The Role of Angiotensin-Converting Enzyme (ACE) Polymorphisms in the Risk of Development and Treatment of Diabetic Nephropathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Groups
2.2. Methods
2.2.1. Determination of ACE Activity, and ACE, Glucose, Creatinine, eGFR, and CRP Concentrations
2.2.2. Determination of Metal Concentrations
2.2.3. Genotyping Analysis
2.2.4. Molecular Docking
2.2.5. Statistical Analysis
3. Results
3.1. Concentrations of the Selected Parameters and ACE Activity in the Studied Groups
3.2. The Influence of the rs4343 and the rs4646994 Polymorphisms in ACE on the Concentrations of the Selected Parameters and on ACE Activity
3.2.1. The Influence of the rs4343 Polymorphism on the Concentrations of the Selected Parameters and on ACE Activity
3.2.2. The Influence of the rs4646994 Polymorphism on the Concentrations of the Selected Parameter and on ACE Activity
3.2.3. The Influence of the rs4646994 Polymorphism on the Concentration of Selected Parameters and ACE Activity in a Group of Patients Using Ramipril
3.3. The Influence of ACE Polymorphisms on the Risk of Occurrence of Diabetic Nephropathy or the Likelihood of Renal Replacement Therapy
3.4. Interaction of ACE with Selected Drugs (Benazepril, Enalapril, Lisinopril and Ramipril)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Madziarska, K.; Banasik, M. Chorzy na cukrzycę w programach hemodializy i dializy otrzewnowej—Zagrożenia, których można uniknąć [Diabetes patients in hemodialysis and peritoneal dialysis programs—risks that can be avoided]. Probl. Lek. 2006, 45, 240–241. [Google Scholar]
- Sulaiman, M.K. Diabetic nephropathy: Recent advances in pathophysiology and challenges in dietary management. Diabetol. Metab. Syndr. 2019, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, Z. ACE insertion/deletion (I/D) polymorphism and diabetic nephropathy. J. Nephropathol. 2012, 1, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.Y.; Chen, L.S.; Zhang, L.C.; Zhou, T.B. Meta-analysis of the relationship between ACE I/D gene polymorphism and end-stage renal disease in patients with diabetic nephropathy. Nephrology 2012, 17, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wu, B.; Tao, J.; Han, Z.; Yang, X.; Zhang, L.; Liu, X.; Wang, Z.; Tan, R.; Gu, M.; et al. Association between Angiotensin I-Converting Enzyme Insertion/Deletion Polymorphism and Prognosis of Kidney Transplantation: A Meta-Analysis. PLoS ONE 2015, 10, e0127320. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rahimi, Z.; Hasanvand, A.; Felehgari, V. Interaction of MTHFR 1298C with ACE D allele augments the risk of diabetic nephropathy in Western Iran. DNA Cell Biol. 2012, 31, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Ruggenenti, P.; Bettinaglio, P.; Pinares, F.; Remuzzi, G. Angiotensin converting enzyme insertion/deletion polymorphism and renoprotection in diabetic and nondiabetic nephropathies. Clin. J. Am. Soc. Nephrol. 2008, 3, 1511–1525. [Google Scholar] [CrossRef] [PubMed]
