]
- Ambale-Venkatesh, B.; Yang, X.; Wu, C.O.; Liu, K.; Hundley, W.G.; McClelland, R.; Gomes, A.S.; Folsom, A.R.; Shea, S.; Guallar, E.; et al. Cardiovascular Event Prediction by Machine Learning: The Multi-Ethnic Study of Atherosclerosis. Circ. Res. 2017, 121, 1092–1101. [Google Scholar] [CrossRef]
- Christopoulos, G.; Graff-Radford, J.; Lopez, C.L.; Yao, X.; Attia, Z.I.; Rabinstein, A.A.; Petersen, R.C.; Knopman, D.S.; Mielke, M.M.; Kremers, W.; et al. Artificial Intelligence-Electrocardiography to Predict Incident Atrial Fibrillation: A Population-Based Study. Circulation. Arrhythmia Electrophysiol. 2020, 13, e009355. [Google Scholar] [CrossRef]
- Hill, N.R.; Ayoubkhani, D.; McEwan, P.; Sugrue, D.M.; Farooqui, U.; Lister, S.; Lumley, M.; Bakhai, A.; Cohen, A.T.; O’Neill, M.; et al. Predicting atrial fibrillation in primary care using machine learning. PLoS ONE 2019, 14, e0224582. [Google Scholar] [CrossRef]
- Hirota, N.; Suzuki, S.; Arita, T.; Yagi, N.; Otsuka, T.; Kishi, M.; Semba, H.; Kano, H.; Matsuno, S.; Kato, Y.; et al. Prediction of current and new development of atrial fibrillation on electrocardiogram with sinus rhythm in patients without structural heart disease. Int. J. Cardiol. 2021, 327, 93–99. [Google Scholar] [CrossRef]
- Hu, W.S.; Hsieh, M.H.; Lin, C.L. A novel atrial fibrillation prediction model for Chinese subjects: A nationwide cohort investigation of 682 237 study participants with random forest model. Europace 2019, 21, 1307–1312. [Google Scholar] [CrossRef]
- Joo, G.; Song, Y.; Im, H.; Park, J. Clinical Implication of Machine Learning in Predicting the Occurrence of Cardiovascular Disease Using Big Data (Nationwide Cohort Data in Korea). IEEE Access 2020, 8, 157643–157653. [Google Scholar] [CrossRef]
- Kaminski, A.E.; Albus, M.L.; Ball, C.T.; White, L.J.; Sheele, J.M.; Attia, Z.I.; Friedman, P.A.; Adedinsewo, D.A.; Noseworthy, P.A. Evaluating atrial fibrillation artificial intelligence for the ED: Statistical and clinical implications. Am. J. Emerg. Med. 2022, 57, 98–102. [Google Scholar] [CrossRef]
- Khurshid, S.; Friedman, S.; Reeder, C.; Di Achille, P.; Diamant, N.; Singh, P.; Harrington, L.X.; Wang, X.; Al-Alusi, M.A.; Sarma, G.; et al. ECG-Based Deep Learning and Clinical Risk Factors to Predict Atrial Fibrillation. Circulation 2022, 145, 122–133. [Google Scholar] [CrossRef]
- Kim, I.-S.; Yang, P.-S.; Jang, E.; Jung, H.; You, S.C.; Yu, H.T.; Kim, T.-H.; Uhm, J.-S.; Pak, H.-N.; Lee, M.-H.; et al. Long-term PM2.5 exposure and the clinical application of machine learning for predicting incident atrial fibrillation. Sci. Rep. 2020, 10, 16324. [Google Scholar] [CrossRef]
- Kim, K.; Park, S.M. Artificial neural networks to compare the contribution of basic clinical factors, ESC SCORE, and multidimensional risk factors for cardiovascular event prediction performance: An observational study. Eur. Heart J. 2020, 41, 2897. [Google Scholar] [CrossRef]
- Lip, G.Y.H.; Genaidy, A.; Tran, G.; Marroquin, P.; Estes, C. Incidence and Complications of Atrial Fibrillation in a Low Socioeconomic and High Disability United States (US) Population: A Combined Statistical and Machine Learning Approach. Int. J. Clin. Pract. 2022, 2022, 8649050. [Google Scholar] [CrossRef]
- Raghunath, S.; Pfeifer, J.M.; Ulloa-Cerna, A.E.; Nemani, A.; Carbonati, T.; Jing, L.; vanMaanen, D.P.; Hartzel, D.N.; Ruhl, J.A.; Lagerman, B.F.; et al. Deep Neural Networks Can Predict New-Onset Atrial Fibrillation From the 12-Lead ECG and Help Identify Those at Risk of Atrial Fibrillation-Related Stroke. Circulation 2021, 143, 1287–1298. [Google Scholar] [CrossRef]
- Schnabel, R.B.; Witt, H.; Walker, J.; Ludwig, M.; Geelhoed, B.; Kossack, N.; Schild, M.; Miller, R.; Kirchhof, P. Machine learning-based identification of risk-factor signatures for undiagnosed atrial fibrillation in primary prevention and post-stroke in clinical practice. Eur. Heart J. Qual. Care Clin. Outcomes 2022, 9, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Sekelj, S.; Sandler, B.; Johnston, E.; Pollock, K.G.; Hill, N.R.; Gordon, J.; Tsang, C.; Khan, S.; Ng, F.S.; Farooqui, U. Detecting undiagnosed atrial fibrillation in UK primary care: Validation of a machine learning prediction algorithm in a retrospective cohort study. Eur. J. Prev. Cardiol. 2021, 28, 598–605. [Google Scholar] [CrossRef]
