Associations Between Clinical Manifestations of SARS-CoV-2 Infection and HLA Alleles in a Caucasian Population: A Molecular HLA Typing Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Groups and Control Group
2.2. SARS-CoV-2 Infection Diagnosis Procedure
2.3. HLA Typing Procedures
2.4. Statistical Analysis
2.5. Ethics Procedures
3. Results
3.1. Comparison of COVID-19 Patients and Control Group
3.2. Comparison of Severe COVID-19 and Non-Severe COVID-19 Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Available online: https://wwwwhoint/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing---5-may-20232023 (accessed on 20 July 2024).
- Wen, W.; Cai, S.; Li, Y.; Wu, X. Risk factors and prognosis for coronavirus disease 2019 among 131 hemodialysis patients during the Omicron variant epidemic. Ren. Fail. 2023, 45, 2228924. [Google Scholar] [CrossRef] [PubMed]
- Cancarevic, I.; Nassar, M.; Daoud, A.; Ali, H.; Nso, N.; Sanchez, A.; Parikh, A.; Ul Hosna, A.; Devanabanda, B.; Ahmed, N.; et al. Mortality rate of COVID-19 infection in end stage kidney disease patients on maintenance hemodialysis: A systematic review and meta-analysis. World J. Virol. 2022, 11, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Reyes, L.F.; Garcia-Gallo, E.; Murthy, S.; Fuentes, Y.V.; Serrano, C.C.; Ibáñez-Prada, E.D.; Lee, J.; Rojek, A.; Citarella, B.W.; Gonçalves, B.P.; et al. Major adverse cardiovascular events (MACE) in patients with severe COVID-19 registered in the ISARIC WHO clinical characterization protocol: A prospective, multinational, observational study. J. Crit. Care 2023, 77, 154318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pang, Q.; Zhou, T.; Meng, J.; Dong, X.; Wang, Z.; Zhang, A. Risk factors for acute kidney injury in COVID-19 patients: An updated systematic review and meta-analysis. Ren. Fail. 2023, 45, 2170809. [Google Scholar] [CrossRef]
- Kousathanas, A.; Pairo-Castineira, E.; Rawlik, K.; Stuckey, A.; Odhams, C.A.; Walker, S.; Russell, C.D.; Malinauskas, T.; Wu, Y.; Millar, J.; et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature 2022, 607, 97–103. [Google Scholar]
- COVID-19 Host Genetics Initiative. A first update on mapping the human genetic architecture of COVID-19. Nature 2022, 608, E1–E10. [Google Scholar] [CrossRef]
- Shelton, J.F.; Shastri, A.J.; Ye, C.; Weldon, C.H.; Filshtein-Sonmez, T.; Coker, D.; Symons, A.; Esparza-Gordillo, J.; The 23andMe COVID-19 Team; Aslibekyan, S.; et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021, 53, 801–808. [Google Scholar] [CrossRef]
- Acar, A. Pan-Cancer Analysis of the COVID-19 Causal Gene SLC6A20. ACS Omega 2023, 8, 13153–13161. [Google Scholar] [CrossRef]
- Severe COVID-19 GWAS Group; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; et al. Genomewide association study of severe COVID-19 with respiratory failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Angulo-Aguado, M.; Corredor-Orlandelli, D.; Carrillo-Martínez, J.C.; Gonzalez-Cornejo, M.; Pineda-Mateus, E.; Rojas, C.; Triana-Fonseca, P.; Contreras Bravo, N.C.; Morel, A.; Parra Abaunza, K.; et al. Association Between the LZTFL1 rs11385942 Polymorphism and COVID-19 Severity in Colombian Population. Front. Med. 2022, 9, 910098. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Anisul, M.; Shilts, J.; Schwartzentruber, J.; Hayhurst, J.; Buniello, A.; Shaikho Elhaj Mohammed, E.; Zheng, J.; Holmes, M.; Ochoa, D.; Carmona, M.; et al. A proteome-wide genetic investigation identifies several SARS-CoV-2-exploited host targets of clinical relevance. eLife 2021, 10, e69719. [Google Scholar] [CrossRef] [PubMed]