- Alimoradi, N.; Sharqi, M.; Firouzabadi, D.; Sadeghi, M.M.; Moezzi, M.I.; Firouzabadi, N. SNPs of ACE1 (rs4343) and ACE2 (rs2285666) genes are linked to SARS-CoV-2 infection but not with the severity of disease. Virol. J. 2022, 19, 48. [Google Scholar] [CrossRef]
- Moradzadegan, A.; Vaisi-Raygani, A.; Nikzamir, A.; Rahimi, Z. Angiotensin converting enzyme insertion/deletion (I/D) (rs4646994) and Vegf polymorphism (+405G/C; rs2010963) in type II diabetic patients: Association with the risk of coronary artery disease. J. Renin-Angiotensin-Aldosterone Syst. 2015, 16, 672–680. [Google Scholar] [CrossRef]
- Ismail, M.F.; Shaker, O.G.; Ashour, E.; Yousif, H.M.; Afify, M.; Gouda, W. The combined effect of ACE, TCF7L2, and PPARGC1A gene polymorphisms in diabetic nephropathy. Egypt. J. Chem. 2017, 60, 869–881. [Google Scholar]
- Aggarwal, N.; Kare, P.K.; Varshney, P.; Kalra, O.P.; Madhu, S.V.; Banerjee, B.D.; Yadav, A.; Raizada, A.; Tripathi, A.K. Role of angiotensin converting enzyme and angiotensinogen gene polymorphisms in angiotensin converting enzyme inhibitor-mediated antiproteinuric action in type 2 diabetic nephropathy patients. World J. Diabetes 2017, 8, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Widodo, W.; Wisnasari, S.; Saifur Rohman, M.; Yunita, L.; Lukitasari, M.; Nuril, M.; Holil, K.; Laila Purwaningroom, D. Alu insertion/deletion of ACE gene polymorphism might not affect significantly the serum bradykinin level in hypertensive patients taking ACE inhibitors. Egypt. J. Med. Hum. Genet. 2017, 18, 187–191. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Fragkou, P.C.; Maratou, E.; Dimopoulou, D.; Kominakis, A.; Kokkinopoulou, I.; Kroupis, C.; Nikolaidou, A.; Antonakos, G.; Papaevangelou, V.; et al. Angiotensin-converting-enzyme insertion/deletion polymorphism, ACE activity, and COVID19: A rather controversial hypothesis. A case-control study. J. Med. Virol. 2022, 94, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Hye Khan, M.A.; Imig, J.D. Antihypertensive Drugs. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Ceconi, C.; Francolini, G.; Olivares, A.; Comini, L.; Bachetti, T.; Ferrari, R. Angiotensin converting enzyme (ACE) inhibitors have different selectivity for bradykinin binding sites of human somatic ACE. Eur. J. Pharmacol. 2007, 577, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.H.; Hayashi, M.A.; Camargo, A.C.; Neshich, G. Structural basis of the lisinopril-binding specificity in N- and C-domains of human somatic ACE. Biochem. Biophys. Res. Commun. 2003, 308, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Denti, P.; Sharp, S.K.; Kröger, W.L.; Schwager, S.L.; Mahajan, A.; Njoroge, M.; Gibhard, L.; Smit, I.; Chibale, K.; Wiesner, L.; et al. Pharmacokinetic evaluation of lisinopril-tryptophan, a novel C-domain ACE inhibitor. Eur. J. Pharm. Sci. 2014, 56, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, H.; Jing, Z.; Wang, Y.; Wang, W.; Jiang, Y.; Sun, W. Relationships of the Trace Elements Zinc and Magnesium with Diabetic Nephropathy-Associated Renal Functional Damage in Patients with Type 2 Diabetes Mellitus. Front. Med. 2021, 8, 626909. [Google Scholar] [CrossRef]