- Tiwari, P.; Colborn, K.L.; Smith, D.E.; Xing, F.; Ghosh, D.; Rosenberg, M.A. Assessment of a Machine Learning Model Applied to Harmonized Electronic Health Record Data for the Prediction of Incident Atrial Fibrillation. JAMA Netw. Open 2020, 3, e1919396. [Google Scholar] [CrossRef]
- Attia, Z.I.; Noseworthy, P.A.; Lopez-Jimenez, F.; Asirvatham, S.J.; Deshmukh, A.J.; Gersh, B.J.; Carter, R.E.; Yao, X.; Rabinstein, A.A.; Erickson, B.J.; et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction. Lancet 2019, 394, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Reinke, F.; Bettin, M.; Ross, L.S.; Kochhäuser, S.; Kleffner, I.; Ritter, M.; Minnerup, J.; Dechering, D.; Eckardt, L.; Dittrich, R. Refinement of detecting atrial fibrillation in stroke patients: Results from the TRACK-AF Study. Eur. J. Neurol. 2018, 25, 631–636. [Google Scholar] [CrossRef]
- Shan, S.M.; Tang, S.C.; Huang, P.W.; Lin, Y.M.; Huang, W.H.; Lai, D.M.; Wu, A.Y. Reliable PPG-based Algorithm in Atrial Fibrillation Detection. In Proceedings of the 2016 IEEE Biomedical Circuits and Systems Conference (BioCAS), Shanghai, China, 17–19 October 2016; pp. 340–343. [Google Scholar]
- Rabinstein, A.A.; Yost, M.D.; Faust, L.; Kashou, A.H.; Latif, O.S.; Graff-Radford, J.; Attia, I.Z.; Yao, X.; Noseworthy, P.A.; Friedman, P.A. Artificial Intelligence-Enabled ECG to Identify Silent Atrial Fibrillation in Embolic Stroke of Unknown Source. J. Stroke Cerebrovasc. Dis. Off. J. Natl. Stroke Assoc. 2021, 30, 105998. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.R.; Leussler, D.; Rosin, L.; Pittrow, D.; Hepp, T. Improved Detection of Paroxysmal Atrial Fibrillation Utilizing a Software-Assisted Electrocardiogram Approach. PLoS ONE 2014, 9, e89328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Konečný, J.; Brendan McMahan, H.; Yu, F.X.; Richtárik, P.; Theertha Suresh, A.; Bacon, D. Federated Learning: Strategies for Improving Communication Efficiency. arXiv 2016, arXiv:1610.05492. [Google Scholar] [CrossRef]
- Warnat-Herresthal, S.; Schultze, H.; Shastry, K.L.; Manamohan, S.; Mukherjee, S.; Garg, V.; Sarveswara, R.; Händler, K.; Pickkers, P.; Aziz, N.A.; et al. Swarm Learning for decentralized and confidential clinical machine learning. Nature 2021, 594, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Voss, E.A.; Makadia, R.; Matcho, A.; Ma, Q.; Knoll, C.; Schuemie, M.; DeFalco, F.J.; Londhe, A.; Zhu, V.; Ryan, P.B. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J. Am. Med. Inform. Assoc. 2015, 22, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Forrest, C.B.; McTigue, K.M.; Hernandez, A.F.; Cohen, L.W.; Cruz, H.; Haynes, K.; Kaushal, R.; Kho, A.N.; Marsolo, K.A.; Nair, V.P.; et al. PCORnet® 2020: Current state, accomplishments, and future directions. J. Clin. Epidemiol. 2021, 129, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Brandes, A.; Stavrakis, S.; Freedman, B.; Antoniou, S.; Boriani, G.; Camm, A.J.; Chow, C.K.; Ding, E.; Engdahl, J.; Gibson, M.M.; et al. Consumer-Led Screening for Atrial Fibrillation: Frontier Review of the AF-SCREEN International Collaboration. Circulation 2022, 146, 1461–1474. [Google Scholar] [CrossRef]
- Price, W.N., II; Gerke, S.; Cohen, I.G. Potential Liability for Physicians Using Artificial Intelligence. JAMA 2019, 322, 1765–1766. [Google Scholar] [CrossRef]
- Mannhart, D.; Lischer, M.; Knecht, S.; du Fay de Lavallaz, J.; Strebel, I.; Serban, T.; Vögeli, D.; Schaer, B.; Osswald, S.; Mueller, C.; et al. Clinical Validation of 5 Direct-to-Consumer Wearable Smart Devices to Detect Atrial Fibrillation: BASEL Wearable Study. JACC Clin. Electrophysiol. 2023, 9, 232–242. [Google Scholar] [CrossRef]