- Pietzner, M.; Chua, R.L.; Wheeler, E.; Jechow, K.; Willett, J.D.S.; Radbruch, H.; Trump, S.; Heidecker, B.; Zeberg, H.; Heppner, F.L.; et al. ELF5 is a potential respiratory epithelial cell-specific risk gene for severe COVID-19. Nat. Commun. 2022, 13, 4484. [Google Scholar] [CrossRef]
- Huffman, J.E.; Butler-Laporte, G.; Khan, A.; Pairo-Castineira, E.; Drivas, T.G.; Peloso, G.M.; Nakanishi, T.; COVID-19 Host Genetics Initiative; Ganna, A.; Verma, A.; et al. Multi-ancestry fine mapping implicates OAS1 splicing in risk of severe COVID-19. Nat. Genet. 2022, 54, 125–127. [Google Scholar] [CrossRef]
- Horowitz, J.E.; Kosmicki, J.A.; Damask, A.; Sharma, D.; Roberts, G.H.L.; Justice, A.E.; Banerjee, N.; Coignet, M.V.; Yadav, A.; Leader, J.B.; et al. Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat. Genet. 2022, 54, 382–392. [Google Scholar] [CrossRef]
- Asano, T.; Boisson, B.; Onodi, F.; Matuozzo, D.; Moncada-Velez, M.; Maglorius Renkilaraj, M.R.L.; Zhang, P.; Meertens, L.; Bolze, A.; Materna, M.; et al. X-linked recessive TLR7 deficiency in~ 1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 2021, 6, eabl4348. [Google Scholar] [CrossRef]
- Fallerini, C.; Daga, S.; Mantovani, S.; Benetti, E.; Picchiotti, N.; Francisci, D.; Paciosi, F.; Schiaroli, E.; Baldassarri, M.; Fava, F.; et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: Findings from a nested case-control study. eLife 2021, 10, e67569. [Google Scholar] [CrossRef]
- van der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; van den Heuvel, G.; Mantere, T.; Kersten, S.; van Deuren, R.C.; Steehouwer, M.; van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of genetic variants among young men with severe COVID-19. JAMA 2020, 324, 663–673. [Google Scholar] [CrossRef]
- Abolnezhadian, F.; Iranparast, S.; Shohan, M.; Shokati Eshkiki, Z.; Hamed, M.; Seyedtabib, M.; Nashibi, R.; Assarehzadegan, M.A.; Mard, S.A.; Shayesteh, A.A.; et al. Evaluation the frequencies of HLA alleles in moderate and severe COVID-19 patients in Iran: A molecular HLA typing study. Heliyon 2024, 10, e28528. [Google Scholar] [CrossRef]
- Castro-Santos, P.; Rojas-Martinez, A.; Riancho, J.A.; Lapunzina, P.; Flores, C.; Carracedo, Á.; Díaz-Peña, R.; Scourge Cohort Group. HLA-A*11:01 and HLA-C*04:01 are associated with severe COVID-19. HLA 2023, 102, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.; Suwalski, P.; Holtgrewe, M.; Rakitko, A.; Thibeault, C.; Müller, M.; Patriki, D.; Quedenau, C.; Krüger, U.; Ilinsky, V.; et al. Increased risk of severe clinical course of COVID-19 in carriers of HLA-C*04:01. eClinicalMedicine 2021, 40, 101099. [Google Scholar] [CrossRef] [PubMed]
- Hajeer, A.; Jawdat, D.; Massadeh, S.; Aljawini, N.; Abedalthagafi, M.S.; Arabi, Y.M.; Alaamery, M. Association between human leukocyte antigen alleles and COVID-19 disease severity. J. Infect. Public Health 2024, 17, 102498. [Google Scholar] [CrossRef] [PubMed]