- Ming, J.; Sana, S.R.G.L.; Deng, X. Identification of copper-related biomarkers and potential molecule mechanism in diabetic nephropathy. Front. Endocrinol. 2022, 13, 978601. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D.S. BIOVIA Discovery Studio Visualizer (2D Diagram and Scheme of the Interactions with Amino Acids); BIOVIA Workbook: San Diego, CA, USA, 2020. [Google Scholar]
- Sagoo, M.K.; Gnudi, L. Diabetic Nephropathy: An Overview. Methods Mol. Biol. 2020, 2067, 3–7. [Google Scholar] [PubMed]
- Kobori, H.; Kamiyama, M.; Harrison-Bernard, L.M.; Navar, L.G. Cardinal role of the intrarenal renin-angiotensin system in the pathogenesis of diabetic nephropathy. J. Investig. Med. 2013, 61, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Kim, J.H.; Kim, I.J. Current Challenges in Diabetic Nephropathy: Early Diagnosis and Ways to Improve Outcomes. Endocrinol. Metab. 2016, 31, 245–253. [Google Scholar] [CrossRef]
- ra Lee, S.; Moon, J.Y.; Lee, S.H.; Ihm, C.G.; Lee, T.W.; Kim, S.K.; Chung, J.H.; Kang, S.W.; Kim, T.H.; Park, S.J.; et al. Angiotensinogen polymorphisms and post-transplantation diabetes mellitus in Korean renal transplant subjects. Kidney Blood Press. Res. 2013, 37, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Azmandian, J.; Mohamadifar, M.; Rahmanian-Koshkaki, S.; Mehdipoor, M.; Nematollahi, M.H.; Saburi, A.; Mandegary, A. Study of the association between the donors and recipients angiotensin-converting enzyme insertion/deletion gene polymorphism and the acute renal allograft rejection. J. Nephropathol. 2015, 4, 62–68. [Google Scholar] [PubMed]
- Bertoncello, N.; Moreira, R.P.; Arita, D.Y.; Aragão, D.S.; Watanabe, I.K.; Dantas, P.S.; Santos, R.; Mattar-Rosa, R.; Yokota, R.; Cunha, T.S.; et al. Diabetic Nephropathy Induced by Increased Ace Gene Dosage Is Associated with High Renal Levels of Angiotensin (1-7) and Bradykinin. J. Diabetes Res. 2015, 2015, 674047. [Google Scholar] [CrossRef]
- de Alcantara Santos, R.; Guzzoni, V.; Silva, K.A.S.; Aragão, D.S.; de Paula Vieira, R.; Bertoncello, N.; Schor, N.; Aimbire, F.; Casarini, D.E.; Cunha, T.S. Resistance exercise shifts the balance of renin-angiotensin system toward ACE2/Ang 1-7 axis and reduces inflammation in the kidney of diabetic rats. Life Sci. 2021, 287, 120058. [Google Scholar] [CrossRef]
- Al-Timimi, D.J.; Sulieman, D.M.; Hussen, K.R. Zinc status in type 2 diabetic patients: Relation to the progression of diabetic nephropathy. J. Clin. Diagn. Res. 2014, 8, CC04–CC08. [Google Scholar] [CrossRef]
- Nie, P.; Lou, Y.; Bai, X.; Zhu, Y.; Guo, Q.; Luo, P.; Zhang, W.; Li, B. Influence of zinc levels and Nrf2 expression in the clinical and pathological changes in patients with diabetic nephropathy. Nutr. Diabetes 2022, 12, 37. [Google Scholar] [CrossRef]
