Machine Learning Model Based on Optimized Radiomics Feature from 18F-FDG-PET/CT and Clinical Characteristics Predicts Prognosis of Multiple Myeloma: A Preliminary Study
Abstract
1. Introduction
2. Methods
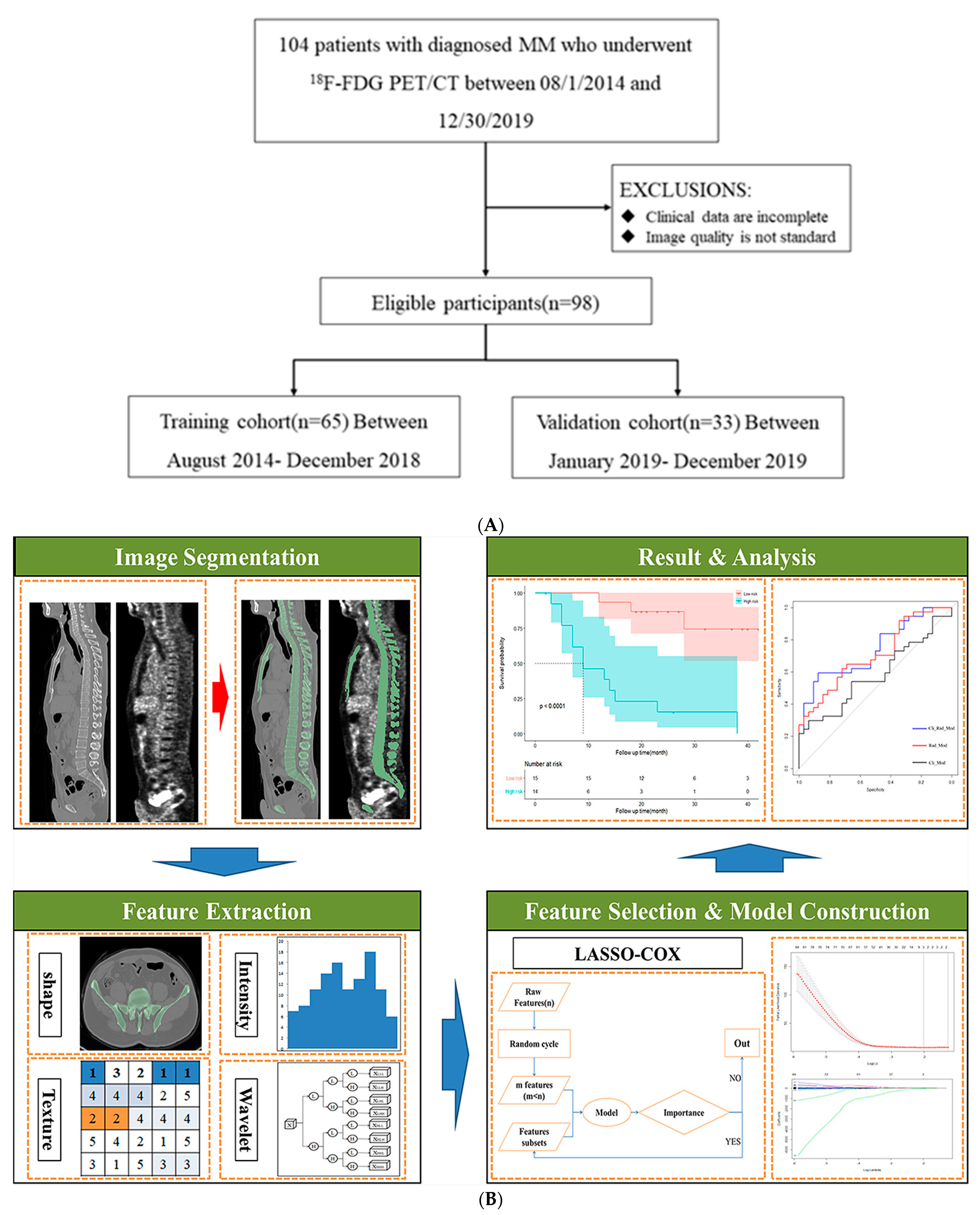
2.1. Study Design and PATIENTS
2.2. Data Collection
2.3. PET/CT Image Acquisition
2.4. Image Preprocessing
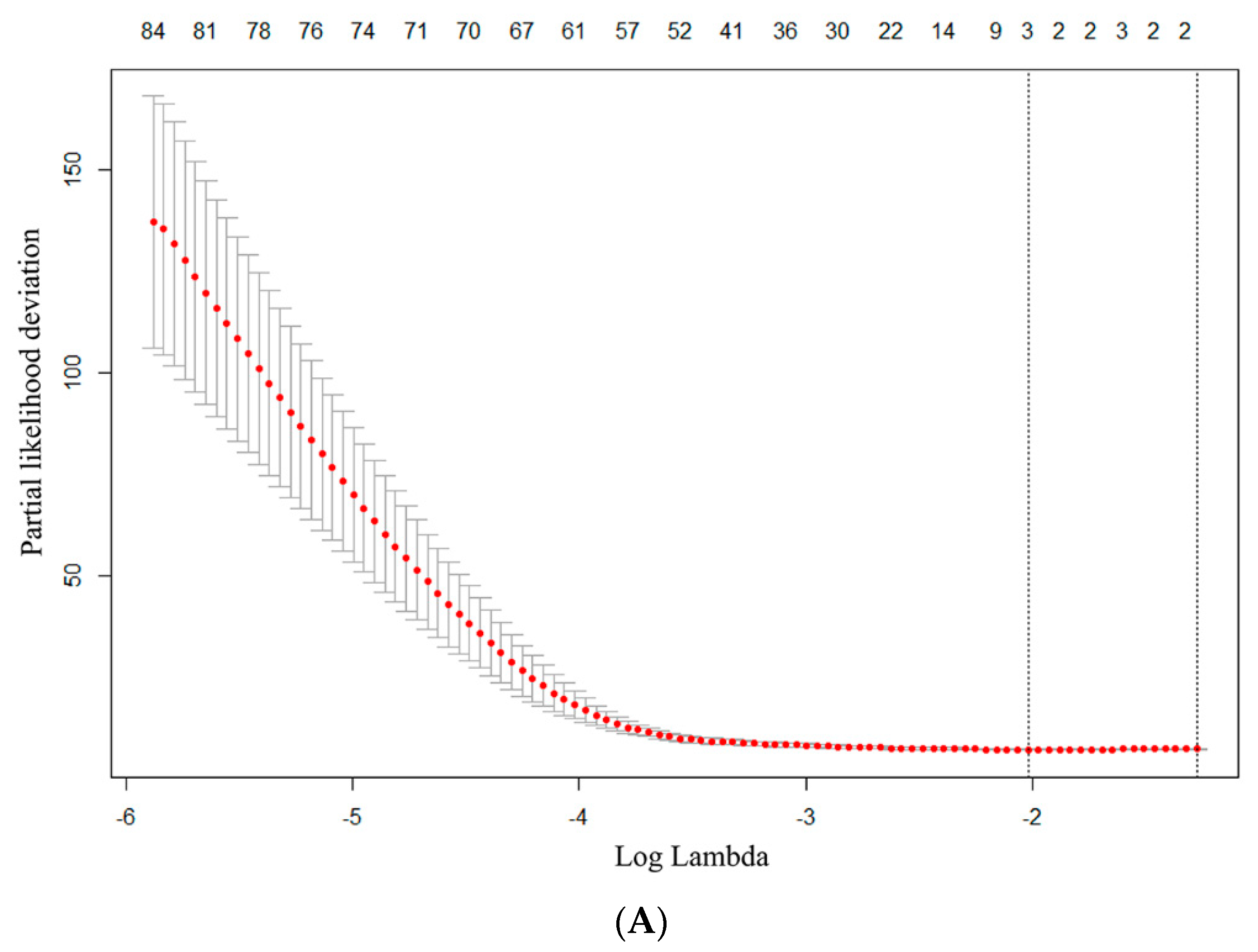
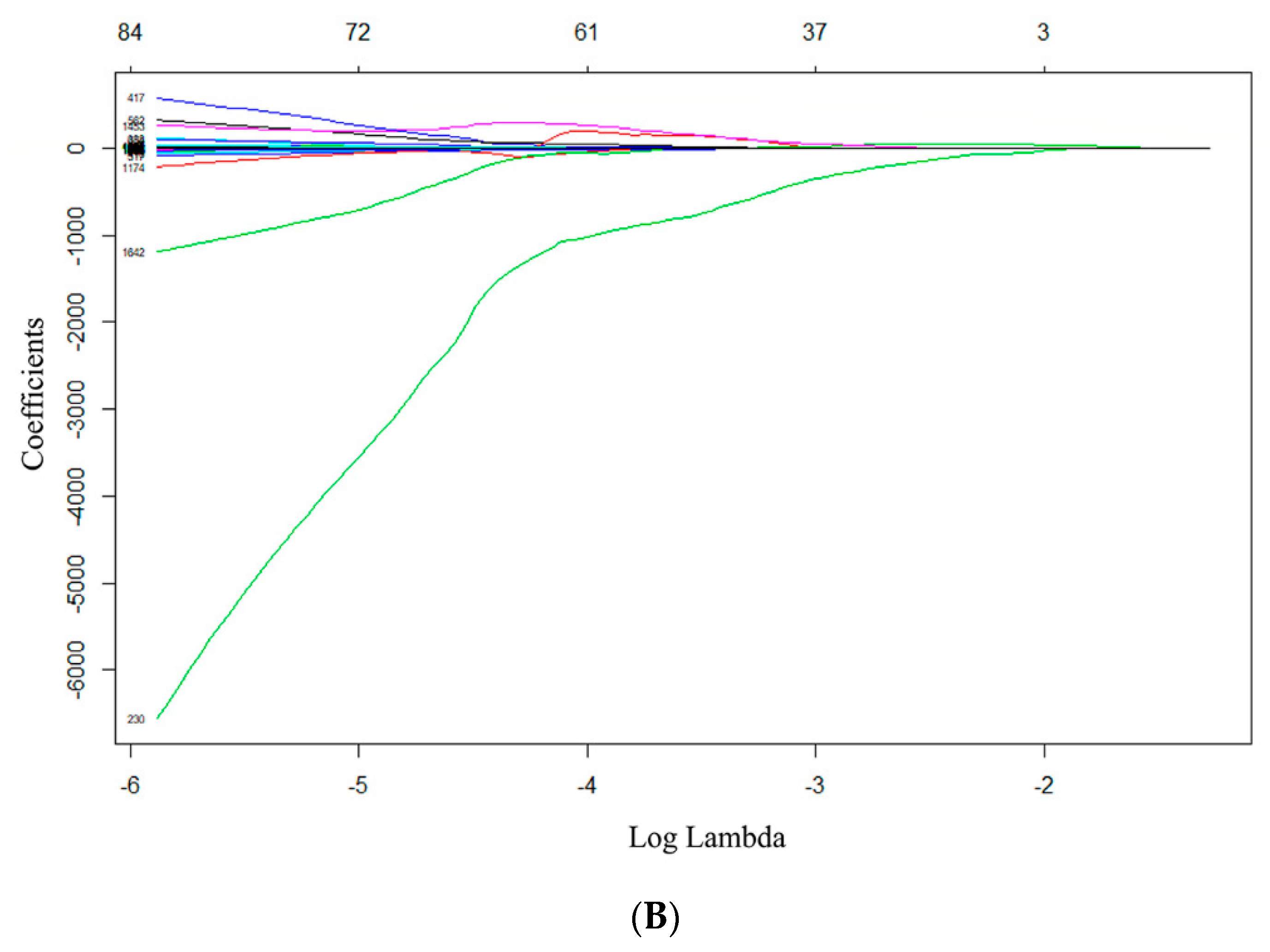
2.5. Radiomics Features Extraction and Selection
2.6. Predictive Model Establishment and Statistical Analysis
3. Result
3.1. Baseline Clinical Characteristics
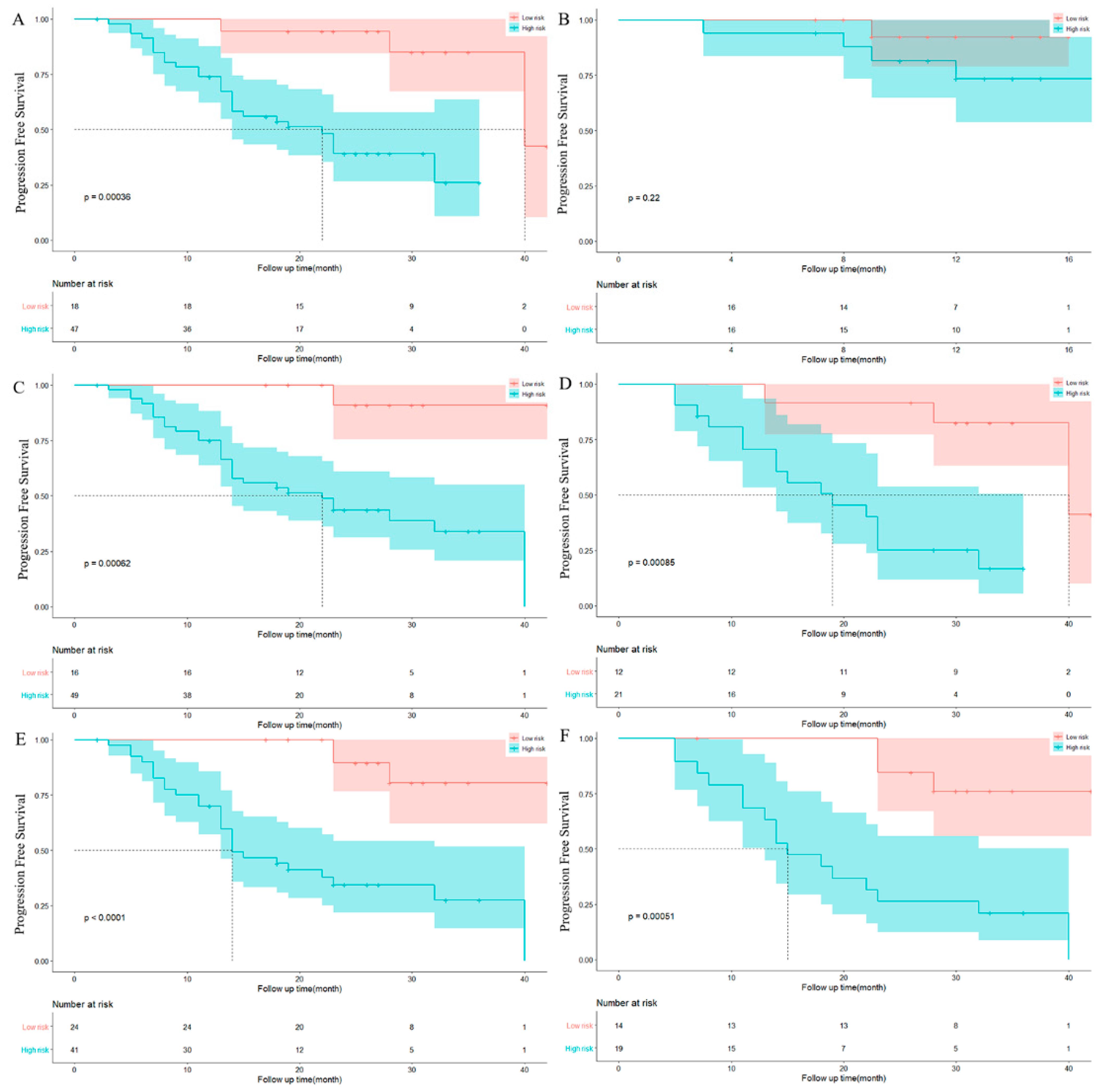
3.2. Feature Selection and Model Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smith, D.; Yong, K. Advances in understanding prognosis in myeloma. Br. J. Haematol. 2016, 175, 367–380. [Google Scholar] [CrossRef]
- Durie, B.G.; Salmon, S.E. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer 1975, 36, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Greipp, P.R.; Miguel, J.S.; Durie, B.G.; Crowley, J.J.; Barlogie, B.; Bladé, J.; Boccadoro, M.; Child, J.A.; Avet-Loiseau, H.; Kyle, R.A.; et al. International staging system for multiple myeloma. J. Clin. Oncol. 2005, 23, 3412–3420. [Google Scholar] [CrossRef]