- Hill, N.R.; Groves, L.; Dickerson, C.; Boyce, R.; Lawton, S.; Hurst, M.; Pollock, K.G.; Sugrue, D.M.; Lister, S.; Arden, C.; et al. Identification of undiagnosed atrial fibrillation using a machine learning risk prediction algorithm and diagnostic testing (PULsE-AI) in primary care: Cost-effectiveness of a screening strategy evaluated in a randomized controlled trial in England. J. Med. Econ. 2022, 25, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Joglar, J.A.; Chung, M.K.; Armbruster, A.L.; Benjamin, E.J.; Chyou, J.Y.; Cronin, E.M.; Deswal, A.; Eckhardt, L.L.; Goldberger, Z.D.; Gopinathannair, R.; et al. 2023 ACC/AHA/ACCP/HRS Guideline for the Diagnosis and Management of Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2024, 149, e1–e156. [Google Scholar] [CrossRef] [PubMed]
| Key Question 1 | Key Question 2 | |
|---|---|---|
| Population | Adult patients without a known history of stroke or atrial fibrillation or cardiovascular comorbidities | Adult patients with a previous history of stroke |
| Interventions | ML models to predict incidence of atrial fibrillation | ML models to detect atrial fibrillation |
| Comparators | None | None |
| Outcomes |
|
|
| Timing | Any observational cohort study | Any observational cohort study |
| Setting | Any setting | Any setting |
| Study (Original Study Proposing Model If Validation Study) | Limitations of the Study Suggested by the Authors | Additional Limitations |
|---|---|---|
| Ahmad et al., 2020 [13] | None listed |
|
| Ambale-Venkatesh et al., 2017 [14] |
|
|
| ECG-AI (Attia et al., 2019 [29], Christopoulos et al., 2020 [15], Kaminski et al., 2022 [20]) |
|
|
| Hill et al., 2019 [16], Sekelj et al., 2021 [27] |
|
|
| Hirota et al., 2021 [17] |
|
|
| Hu et al., 2019 [18] |
|
|
| Joo et al., 2020 [19] | None listed |
|
| Khurshid et al., 2022 [21] |
|
|
| Kim et al., 2020 [22] |
| |
| Kim et al., 2020 [23] |
|
|
| Lip et al., 2022 [24] |
|
|
| Raghunath et al., 2021 [25] |
|
|
| Schnabel et al., 2023 [26] |
|
|
| Tiwari et al., 2020 [28] |
|
| Selected Study (Original Study Proposing Model If Validation Study) | Input Data | Data Source/Data Curated for Approved Access? | Model Architecture/Validation | Results | Model Interpretation | Code or Model Available/Reported Handling of Sparse Data | Model Currently Available for Clinical Use? |
|---|---|---|---|---|---|---|---|
| Rabinstein et al., 2021 [32] (ECG-AI [29]) | ECG trace | Prospective; local EHR/no | CNN/External | Sn: 63% Sp: 75% PPV: 23% NPV: 94% | No | Neither/No | No |
| Reinke et al., 2018 [30]/ (Schaefer et al., 2014 [33]) | ECG parameters | Prospective; local EHR/no | SVM (Proprietary model)/External | Sn: 95% Sp: 35% PPV: 27% NPV: 96% | No | Neither/No | Yes |
| Shan et al., 2014 [31] | Photoplethysmogram data | Prospective; local EHR/no | SVM/Internal | Acc: 96% Sn: 94% Sp: 96% AUC: 0.97 | No | Neither/No | No |
| Study | Limitations of the Study Suggested by the Authors | Additional Limitations |
|---|---|---|
| Rabinstein et al., 2021 [32] |
|
|
| Reinke et al., 2018 [30] |
|
|
| Shan et al., 2014 [31] | None listed |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawamura, Y.; Vafaei Sadr, A.; Abedi, V.; Zand, R. Many Models, Little Adoption—What Accounts for Low Uptake of Machine Learning Models for Atrial Fibrillation Prediction and Detection? J. Clin. Med. 2024, 13, 1313. https://doi.org/10.3390/jcm13051313
Kawamura Y, Vafaei Sadr A, Abedi V, Zand R. Many Models, Little Adoption—What Accounts for Low Uptake of Machine Learning Models for Atrial Fibrillation Prediction and Detection? Journal of Clinical Medicine. 2024; 13(5):1313. https://doi.org/10.3390/jcm13051313
Chicago/Turabian StyleKawamura, Yuki, Alireza Vafaei Sadr, Vida Abedi, and Ramin Zand. 2024. "Many Models, Little Adoption—What Accounts for Low Uptake of Machine Learning Models for Atrial Fibrillation Prediction and Detection?" Journal of Clinical Medicine 13, no. 5: 1313. https://doi.org/10.3390/jcm13051313
APA StyleKawamura, Y., Vafaei Sadr, A., Abedi, V., & Zand, R. (2024). Many Models, Little Adoption—What Accounts for Low Uptake of Machine Learning Models for Atrial Fibrillation Prediction and Detection? Journal of Clinical Medicine, 13(5), 1313. https://doi.org/10.3390/jcm13051313