- Nikoobakht, M.R.; Pourmand, G.; Allameh, F.; Dialameh, H.; Sharifi, A.; Hashemiaghdam, A. Serum trace elements before and 3 months after renal transplantation in kidney recipients: An Iranian study. Indian J. Transplant. 2014, 8, 8–11. [Google Scholar] [CrossRef]
- Muralidhara, K.; Kannan, S.; Ahamed, I.; Kishore, K.; Vincent, L.; Hegde, N. A prospective observational study on the traditional and novel risk factors associated with post-transplant diabetes mellitus in renal transplant recipients. Biomedicine 2021, 41, 482–488. [Google Scholar] [CrossRef]
- Schüler, R.; Osterhoff, M.A.; Frahnow, T.; Möhlig, M.; Spranger, J.; Stefanovski, D.; Bergman, R.N.; Xu, L.; Seltmann, A.C.; Kabisch, S.; et al. Dietary Fat Intake Modulates Effects of a Frequent ACE Gene Variant on Glucose Tolerance with Association to Type 2 Diabetes. Sci. Rep. 2017, 7, 9234. [Google Scholar] [CrossRef]
- ALkafaji, M.J.M.; Shehab, A.F.; Omair, H.A. ACE Polymorphisms Rs (4343) and Its Association with Ace Enzyme in Some of Patients with Diabetic Nephropathy in Some Population of Salah Al-Din Governorate in Iraq. Pak. Heart J. 2023, 56, 193–199. [Google Scholar]
- Raza, S.T.; Abbas, S.; Siddiqi, Z.; Mahdi, F. Association between ACE (rs4646994), FABP2 (rs1799883), MTHFR (rs1801133), FTO (rs9939609) Genes Polymorphism and Type 2 Diabetes with Dyslipidemia. Int. J. Mol. Cell. Med. 2017, 6, 121–130. [Google Scholar] [PubMed]
- Zhao, Y.; Zhu, R.; Wang, D.; Liu, X. Genetics of diabetic neuropathy: Systematic review, meta-analysis and trial sequential analysis. Ann. Clin. Transl. Neurol. 2019, 6, 1996–2013. [Google Scholar] [CrossRef]
- Bayoumy, N.; El-Shabrawi, M.M.; Leheta, O.F.; Omar, H.H. ACE (rs4646994) and MTHFR (rs1801133) single nucleotide polymorphisms in type 2 Diabetes Mellitus patients with dyslipidemia. Rom. J. Diabetes Nutr. Metab. Dis. 2022, 29, 401–406. [Google Scholar]
- Sembach, F.E.; Fink, L.N.; Johansen, T.; Boland, B.B.; Secher, T.; Thrane, S.T.; Nielsen, J.C.; Fosgerau, K.; Vrang, N.; Jelsing, J.; et al. Impact of sex on diabetic nephropathy and the renal transcriptome in UNx db/db C57BLKS mice. Physiol. Rep. 2019, 7, e14333. [Google Scholar] [CrossRef]
- Duan, J.; Wang, C.; Liu, D.; Qiao, Y.; Pan, S.; Jiang, D.; Zhao, Z.; Liang, L.; Tian, F.; Yu, P.; et al. Prevalence and risk factors of chronic kidney disease and diabetic kidney disease in Chinese rural residents: A cross-sectional survey. Sci. Rep. 2019, 9, 10408. [Google Scholar] [CrossRef]
- Hussain, S.; Jamali, M.C.; Habib, A.; Hussain, M.S.; Akhtar, M.; Najmi, A.K. Diabetic kidney disease: An overview of prevalence, risk factors, and biomarkers. Clin. Epidemiol. Glob. Health 2021, 9, 2–6. [Google Scholar] [CrossRef]
- Selmi, A.; Aydi, R.; Kammoun, O.; Bougatef, H.; Bougatef, A.; Miled, N.; Alghamdi, O.A.; Kammoun, M. Synthesis, crystal structure, molecular docking studies and biological evaluation of aryl substituted dihydroisoquinoline imines as a potent angiotensin converting enzyme inhibitor. J. Mol. Struct. 2021, 1235, 130230. [Google Scholar] [CrossRef]