- Mikhael, J.R.; Dingli, D.; Roy, V.; Reeder, C.B.; Buadi, F.K.; Hayman, S.R.; Dispenzieri, A.; Fonseca, R.; Sher, T.; Kyle, R.A.; et al. Management of newly diagnosed symptomatic multiple myeloma: Updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin. Proc. 2013, 88, 360–376. [Google Scholar] [CrossRef]
- Zamagni, E.; Patriarca, F.; Nanni, C.; Zannetti, B.; Englaro, E.; Pezzi, A.; Tacchetti, P.; Buttignol, S.; Perrone, G.; Brioli, A.; et al. Prognostic relevance of 18-F FDG PET/CT in newly diagnosed multiple myeloma patients treated with up-front autologous transplantation. Blood J. Am. Soc. Hematol. 2011, 118, 5989–5995. [Google Scholar] [CrossRef]
- Usmani, S.Z.; Mitchell, A.; Waheed, S.; Crowley, J.; Hoering, A.; Petty, N.; Brown, T.; Bartel, T.; Anaissie, E.; van Rhee, F.; et al. Prognostic implications of serial 18-fluoro-deoxyglucose emission tomography in multiple myeloma treated with total therapy 3. Blood J. Am. Soc. Hematol. 2013, 121, 1819–1823. [Google Scholar] [CrossRef]
- Fonti, R.; Larobina, M.; Del Vecchio, S.; De Luca, S.; Fabbricini, R.; Catalano, L.; Pane, F.; Salvatore, M.; Pace, L. Metabolic tumor volume assessed by 18F-FDG PET/CT for the prediction of outcome in patients with multiple myeloma. J. Nucl. Med. 2012, 53, 1829–1835. [Google Scholar] [CrossRef]