- Hovhannisyan, A.; Madelian, V.; Avagyan, S.; Nazaretyan, M.; Hyussyan, A.; Sirunyan, A.; Arakelyan, R.; Manukyan, Z.; Yepiskoposyan, L.; Mayilyan, K.R.; et al. HLA-C*04:01 Affects HLA Class I Heterozygosity and Predicted Affinity to SARS-CoV-2 Peptides, and in Combination With Age and Sex of Armenian Patients Contributes to COVID-19 Severity. Front. Immunol. 2022, 13, 769900. [Google Scholar] [CrossRef]
- Naemi, F.M.A.; Al-Adwani, S.; Al-Khatabi, H.; Al-Nazawi, A. Association between the HLA genotype and the severity of COVID-19 infection among South Asians. J. Med. Virol. 2021, 93, 4430–4437. [Google Scholar] [CrossRef]
- Marchal, A.; Cirulli, E.T.; Neveux, I.; Bellos, E.; Thwaites, R.S.; Schiabor Barrett, K.M.; Zhang, Y.; Nemes-Bokun, I.; Kalinova, M.; Catchpole, A.; et al. Lack of association between classical HLA genes and asymptomatic SARS-CoV-2 infection. HGG Adv. 2024, 5, 100300. [Google Scholar] [CrossRef]
- Lorente, L.; Martín, M.M.; Franco, A.; Barrios, Y.; Cáceres, J.J.; Solé-Violán, J.; Perez, A.; Marcos, Y.; Ramos, J.A.; Ramos-Gómez, L.; et al. HLA genetic polymorphisms and prognosis of patients with COVID-19. Med. Intensiv. 2021, 45, 96–103. [Google Scholar] [CrossRef]
- Hai, N.T.T.; Nhung, V.P.; Tam, N.T.T.; Ngoc, T.T.B.; Thuong, M.T.H.; Dai, H.V.; Duong, N.T.; Hai, N.V.; Ton, N.D.; Thach, P.N.; et al. HLA alleles associated with susceptibility and severity of the COVID-19 in Vietnamese. Hum. Immunol. 2024, 85, 110796. [Google Scholar] [CrossRef]
- Hosseini, E.; Minagar, A.; Ghasemzadeh, M.; Arabkhazaeli, A.; Ghasemzadeh, A. HLA-E*01:01 + HLA-E*01:01 genotype confers less susceptibility to COVID-19, while HLA-E*01:03 + HLA-E*01:03 genotype is associated with more severe disease. Hum. Immunol. 2023, 84, 263–271. [Google Scholar] [CrossRef]
- Wang, F.; Huang, S.; Gao, R.; Zhou, Y.; Lai, C.; Li, Z.; Xian, W.; Qian, X.; Li, Z.; Huang, Y.; et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020, 6, 83. [Google Scholar] [CrossRef]
- Augusto, D.G.; Yusufali, T.; Sabatino, J.J., Jr.; Peyser, N.D.; Murdolo, L.D.; Butcher, X.; Murray, V.; Pae, V.; Sarvadhavabhatla, S.; Beltran, F.; et al. A common allele of HLA mediates asymptomatic SARS-CoV-2 infection. Nature 2023, 620, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Langton, D.J.; Bourke, S.C.; Lie, B.A.; Reiff, G.; Natu, S.; Darlay, R.; Burn, J.; Echevarria, C. The influence of HLA genotype on the severity of COVID-19 infection. HLA 2021, 98, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Schindler, E.; Dribus, M.; Duffy, B.F.; Hock, K.; Farnsworth, C.W.; Gragert, L.; Liu, C. HLA genetic polymorphism in patients with Coronavirus Disease 2019 in Midwestern United States. HLA 2021, 98, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Novelli, A.; Andreani, M.; Biancolella, M.; Liberatoscioli, L.; Passarelli, C.; Colona, V.L.; Rogliani, P.; Leonardis, F.; Campana, A.; Carsetti, R.; et al. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA 2020, 96, 610–614. [Google Scholar] [CrossRef]
- Littera, R.; Campagna, M.; Deidda, S.; Angioni, G.; Cipri, S.; Melis, M.; Firinu, D.; Santus, S.; Lai, A.; Porcella, R.; et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Front. Immunol. 2020, 11, 605688. [Google Scholar] [CrossRef]
- Khor, S.S.; Omae, Y.; Nishida, N.; Sugiyama, M.; Kinoshita, N.; Suzuki, T.; Suzuki, M.; Suzuki, S.; Izumi, S.; Hojo, M.; et al. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, Age and Sex Are Associated with Severity of Japanese COVID-19 with Respiratory Failure. Front. Immunol. 2021, 12, 658570. [Google Scholar] [CrossRef]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef]