- Dey, T.K.; Chatterjee, R.; Mandal, R.S.; Roychoudhury, A.; Paul, D.; Roy, S.; Pateiro, M.; Das, A.K.; Lorenzo, J.M.; Dhar, P. ACE Inhibitory Peptides from Bellamya bengalensis Protein Hydrolysates: In Vitro and In Silico Molecular Assessment. Processes 2021, 9, 1316. [Google Scholar] [CrossRef]
- Januszewicz, A.; Januszewicz, W.; Szczepańska-Sadowska, E.; Sznajderman, M. Nadciśnienie Tętnicze [Hypertension]; Wydawnictwo Medycyna Praktyczna [Practical Medicine Publishing House]: Kraków, Poland, 2007; pp. 1079–1080. [Google Scholar]
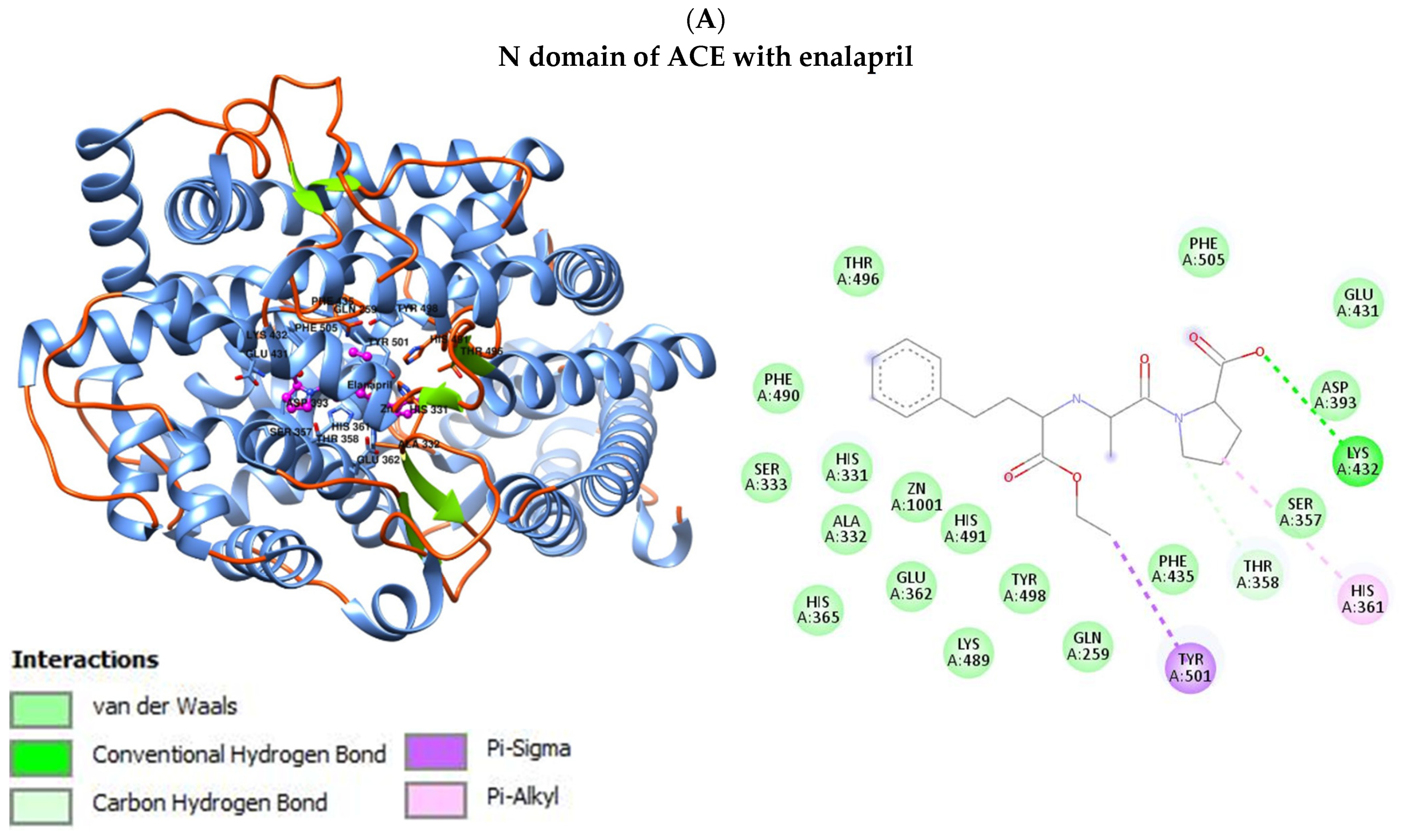
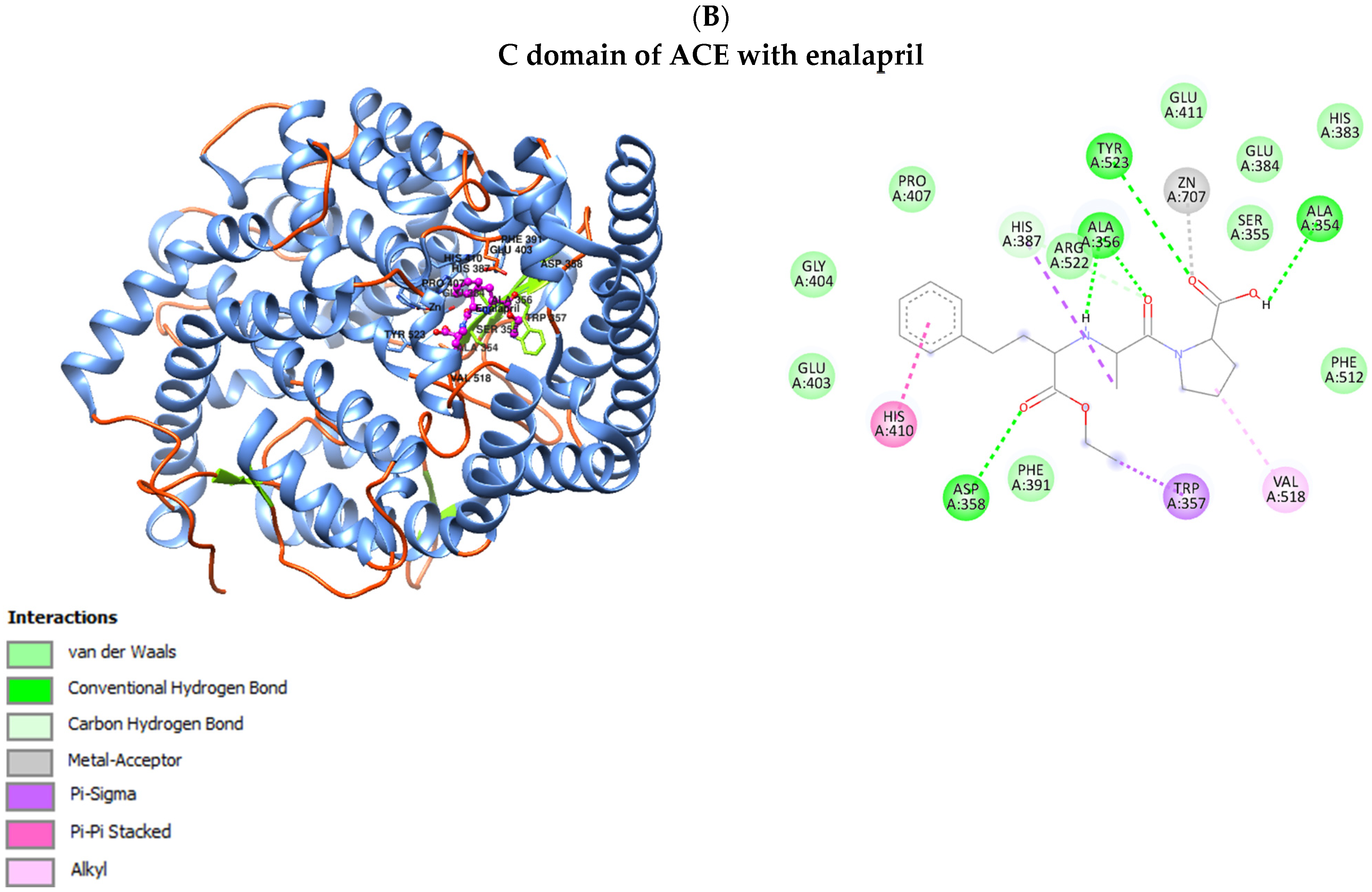
| Parameter | Control Group (N = 50) | Diabetic Nephropathy Group (N = 81) | Kidney Transplant Diabetic Nephropathy Group (N = 94) | p |
|---|---|---|---|---|
| Age (years) | {25; 34; 47} | {65; 71; 78} * | {55; 62; 69} *,** | <0.001 |
| Sex | Men: 21 Women: 29 | Men: 42 Women: 39 | Men: 47 Women: 47 | 0.540 |
| BMI (kg/m2) | 23.83 ± 3.37 | 30.02 ± 5.33 * | 27.05 ± 4.76 *,** | <0.001 |
| Glucose (mg/dL) | {81.00; 85.50; 88.92} | {106.00; 139.50; 178.00} * | {113.00; 139.50; 173.00} * | <0.001 |
| Creatinine (mg/dL) | - | {1.14; 1.36; 1.72} | {1.14; 1.30; 1.70} | 0.405 |
| eGFR (mL/min/1.73 m2) | - | {35.00; 48.00; 58.00} | {42.00; 53.50; 63.00} | 0.086 |
| CRP (mg/L) | {0.33; 0.65; 1.10} | {0.72; 1.91; 4.88} * | {0.97; 2.66; 4.11} * | <0.001 |
| SNP | Primers | PCR-RFLP Conditions |
|---|---|---|
| rs4343 | Forward primer—5′ CTG ACG AAT GTG ATG GCC GC 3′ Reverse primer—5′ TGA TGA GTT CCA CGT ATT TCG 3′ | the initial denaturation—95 °C for 5 min denaturation—95 °C for 40 s annealing—58.4 °C for 35 s elongation—72 °C for 40 s the final elongation—72 °C for 10 min |
| Restriction enzyme | Restriction enzyme digestion conditions | |
| BstUI | 37 °C for 16 h | |
| rs4646994 | Forward primer—5′ CTG GAG ACC ACT CCC ATC CTT TCT 3′ Reverse primer—5′ GAT GTG GCC ATC ACA TTC GTC AGA T 3′ | the initial denaturation—95 °C for 5 min denaturation—95 °C for 40 s annealing—60 °C for 35 s elongation—72 °C for 40 s the final elongation—72 °C for 10 min |
| Parameter | Control Group (N = 50) | Diabetic Nephropathy Group (N = 81) | Kidney Transplant Diabetic Nephropathy Group (N = 94) | p |
|---|---|---|---|---|
| ACE (ng/mL) | {42.64; 71.97; 99.30} | {79.06; 89.52; 101.72} * | {81.25; 89.64; 100.38} * | 0.005 |
| ACE (mU/mL) | {0.066; 0.079; 0.092} | {0.026; 0.052; 0.071} * | {0.045; 0.063; 0.076} * | <0.001 |
| Zn (µg/L) | {755.00; 830.00; 913.50} | {720.00; 804.50; 880.00} * | {854.21; 946.55; 1031.87} * | <0.001 |
| Cu (µg/L) | {880.00; 1019.00; 1151.50} | {909.00; 1084.00; 1200.00} | {920.91; 1012.89; 1181.65} | 0.293 |
| SNP | Groups (N) | Genotype Frequencies (%) | ||
|---|---|---|---|---|
| G/G | G/A | A/A | ||