- McDonald, J.E.; Kessler, M.M.; Gardner, M.W.; Buros, A.F.; Ntambi, J.A.; Waheed, S.; van Rhee, F.; Zangari, M.; Heuck, C.J.; Petty, N.; et al. Assessment of Total Lesion Glycolysis by 18F FDG PET/CT Significantly Improves Prognostic Value of GEP and ISS in Myeloma. Clin. Cancer Res. 2017, 23, 1981–1987. [Google Scholar] [CrossRef]
- Cavo, M.; Terpos, E.; Nanni, C.; Moreau, P.; Lentzsch, S.; Zweegman, S.; Hillengass, J.; Engelhardt, M.; Usmani, S.Z.; Vesole, D.H.; et al. Role of 18F-FDG PET/CT in the diagnosis and management of multiple myeloma and other plasma cell disorders: A consensus statement by the International Myeloma Working Group. Lancet Oncol. 2017, 18, e206–e217. [Google Scholar] [CrossRef] [PubMed]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P. Introduction to Radiomics. J Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Chetan, M.R.; Gleeson, F.V. Radiomics in predicting treatment response in non-small-cell lung cancer: Current status, challenges and future perspectives. Eur. Radiol. 2021, 31, 1049–1058. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, Y.; Wang, Y.; Jiang, S.; Fan, R.; Zhang, H.; Jiang, W. Application of radiomics and machine learning in head and neck cancers. Int. J. Biol. Sci. 2021, 17, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, L.; Liu, S.; You, J.; Chen, L.; Jin, Z.; Zhang, S.; Zhang, B. Radiomics in precision medicine for gastric cancer: Opportunities and challenges. Eur. Radiol. 