- Vishnubhotla, R.; Sasikala, M.; Ketavarapu, V.; Reddy, D.N. High-resolution HLA genotyping identifies alleles associated with severe COVID-19: A preliminary study from India. Immun. Inflamm. Dis. 2021, 9, 1781–1785. [Google Scholar] [CrossRef]
- Anzurez, A.; Naka, I.; Miki, S.; Nakayama-Hosoya, K.; Isshiki, M.; Watanabe, Y.; Nakamura-Hoshi, M.; Seki, S.; Matsumura, T.; Takano, T.; et al. Association of HLA-DRB1*09:01 with severe COVID-19. HLA 2021, 98, 37–42. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, L.; Chen, J.; Gao, Y. Targeted capture enrichment and sequencing identifies HLA variants associated with the severity of COVID-19. Genes Genom. 2023, 45, 451–456. [Google Scholar] [CrossRef]
- Tanaka, K.; Meguro, A.; Hara, Y.; Endo, L.; Izawa, A.; Muraoka, S.; Kaneko, A.; Somekawa, K.; Hirata, M.; Otsu, Y.; et al. HLA-DQA1*01:03 and DQB1*06:01 are risk factors for severe COVID-19 pneumonia. HLA 2024, 104, e15609. [Google Scholar] [CrossRef] [PubMed]
- Metlay, J.P.; Waterer, G.W.; Long, A.C.; Anzueto, A.; Brozek, J.; Crothers, K.; Cooley, L.A.; Dean, N.C.; Fine, M.J.; Flanders, S.A.; et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am. J. Respir. Crit. Care Med. 2019, 200, e45–e67. [Google Scholar] [CrossRef] [PubMed]
- Tymoniuk, B.; Zmora, P.; Latowska, J.; Grabowska, A.; Ciesiołka, A.; Joachimiak, P.; Figura, G.; Borowiec, M.; Rolle, K.; Handschuh, L.; et al. Genetic tests based on the RT-PCR reaction in the diagnostics of SARS-CoV-2 infection. Przegl. Epidemiol. 2021, 75, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Hollenbach, J.; Shi, X.; Shi, W.; Chopek, M.; Fernández-Viña, M.A. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 2001, 62, 1009–1030. [Google Scholar] [CrossRef]
- Correale, P.; Mutti, L.; Pentimalli, F.; Baglio, G.; Saladino, R.E.; Sileri, P.; Giordano, A. HLA-B*44 and C*01 Prevalence Correlates with COVID-19 Spreading across Italy. Int. J. Mol. Sci. 2020, 21, 5205. [Google Scholar] [CrossRef]
- Stasiak, M.; Tymoniuk, B.; Michalak, R.; Stasiak, B.; Kowalski, M.L.; Lewiński, A. Subacute Thyroiditis is Associated with HLA-B*18:01, -DRB1*01 and -C*04:01—The Significance of the New Molecular Background. J. Clin. Med. 2020, 9, 534. [Google Scholar] [CrossRef]
- Stasiak, M.; Lewiński, A. New aspects in the pathogenesis and management of subacute thyroiditis. Rev. Endocr. Metab. Disord. 2021, 22, 1027–1039. [Google Scholar] [CrossRef]
- Stasiak, M.; Zawadzka-Starczewska, K.; Lewiński, A. Clinical Manifestation of Subacute Thyroiditis Triggered by SARS-CoV-2 Infection Can Be HLA-Dependent. Viruses 2021, 13, 2447. [Google Scholar] [CrossRef]
- Stasiak, M.; Zawadzka-Starczewska, K.; Lewiński, A. Significance of HLA Haplotypes in Two Patients with Subacute Thyroiditis Triggered by mRNA-Based COVID-19 Vaccine. Vaccines 2022, 10, 280. [Google Scholar] [CrossRef]
- Şendur, S.N.; Özmen, F.; Oğuz, S.H.; İremli, B.G.; Malkan, Ü.Y.; Gürlek, A.; Erbas, T.; Ünlütürk, U. Association of Human Leukocyte Antigen Genotypes with Severe Acute Respiratory Syndrome Coronavirus 2 Vaccine-Induced Subacute Thyroiditis. Thyroid 2022, 32, 640–647. [Google Scholar] [CrossRef]