| rs4343 | Control (N = 49) | N = 11 (22.45%) | N = 4 (8.16%) | N = 34 (69.39%) |
| Diabetic Nephropathy (N = 84) | N = 16 (19.05%) | N = 42 (50.00%) | N = 26 (30.95%) | |
| Kidney Transplant Diabetic Nephropathy (N = 93) | N = 26 (27.96%) | N = 43 (46.24%) | N = 24 (25.80%) | |
| SNP | Groups (N) | Genotype Frequencies (%) | ||
| I/I | I/D | D/D | ||
| rs4646994 | Control (N = 49) | N = 0 (0.00%) | N = 42 (84.00%) | N = 8 (16.00%) |
| Diabetic Nephropathy (N = 81) | N = 11 (13.58%) | N = 51 (62.96%) | N = 19 (23.46%) | |
| Kidney Transplant Diabetic Nephropathy (N = 93) | N = 13 (13.98%) | N = 59 (63.44%) | N = 21 (22.58) | |
| Parameter | Control Group (N = 49) | Diabetic Nephropathy Group (N = 84) | Kidney Transplant Diabetic Nephropathy Group (N = 93) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| G/G (N = 11) | G/A (N = 4) | A/A (N = 34) | G/G (N = 16) | G/A (N = 42) | A/A (N = 26) | G/G (N = 26) | G/A (N = 43) | A/A (N = 24) | |
| ACE (ng/mL) | {67.86; 83.77; 88.92} | {51.04; 60.85; 90.94} | {41.62; 78.13; 101.64} | {81.66; 88.36; 104.25} | {79.39; 90.71; 100.17} | {77.10; 86.63; 101.97} | {79.55; 84.74; 97.05} | {81.02; 90.20; 101.21} | {85.90; 94.63; 100.64} |
| ACE (mU/mL) | {0.073; 0.078; 0.106} | {0.079; 0.085; 0.088} | {0.063; 0.079; 0.093} | {0.050; 0.063; 0.077} | {0.022; 0.049; 0.074} | {0.025; 0.051; 0.066} ** | {0.054; 0.063; 0.073] | {0.040; 0.063; 0.076} | {0.037; 0.064; 0.078} |
| Glucose (mg/dL) | {81.00; 82.98; 88.92} | {85.50; 90.00; 94.50} | {79.92; 84.47; 88.92} | {102.00; 124.50; 154.00} | {106.00; 139.00; 183.00} | {105.00; 141.00; 163.00} ** | {114.00; 126.00; 143.00} * | {111.00; 149.00; 184.00} | {113.00; 155.00; 183.00} |
| Creatinine (mg/dL) | - | - | - | {1.06; 1.35; 1.46} | {1.21; 1.42; 2.00} | {1.05; 1.24; 1.60} | {1.08; 1.26; 1.41} | {1.14; 1.31; 1.80} | {1.17; 1.30; 1.70} |
| eGFR (mL/min/1.73 m2) | - | - | - | {48.50; 52.00; 59.50} | {29.00; 44.00; 54.00} | {44.00; 56.00; 64.00} | {43.00; 56.00; 61.00} | {36.00; 53.00; 63.00} | {45.00; 54.00; 66.00} |
| CRP (mg/L) | {0.27; 0.64; 1.01} | {0.37; 0.99; 3.12} | {0.33; 0.65; 1.10} | {0.60; 2.55; 4.49} | {1.29; 2.38; 5.35} | {0.23; 0.72; 1.24} | {1.21; 2.79; 4.07} | {1.49; 2.26; 4.33} | {0.70; 0.96; 4.10} |
| Zn (µg/L) | {845.49; 895.37; 1019.99} | {823.33; 933.47; 1075.62} | {901.82; 972.73; 1043.34} | {817.00; 855.00; 922.00} | {752.00; 800.50; 884.00} | {729.00; 855.50; 937.00} ** | {749.00; 825.00; 940.00} | {721.00; 807.00; 887.00} | {672.00; 753.50; 833.00} ** |
| Cu (µg/L) | {986.90; 1046.20; 1184.39} | {1006.96; 1251.14; 2039.81} | {868.53; 998.18; 1155.12} | {896.00; 1009.00; 1205.00} | {884.00; 1002.00; 1168.00} | {859.00; 1058.00; 1129.00} | {904.00; 1047.00; 1158.00} | {911.00; 1092.00; 1254.00} | {900.00; 1074.00; 1225.00} |
| Parameter | Control Group (N = 50) | Diabetic Nephropathy Group (N = 81) | Kidney Transplant Diabetic Nephropathy Group (N = 93) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| I/I (N = 0) | I/D (N = 42) | D/D (N = 8) | I/I (N = 11) | I/D (N = 51) | D/D (N = 19) | I/I (N = 13) | I/D (N = 59) | D/D (N = 21) | |