2022, 32, 5852–5868. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zeng, P.; Guo, W.; Wang, C.; Geng, Y.; Lang, N.; Yuan, H. Prediction of High-Risk Cytogenetic Status in Multiple Myeloma Based on Magnetic Resonance Imaging: Utility of Radiomics and Comparison of Machine Learning Methods. J. Magn. Reson. Imaging 2021, 54, 1303–1311. [Google Scholar] [CrossRef] [PubMed]
- Moulopoulos, L.A.; Dimopoulos, M.A.; Kastritis, E.; Christoulas, D.; Gkotzamanidou, M.; Roussou, M.; Koureas, A.; Migkou, M.; Gavriatopoulou, M.; Eleutherakis-Papaiakovou, E.; et al. Diffuse pattern of bone marrow involvement on magnetic resonance imaging is associated with highrisk cytogenetics and poor outcome in newly diagnosed, symptomatic patients with multiple myeloma: A single center experience on 228 patients. Am. J. Hematol. 2012, 87, 861–864. [Google Scholar] [CrossRef]
- Milara, E.; Gómez-Grande, A.; Tomás-Soler, S.; Seiffert, A.P.; Alonso, R.; Gómez, E.J.; Martínez-López, J.; Sánchez-González, P. Bone marrow segmentation and radiomics analysis of [18F]FDG PET/CT images for measurable residual disease assessment in multiple myeloma. Comput. Methods Programs Biomed. 2022, 225, 107083. [Google Scholar] [CrossRef]