- Wang, L.; Zou, Z.Q.; Wang, K. Clinical Relevance of HLA Gene Variants in HBV Infection. J. Immunol. Res. 2016, 2016, 9069375. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, A.; Magistroni, P.; Vespasiano, F.; Bella, A.; Bellino, S.; Puoti, F.; Alizzi, S.; Vaisitti, T.; Boros, S.; Grossi, P.A.; et al. HLA and AB0 Polymorphisms May Influence SARS-CoV-2 Infection and COVID-19 Severity. Transplantation 2021, 105, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka-Starczewska, K.; Tymoniuk, B.; Stasiak, B.; Lewiński, A.; Stasiak, M. Actual Associations Between HLA Haplotype and Graves’ Disease Development. J. Clin. Med. 2022, 11, 2492. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Spaepen, M.; Bex, M.; Bouillon, R.; Cassiman, J.J. Primary role of the HLA class II DRB1*0301 allele in Graves disease. Am. J. Med. Genet. 2000, 95, 432–437. [Google Scholar] [CrossRef]
- Stasiak, M.; Zawadzka-Starczewska, K.; Tymoniuk, B.; Stasiak, B.; Lewiński, A. Significance of HLA in the development of Graves’ orbitopathy. Genes. Immun. 2023, 24, 32–38. [Google Scholar] [CrossRef]
- Zeitlin, A.A.; Heward, J.M.; Newby, P.R.; Carr-Smith, J.D.; Franklyn, J.A.; Gough, S.C.; Simmonds, M.J. Analysis of HLA class II genes in Hashimoto’s thyroiditis reveals differences compared to Graves’ disease. Genes. Immun. 2008, 9, 358–363. [Google Scholar] [CrossRef]
- Baker, P.R.; Baschal, E.E.; Fain, P.R.; Triolo, T.M.; Nanduri, P.; Siebert, J.C.; Armstrong, T.K.; Babu, S.R.; Rewers, M.J.; Gottlieb, P.A.; et al. Haplotype analysis discriminates genetic risk for DR3-associated endocrine autoimmunity and helps define extreme risk for Addison’s disease. J. Clin. Endocrinol. Metab. 2010, 95, E263–E270. [Google Scholar] [CrossRef]
- Giraud, M.; Beaurain, G.; Yamamoto, A.M.; Eymard, B.; Tranchant, C.; Gajdos, P.; Garchon, H.J. Linkage of HLA to myasthenia gravis and genetic heterogeneity depending on anti-titin antibodies. Neurology 2001, 57, 1555–1560. [Google Scholar] [CrossRef]
- Hasan, Z.N.; Zalzala, H.H.; Mohammedsalih, H.R.; Mahdi, B.M.; Abid, L.A.; Shakir, Z.N.; Fadhel, M.J. Association between human leukocyte antigen-DR and demylinating Guillain-Barre syndrome. Neurosciences 2014, 19, 301–305. [Google Scholar]
- Rood, M.J.; van Krugten, M.V.; Zanelli, E.; van der Linden, M.W.; Keijsers, V.; Schreuder, G.M.; Verduyn, W.; Westendorp, R.G.; de Vries, R.R.; Breedveld, F.C.; et al. TNF-308A and HLA-DR3 alleles contribute independently to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2000, 43, 129–134. [Google Scholar] [CrossRef]
- Khan, F.; Sharma, P.; Pandey, S.; Sharma, D.; V, V.; Kumar, N.; Shukla, S.; Dandu, H.; Jain, A.; Garg, R.K.; et al. COVID-19-associated Guillain-Barre syndrome: Postinfectious alone or neuroinvasive too? J. Med. Virol. 2021, 93, 6045–6049. [Google Scholar] [CrossRef] [PubMed]
- Price, P.; Witt, C.; Allcock, R.; Sayer, D.; Garlepp, M.; Kok, C.C.; French, M.; Mallal, S.; Christiansen, F. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 1999, 167, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Lapa, D.; Trimarchi, F.; Vita, G.; Fallahi, P.; Antonelli, A.; Benvenga, S. Certain HLA alleles are associated with stress-triggered Graves’ disease and influence its course. Endocrine 2017, 55, 93–100. [Google Scholar] [CrossRef]
- Heward, J.M.; Allahabadia, A.; Daykin, J.; Carr-Smith, J.; Daly, A.; Armitage, M.; Dodson, P.M.; Sheppard, M.C.; Barnett, A.H.; Franklyn, J.A.; et al. Linkage disequilibrium between the human leukocyte antigen class II region of the major histocompatibility complex and Graves’ disease: Replication using a population case control and family-based study. J. Clin. Endocrinol. Metab. 1998, 83, 3394–3397. [Google Scholar]