| ACE (ng/mL) | - | {42.64; 71.51; 89.32} | {60.50; 88.52; 99.30} | {80.05; 96.22; 101.98} | {76.48; 87.19; 100.17} | {83.22; 88.32; 101.72} | {86.33; 93.61; 96.98} | {80.05; 89.64; 101.02} | {76.15; 86.66; 100.49} |
| ACE (mU/mL) | - | {0.063; 0.079; 0.092} | {0.074; 0.077; 0.086} | {0.016; 0.036; 0.070} | {0.022; 0.051; 0.074} * | {0.045; 0.057; 0.073} | {0.027; 0.064; 0.069} | {0.040; 0.062; 0.076} | {0.054; 0.063; 0.073} |
| Glucose (mg/dL) | - | {81.00; 85.50; 88.92} | {81.99; 85.95; 92.52} | {103.00; 110.00; 146.00} | {107.00; 141.00; 183.00} * | {94.50; 152.00; 202.50} | {130.50; 161.00; 178.00} | {113.00; 139.00; 170.00} * | {112.00; 122.00; 146.00} |
| Creatinine (mg/dL) | - | - | - | {1.12; 1.24; 1.34} | {1.18; 1.39; 1.80} | {1.14; 1.41; 1.48} | {1.12; 1.27; 1.54} | {1.14; 1.41; 1.80} | {1.06; 1.22; 1.31} |
| eGFR (mL/min/1.73 m2) | - | - | - | {54.00; 56.00; 61.00} | {34.00; 45.00; 55.50} | {36.00; 50.00; 61.00} | {45.50; 54.00; 67.00} | {36.00; 51.00; 59.00} | {52.00; 59.00; 64.00} |
| CRP (mg/L) | - | {0.37; 0.72; 1.15} | {0.15; 0.36; 1.03} | {0.71; 0.98; 1.24} | {0.72; 2.13; 5.42} | {0.60; 1.94; 4.49} | {0.77; 0.93; 4.11} | {0.99; 2.86; 4.33} * | {1.21; 2.43; 3.37} |
| Zn (µg/L) | - | {890.78; 972.73; 1043.34} | {824.51; 877.31; 995.86} | {762.00; 879.00; 945.00} | {741.00; 807.00; 898.00} * | {759.00; 855.00; 922.00} | {704.00; 744.00; 798.00} | {718.00; 805.00; 887.00} * | {780.00; 825.00; 880.00} |
| Cu (µg/L) | - | {920.91; 1002.02; 1181.65} | {930.06; 1032.31; 1233.67} | {835.00; 941.00; 1144.00} | {896.00; 1045.00; 1150.00} | {822.00; 962.00; 1205.00} | {820.00; 1097.00; 1225.00} | {911.00; 1087.00; 1248.00} | {934.00; 1088.00; 1139.00} |
| Parameter | I/I (N = 6) | I/D (N = 17) | D/D (N = 8) | p |
|---|---|---|---|---|
| BMI (kg/m2) | {22.54; 32.57; 38.81} | {26.29; 27.92; 33.27} | {29.28; 31.99; 40.39} | 0.186 |
| ACE (ng/mL) | {86.48; 92.68; 97.97} | {83.10; 89.61; 104.70} | {81.20; 92.97; 97.35} | 0.983 |
| ACE (mU/mL) | {0.027; 0.051; 0.066} | {0.018; 0.062; 0.077} | {0.035; 0.069; 0.073} | 0.886 |
| Glucose (mg/dL) | {103.00; 106.00; 110.00} | {133.00; 157.00; 216.00} * | {128.00; 146.50; 224.00} | 0.027 |
| Creatinine (mg/dL) | 1.23 ± 0.21 | 1.26 ± 0.34 | 1.30 ± 0.29 | 0.919 |
| eGFR (mL/min/1.73 m2) | 53.67 ± 9.33 | 61.13 ± 21.36 | 54.88 ± 17.72 | 0.620 |
| CRP (mg/L) | {1.31; 2.72; 5.02} | {0.45; 0.72; 1.54} | - | 0.395 |
| Zn (µg/L) | 778.67 ± 105.56 | 810.47 ± 150.56 | 797.50 ± 60.63 | 0.947 |
| Cu (µg/L) | 1007.83 ± 225.69 | 1067.88 ± 216.30 | 940.25 ± 188.94 | 0.386 |
| SNP (Gene) | Genotype | Diabetic Nephropathy Group | Control Group | p | OR | 95% CI OR |
|---|---|---|---|---|---|---|
| rs4343 (ACE) | G/G | 42 | 11 | 0.014 | 2.675 | 1.216–5.884 |
| G/A | 85 | 4 | <0.001 | 13.894 | 4.662–41.408 | |
| A/A | 50 | 34 | - | 1.000 | - | |