- Li, Y.; Yin, P.; Liu, Y.; Hao, C.; Chen, L.; Sun, C.; Wang, S.; Hong, N. Radiomics Models Based on Magnetic Resonance Imaging for Prediction of the Response to Bortezomib-Based Therapy in Patients with Multiple Myeloma. Biomed. Res. Int. 2022, 2022, 6911246. [Google Scholar] [CrossRef] [PubMed]
- Ekert, K.; Hinterleitner, C.; Baumgartner, K.; Fritz, J.; Horger, M. Extended Texture Analysis of Non-Enhanced Whole-Body MRI Image Data for Response Assessment in Multiple Myeloma Patients Undergoing Systemic Therapy. Cancers 2020, 12, 761. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, X.Y.; Shi, Y.J.; Wang, L.; Zhu, H.T.; Tang, Z.; Wang, S.; Li, X.T.; Tian, J.; Sun, Y.S. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin. Cancer Res. 2017, 23, 7253–7262. [Google Scholar] [CrossRef]
- Sonneveld, P.; Avet-Loiseau, H.; Lonial, S.; Usmani, S.; Siegel, D.; Anderson, K.C.; Chng, W.J.; Moreau, P.; Attal, M.; Kyle, R.A.; et al. Treatment of multiple myeloma with high-risk cytogenetics: A consensus of the International Myeloma Working Group. Blood J. Am. Soc. Hematol. 2016, 127, 2955–2962. [Google Scholar] [CrossRef]
- Mesguich, C.; Hindie, E.; de Senneville, B.D.; Tlili, G.; Pinaquy, J.B.; Marit, G.; Saut, O. Improved 18-FDG PET/CT diagnosis of multiple myeloma diffuse disease by radiomics analysis. Nucl. Med. Commun. 2021, 42, 1135–1143. [Google Scholar] [CrossRef]