- Barlow, A.B.; Wheatcroft, N.; Watson, P.; Weetman, A.P. Association of HLA-DQA1*0501 with Graves’ disease in English Caucasian men and women. Clin. Endocrinol. 1996, 44, 73–77. [Google Scholar] [CrossRef]
- Yanagawa, T.; Mangklabruks, A.; Chang, Y.B.; Okamoto, Y.; Fisfalen, M.E.; Curran, P.G.; DeGroot, L.J. Human histocompatibility leukocyte antigen-DQA1*0501 allele associated with genetic susceptibility to Graves’ disease in a Caucasian population. J. Clin. Endocrinol. Metab. 1993, 76, 1569–1574. [Google Scholar]
- Mangklabruks, A.; Cox, N.; DeGroot, L.J. Genetic factors in autoimmune thyroid disease analyzed by restriction fragment length polymorphisms of candidate genes. J. Clin. Endocrinol. Metab. 1991, 73, 236–244. [Google Scholar] [CrossRef]
- Ehrenstein, M.R.; Evans, J.G.; Singh, A.; Moore, S.; Warnes, G.; Isenberg, D.A.; Mauri, C. Compromised function of regulatory T cells in rheumatoid arthritis and reversal by anti-TNFalpha therapy. J. Exp. Med. 2004, 200, 277–285. [Google Scholar] [CrossRef]
- Ettinger, R.A.; Papadopoulos, G.K.; Moustakas, A.K.; Nepom, G.T.; Kwok, W.W. Allelic variation in key peptide-binding pockets discriminates between closely related diabetes-protective and diabetes-susceptible HLA-DQB1*06 alleles. J. Immunol. 2006, 176, 1988–1998. [Google Scholar] [CrossRef]
- Grommé, M.; Neefjes, J. Antigen degradation or presentation by MHC class I molecules via classical and non-classical pathways. Mol. Immunol. 2002, 39, 181–202. [Google Scholar] [CrossRef]
- Parham, P. Presentation of HLA class I-derived peptides: Potential involvement in allorecognition and HLA-B27-associated arthritis. Immunol. Rev. 1996, 154, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Möller, E. Mechanisms for induction of autoimmunity in humans. Acta Paediatr. Suppl. 1998, 424, 16–20. [Google Scholar] [CrossRef]
- Bowness, P. HLA B27 in health and disease: A double-edged sword? Rheumatology 2002, 41, 857–868. [Google Scholar] [CrossRef]
- Mobasheri, L.; Nasirpour, M.H.; Masoumi, E.; Azarnaminy, A.F.; Jafari, M.; Esmaeili, S.A. SARS-CoV-2 triggering autoimmune diseases. Cytokine 2022, 154, 155873. [Google Scholar] [CrossRef]
- Gusev, E.; Sarapultsev, A.; Solomatina, L.; Chereshnev, V. SARS-CoV-2-Specific Immune Response and the Pathogenesis of COVID-19. Int. J. Mol. Sci. 2022, 23, 1716. [Google Scholar] [CrossRef]
- Winchester, N.; Calabrese, C.; Calabrese, L. The intersection of COVID-19 and autoimmunity: What is our current understanding? Pathog. Immun. 2021, 6, 31. [Google Scholar] [CrossRef]
- Krakauer, T. Staphylococcal Superantigens: Pyrogenic Toxins Induce Toxic Shock. Toxins 2019, 11, 178. [Google Scholar] [CrossRef]
- Alouf, J.E.; Müller-Alouf, H. Staphylococcal and streptococcal superantigens: Molecular, biological and clinical aspects. Int. J. Med. Microbiol. 2003, 292, 429–440. [Google Scholar] [CrossRef]
- Cheng, M.H.; Zhang, S.; Porritt, R.A.; Noval Rivas, M.; Paschold, L.; Willscher, E.; Binder, M.; Arditi, M.; Bahar, I. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. USA 2020, 117, 25254–25262. [Google Scholar] [CrossRef]