| G allele | 169 | 26 | <0.001 | 2.530 | 1.543–4.148 | |
| A allele | 185 | 72 | - | 1.000 | - | |
| rs4646994 (ACE) | I/I | 24 | 0 | - | - | - |
| I/D | 110 | 42 | 0.131 | 0.524 | 0.227–1.211 | |
| D/D | 40 | 8 | - | 1.000 | - | |
| I allele | 168 | 42 | 0.382 | 1.221 | 0.780–1.911 | |
| D allele | 190 | 58 | - | 1.000 | - | |
| Other Variables | Category | Diabetic Nephropathy Group | Control Group | p | OR | 95% CI OR |
| Age | - | - | - | <0.001 | 1.173 | 1.123–1.226 |
| BMI | - | - | - | <0.001 | 1.266 | 1.157–1.386 |
| Sex | Men | 89 | 21 | - | 1.000 | - |
| Women | 85 | 29 | 0.270 | 0.701 | 0.372–1.318 |
| SNP (Gene) | Genotype | Kidney Transplant Diabetic Nephropathy Group | Control Group | p | OR | 95% CI OR |
|---|---|---|---|---|---|---|
| rs4343 (ACE) | G/G | 26 | 11 | 0.007 | 3.348 | 1.392–8.053 |
| G/A | 43 | 4 | <0.001 | 15.229 | 4.821–48.103 | |
| A/A | 24 | 34 | - | 1.000 | - | |
| G allele | 95 | 26 | <0.001 | 2.891 | 1.697–4.925 | |
| A allele | 91 | 72 | - | 1.000 | - | |
| rs4646994 (ACE) | I/I | 13 | 0 | - | - | - |
| I/D | 59 | 42 | 0.176 | 0.535 | 0.216–1.323 | |
| D/D | 21 | 8 | - | 1.000 | - | |
| I allele | 85 | 42 | 0.548 | 1.162 | 0.711–1.899 | |
| D allele | 101 | 58 | - | 1.000 | - | |
| Other Variables | Category | Kidney Transplant Diabetic Nephropathy Group | Control Group | p | OR | 95% CI OR |
| Age | - | - | - | <0.001 | 0.848 | 0.805–0.894 |
| BMI | - | - | - | <0.001 | 0.826 | 0.748–0.911 |
| Sex | Men | 47 | 21 | - | 1.000 | - |
| Women | 46 | 29 | 0.327 | 1.409 | 0.710–2.798 |
| Ligands | The Binding Affinity (kcal/mol) | |
|---|---|---|
| N Domain | C Domain | |
| Enalapril | −7.4 | −7.9 |
| Ramipril | −8.0 | −8.2 |
| Benazepril | −6.6 | −8.3 |
| Lisinopril | −5.8 | −6.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Król-Kulikowska, M.; Abramenko, N.; Jakubek, M.; Banasik, M.; Kepinska, M. The Role of Angiotensin-Converting Enzyme (ACE) Polymorphisms in the Risk of Development and Treatment of Diabetic Nephropathy. J. Clin. Med. 2024, 13, 995. https://doi.org/10.3390/jcm13040995
Król-Kulikowska M, Abramenko N, Jakubek M, Banasik M, Kepinska M. The Role of Angiotensin-Converting Enzyme (ACE) Polymorphisms in the Risk of Development and Treatment of Diabetic Nephropathy. Journal of Clinical Medicine. 2024; 13(4):995. https://doi.org/10.3390/jcm13040995
Chicago/Turabian StyleKról-Kulikowska, Magdalena, Nikita Abramenko, Milan Jakubek, Mirosław Banasik, and Marta Kepinska. 2024. "The Role of Angiotensin-Converting Enzyme (ACE) Polymorphisms in the Risk of Development and Treatment of Diabetic Nephropathy" Journal of Clinical Medicine 13, no. 4: 995. https://doi.org/10.3390/jcm13040995
APA StyleKról-Kulikowska, M., Abramenko, N., Jakubek, M., Banasik, M., & Kepinska, M. (2024). The Role of Angiotensin-Converting Enzyme (ACE) Polymorphisms in the Risk of Development and Treatment of Diabetic Nephropathy. Journal of Clinical Medicine, 13(4), 995. https://doi.org/10.3390/jcm13040995