- Chng, W.J.; Dispenzieri, A.; Chim, C.S.; Fonseca, R.; Goldschmidt, H.; Lentzsch, S.; Munshi, N.; Palumbo, A.; Miguel, J.S.; Sonneveld, P.; et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia 2014, 28, 269–277. [Google Scholar] [CrossRef]
- Munshi, N.C.; Anderson, K.C.; Bergsagel, P.L.; Shaughnessy, J.; Palumbo, A.; Durie, B.; Fonseca, R.; Stewart, A.K.; Harousseau, J.L.; Dimopoulos, M.; et al. Consensus recommendations for risk stratification in multiple myeloma: Report of the International Myeloma Workshop Consensus Panel 2. Blood J. Am. Soc. Hematol. 2011, 117, 4696–4700. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2020, 95, 548–567. [Google Scholar] [CrossRef]
- Wennmann, M.; Klein, A.; Bauer, F.; Chmelik, J.; Grözinger, M.; Uhlenbrock, C.; Lochner, J.; Nonnenmacher, T.; Rotkopf, L.T.; Sauer, S.; et al. Combining Deep Learning and Radiomics for Automated, Objective, Comprehensive Bone Marrow Characterization From Whole-Body MRI: A Multicentric Feasibility Study. Investig. Radiol. 2022, 57, 752–763. [Google Scholar] [CrossRef]
- Wu, Z.; Bian, T.; Dong, C.; Duan, S.; Fei, H.; Hao, D.; Xu, W. Spinal MRI-Based Radiomics Analysis to Predict Treatment Response in Multiple Myeloma. J. Comput. Assist. Tomogr. 2022, 46, 447–454. [Google Scholar] [CrossRef]