- Porritt, R.A.; Paschold, L.; Rivas, M.N.; Cheng, M.H.; Yonker, L.M.; Chandnani, H.; Lopez, M.; Simnica, D.; Schultheiß, C.; Santiskulvong, C.; et al. HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children. J. Clin. Investig. 2021, 131, e146614. [Google Scholar] [CrossRef]
| Risk and Clinical Course | Allele | Sizes of Compared Groups | p-Value | Country [Ref] |
|---|---|---|---|---|
| Protection | A*03:01 | 159 vs. 52 * | 0.035 | Vietnam [29] |
| A*30:01 | 159 vs. 52 * | 0.017 | Vietnam [29] | |
| B*14:02 | 9373 vs. 5943 * | 0.006 | Spain [22] | |
| B*51:01 | 575 vs. 28,927 * | 0.0001 | Saudi Arabia [24] | |
| B*51:01 | 299 vs. 2781 * | <0.036 | Armenia [25] | |
| B*51:01 | 109 vs. 70 * | 0.027 | Iran [21] | |
| C*08:02 | 9373 vs. 5943 * | 0.024 | Spain [22] | |
| E*01:01 + E*01:01 | 122 vs. 68 * | 0.044 | Iran [30] | |
| DPB1*03:01 | 332 + | 0.03 | China [31] | |
| DRB1*11:05 | 109 vs. 70 * | 0.003 | Iran [21] | |
| DRB1*12:01 | 332 + | 0.04 | China [31] | |
| DRB1*13:05 | 109 vs. 70 * | 0.022 | Iran [21] | |
| DRB1*14:01 | 109 vs. 70 * | 0.006 | Iran [21] | |
| DRB1*15:01 | 159 vs. 52 * | 0.009 | Vietnam [29] | |
| DRB5*02:02 | 159 vs. 52 * | 0.004 | Vietnam [29] | |
| DQA1*01:02 | 159 vs. 52 * | 0.031 | Vietnam [29] | |
| Asymptomatic | B*15:01 | 1428 vs. 29,947 * | 0.00006 | USA [32] |
| infection | DRB1*04:01 | 49 vs. 69 a | 0.003 | UK [33] |
| Infection | A*30:02 | 234 vs. 22,000 * | 0.0017 | USA [34] |
| (all clinical courses) | B*07:35 | 109 vs. 70 * | 0.031 | Iran [21] |
| B*27:07 | 99 vs. 1017 * | 0.00001 | Italy [35] | |
| C*04:01 | 182 vs. 619 * | 0.012 | Italy (Sardinia) [36] | |
| DRB1*07:01 | 109 vs. 70 * | 0.003 | Iran [21] | |
| DRB1*08:02 | 234 vs. 22,000 * | 0.01 | USA [34] | |
| DRB1*09:01 | 159 vs. 52 * | 0.044 | Vietnam [29] | |
| DRB1*15:01 | 99 vs. 1017 * | 0.0015 | Italy [35] | |
| DRB1*15:01 | 575 vs. 28,927 * | <0.0001 | Saudi Arabia [24] | |
| DQB1*06:02 | 99 vs. 1017 * | 0.0001 | Italy [35] | |
| Mild disease | A*02:01:01 | 575 vs. 28,927 * | 0.0114 | Saudi Arabia [24] |
| DRB1*05:01 | 159 vs. 52 * | 0.034 | Vietnam [29] | |
| DPA1*02:01 | 159 vs. 52 * | 0.012 | Vietnam [29] | |
| DRB1*09:01 | 159 vs. 52 * | 0.026 | Vietnam [29] | |
| Moderate disease | DPA1*01:03 | 159 vs. 52 * | 0.003 | Vietnam [29] |
| DRB1*04:01 | 109 vs. 70 * | 0.0002 | Iran [21] | |
| Severe disease | A*11:01 | 9373 vs. 5943 * | 0.033 | Spain [22] |
| A*11:01 | 137 vs. 53 s | 0.015 | Japan [37] | |
| A*11:01 | Epi | 0.003 | Japan [38] | |
| A*11:01 | 332 + | 0.008 | China [31] | |
| B*50:01 | 575 vs. 28,927 * | <0.0001 | Saudi Arabia [24] | |
| B*51:01 | 332 + | 0.007 | China [31] | |
| C*04:01 | 9373 vs. 5943 * | 0.045 | Spain [22] | |
| C*04:01 | 299 vs. 2781 * | <0.021 | Armenia [25] | |
| C*04:01 | 435 + | 0.00011 | Germany, Spain, Switzerland, US [23] | |
| C*04:01 | 54 vs. 42 a | 0.02 | India [38] | |
| C*06:02 | 575 vs. 28,927 * | <0.0001 | Saudi Arabia [24] | |
| C*14:02 | 332 + | 0.003 | China [31] | |
| E*01:03 + E01:03 | 122 vs. 68 * | 0.02 | Iran [30] | |
| F*01:01 | 159 vs. 52 * | <0.001 | Vietnam [29] | |
| F*01:03 | 159 vs. 52 * | 0.0028 | Vietnam [29] | |
| DPA1*01:03 | 54 vs. 42 a | 0.001 | India [39] | |
| DPB1*04:01 | 159 vs. 52 * | 0.001 | Vietnam [29] | |
| DRB1*01:01 | 332 + | 0.02 | China [31] | |
| DRB1*04:03 | 109 vs. 70 * | 0.004 | Iran [21] | |
| DRB1*07:01 | 575 vs. 28,927 * | 0.0047 | Saudi Arabia [24] | |
| DRB1*08:01 | 39 vs. 143 | 0.024 | Italy (Sardinia) [36] | |
| DRB1*09:01 | 73 s vs. 105 | 0.003 | Japan [40] | |
| DRB1*11:01 | 109 vs. 70 * | <0.001 | Iran [21] | |
| DRB1*14:04 | 332 + | 0.01 | China [31] | |
| DRB3*01:01 | 8 s vs. 8 | 0.0064 | China [41] | |
| DRB5*01:01 | 54 vs. 42 a | 0.03 | India [38] | |
| DQA1*01:01 | 332 + | 0.04 | China [31] | |
| DQA1*01:02 | 159 vs. 52 * | 0.004 | Vietnam [29] | |
| DQA1*01:03 | 209 + | 0.0013 | Japan [42] | |
| DQA1*03:01 | 54 vs. 42 a | 0.03 | India [38] | |
| DQB1*05:02 | 159 vs. 52 * | 0.008 | Vietnam [29] | |
| DQB1*06:01 | 209 + | 0.013 | Japan [41] |
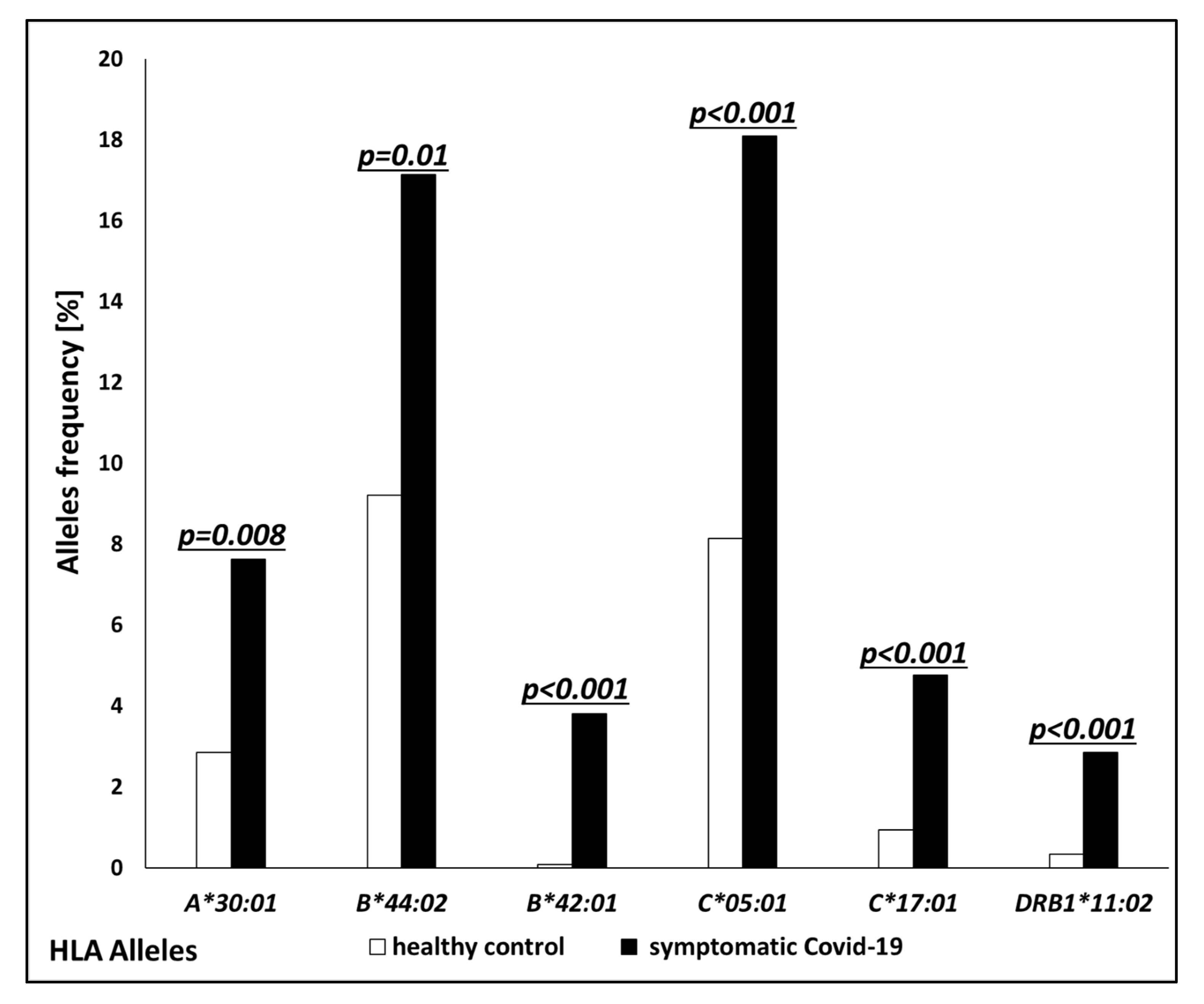
| Allele | Association | Symptomatic COVID-19; n = 105 No. of Carriers (%) | Healthy Control; n = 2217 No. of Carriers (%) | p-Value | Odds Ratio (OR) |
|---|---|---|---|---|---|
| A*30:01 | risk | 8 (7.62%) | 66 (2.84%) | 0.008 | 2.7 |
| B*44:02 | risk | 18 (17.14%) | 214 (9.22%) | 0.01 | 1.9 |
| B*42:01 | risk | 4 (3.81%) | 2 (0.09%) | <0.001 | 43.9 |
| C*05:01 | risk | 19 (18.1%) | 189 (8.14%) | <0.001 | 2.4 |
| C*17:01 | risk | 5 (4.76%) | 22 (0.95%) | <0.001 | 5.0 |
| C*07:04 | protective | 0 (0.0%) | 99 (4.26%) | 0.02 | - |
| DRB1*11:02 | risk | 3 (2.86%) | 8 (0.34%) | <0.001 | 8.1 |
| DQB1*03:03 | protective | 3 (2.86%) | 210 (9.04%) | <0.001 | 0.3 |
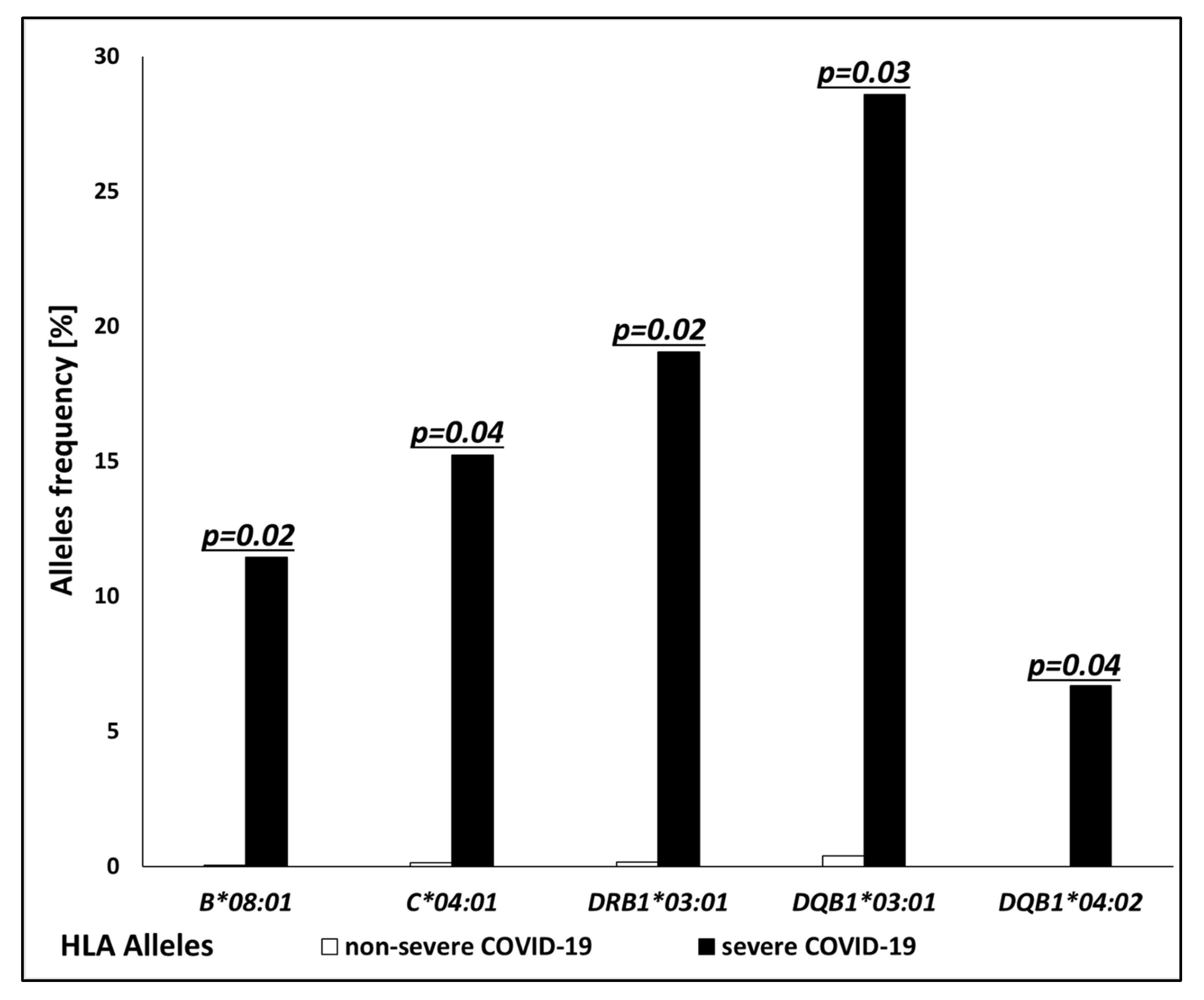
| Allele | Association | Severe COVID-19; n = 67 No. of Carriers (%) | Non-Severe COVID-19; n = 38 No. of Carriers (%) | p-Value | Odds Ratio (OR) |
|---|---|---|---|---|---|
| B*08:01 | risk | 12 (11.43%) | 1 (0.04%) | 0.02 | 8.1 |
| C*04:01 | risk | 16 (15.24%) | 3 (0.13%) | 0.04 | 3.7 |
| DRB1*03:01 | risk | 20 (19.05%) | 4 (0.17%) | 0.02 | 3.6 |
| DQB1*03:01 | risk | 30 (28.57%) | 9 (0.39%) | 0.03 | 2.6 |
| DQB1*04:02 | risk | 7 (6.67%) | 0 (0.0%) | 0.04 | - |
| DRB1*08:01 | protective | 1 (0.22%) | 5 (0.95%) | 0.01 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tymoniuk, B.; Borowiec, M.; Makowska, J.; Holwek, E.; Sarnik, J.; Styrzyński, F.; Dróżdż, I.; Lewiński, A.; Stasiak, M. Associations Between Clinical Manifestations of SARS-CoV-2 Infection and HLA Alleles in a Caucasian Population: A Molecular HLA Typing Study. J. Clin. Med. 2024, 13, 7695. https://doi.org/10.3390/jcm13247695
Tymoniuk B, Borowiec M, Makowska J, Holwek E, Sarnik J, Styrzyński F, Dróżdż I, Lewiński A, Stasiak M. Associations Between Clinical Manifestations of SARS-CoV-2 Infection and HLA Alleles in a Caucasian Population: A Molecular HLA Typing Study. Journal of Clinical Medicine. 2024; 13(24):7695. https://doi.org/10.3390/jcm13247695
Chicago/Turabian StyleTymoniuk, Bogusław, Maciej Borowiec, Joanna Makowska, Emilia Holwek, Joanna Sarnik, Filip Styrzyński, Izabela Dróżdż, Andrzej Lewiński, and Magdalena Stasiak. 2024. "Associations Between Clinical Manifestations of SARS-CoV-2 Infection and HLA Alleles in a Caucasian Population: A Molecular HLA Typing Study" Journal of Clinical Medicine 13, no. 24: 7695. https://doi.org/10.3390/jcm13247695
APA StyleTymoniuk, B., Borowiec, M., Makowska, J., Holwek, E., Sarnik, J., Styrzyński, F., Dróżdż, I., Lewiński, A., & Stasiak, M. (2024). Associations Between Clinical Manifestations of SARS-CoV-2 Infection and HLA Alleles in a Caucasian Population: A Molecular HLA Typing Study. Journal of Clinical Medicine, 13(24), 7695. https://doi.org/10.3390/jcm13247695