- Jamet, B.; Morvan, L.; Nanni, C.; Michaud, A.V.; Bailly, C.; Chauvie, S.; Moreau, P.; Touzeau, C.; Zamagni, E.; Bodet-Milin, C.; et al. Random survival forest to predict transplant-eligible newly diagnosed multiple myeloma outcome including FDG-PET radiomics: A combined analysis of two independent prospective European trials. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1005–1015. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Cea, M.; Rossi, F.; Valdora, F.; Bignotti, B.; Succio, G.; Gualco, S.; Conte, A.; Dominietto, A. Differentiating diffuse from focal pattern on Computed Tomography in multiple myeloma: Added value of a Radiomics approach. Eur. J. Radiol. 2019, 121, 108739. [Google Scholar] [CrossRef]
- Wennmann, M.; Bauer, F.; Klein, A.; Chmelik, J.; Grözinger, M.; Rotkopf, L.T.; Neher, P.; Gnirs, R.; Kurz, F.T.; Nonnenmacher, T.; et al. In Vivo Repeatability and Multiscanner Reproducibility of MRI Radiomics Features in Patients with Monoclonal Plasma Cell Disorders: A Prospective Bi-institutional Study. Investig. Radiol. 2022, 10–1097. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Vallières, M.; Löck, S. Initiative for the IBS. Image Biomarker Standardisation Initiative. 2016. Available online: http://arxiv.org/abs/1612.07003 (accessed on 12 March 2022).
- Stytz, M.R.; Parrott, R.W. Using kriging for 3d medical imaging. Comput. Med. Imaging Graph. 1993, 17, 421–442. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillon-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
| Variables | |
|---|---|
| Median age, years (range) | 65.0 (41.0–86.0) |
| ≥65 years, n (%) | 52 (53.1%) |
| gender, n (%) | |
| Male | 45 (45.9%) |
| Female | 53 (54.1%) |
| Immunoglobulin (Ig) type, n (%) | |
| IgG | 48 (49.0%) |
| IgA | 25 (25.5%) |
| IgD | 7 (7.1%) |
| Light chain only | 18 (18.4%) |
| International Staging System (ISS), n (%) | |
| I | 11 (11.2%) |
| II | 49 (50.0%) |
| III | 38 (38.8%) |
| ECOG PS ≥ 2, n (%) | 22 (22.4%) |
| Bone marrow plasmacyte ratio (BMPC ratio) ≥ 60% | 12 (12.3%) |
| Hemoglobin (g/L) < 100, n (%) | 65 (66.3%) |
| LDH > (1 × ULN), n (%) | 25 (25.5%) |
| β2MG (mg/L) ≥ 5.5 | 55 (56.1%) |
| Albumin (g/L) < 35, n (%) | 58 (59.2%) |
| Calcium (mmol/L) > 2.65 | 12 (12.2%) |
| Creatinine (mg/dL) ≥ 2 | 32 (32.7%) |
| Cytogenetic abnormality (79/98) | |
| High risk | 26 (32.9%) |
| Standard risk | 53 (67.1%) |
| Frontline treatment, n (%) | |
| IMiD-based | 5 (5.1%) |
| Proteasome inhibitor-based | 62 (63.3%) |
| IMiD + proteasome inhibitor | 17 (17.3%) |
| Daratumumab-based | 14 (14.3%) |
| Performance of ASCT, n (%) | 22 (22.4%) |
| Best response (92/98) | |
| CR (complete remission) | 32 (34.8%) |
| Not reach CR | 60 (65.2%) |
| Variable | Univariate Cox Regression | Multivariate Cox Regression | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Gender | 1.353 (0.483, 3.793) | 0.565 | ||
| Age | 1.025 (0.957, 1.097) | 0.479 | ||
| ISS staging | 7.169 (1.141, 45.025) | 0.036 | 1.147 (0.665, 1.978) | 0.621 |
| BMPC ratio | 1.000 (0.975, 1.025) | 0.998 | ||
| β2MG (mg/L) | 1.007 (0.998, 1.016) | 0.113 | ||
| Albumin (g/L) | 1.015 (0.966, 1.066) | 0.560 | ||
| Calcium (mmol/L) | 1.101 (0.380, 3.188) | 0.860 | ||
| Creatinine (mg/dI) | 0.984 (0.953, 1.015) | 0.311 | ||
| High-risk cytogenetics | 1.301 (0.257, 6.600) | 0.750 | ||
| Ki-67 | 1.003 (0.978, 1.028) | 0.844 | ||
| LDH | 1.005 (1.001, 1.010) | 0.028 * | 1.004 (1.000, 1.008) | 0.034 * |
| Bone destruction | 0.188 (0.031, 1.162) | 0.072 | ||
| Focal lesion | 4.603 (1.140, 18.596) | 0.032 * | 1.976 (0.878, 4.447) | 0.100 |
| SUVmax | 1.189 (1.077, 1.314) | 0.001 * | 1.114 (1.043, 1.189) | 0.001 * |
| Orde | Wavelet-Transformation | Imaging Parameter | Radiomics Feature | Feature Type |
|---|---|---|---|---|
| 1 | LHL | PET | Idmn | glcm |
| 2 | LHL | PET | LDLGLE | gldm |
| 3 | LHL | PET | LALGLE | glszm |
| Model | Training Cohort | Validation Cohort | ||
|---|---|---|---|---|
| C-index | (95% CI) | C-Index | (95% CI) | |
| Cli_Mod | 0.736 | 0.401, 1.600 | 0.563 | −0.641, 1.021 |
| Rad_Mod | 0.675 | 0.376, 1.624 | 0.651 | −0.597, 3.270 |
| Cli_Rad_Mod | 0.790 | 0.560, 1.442 | 0.698 | −0.346, 1.048 |
| Model | Validation Cohort (1 Y) NRI (95% CI) | Validation Cohort (3 Y) NRI (95% CI) |
|---|---|---|
| Cli_Mod | Reference | Reference |
| Cli_Rad_Mod | 0.482 (−0.149, 1.131) | 0.497 (0.142, 1.131) |
| Rad_Mod | Reference | Reference |
| Cli_Rad_Mod | 0.739 (0.350, 1.380) | 0.623 (0.360, 1.133) |
| Training Cohort | Validation Cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| AUC | SEN | SPE | p Value | AUC | SEN | SPE | p Value | |
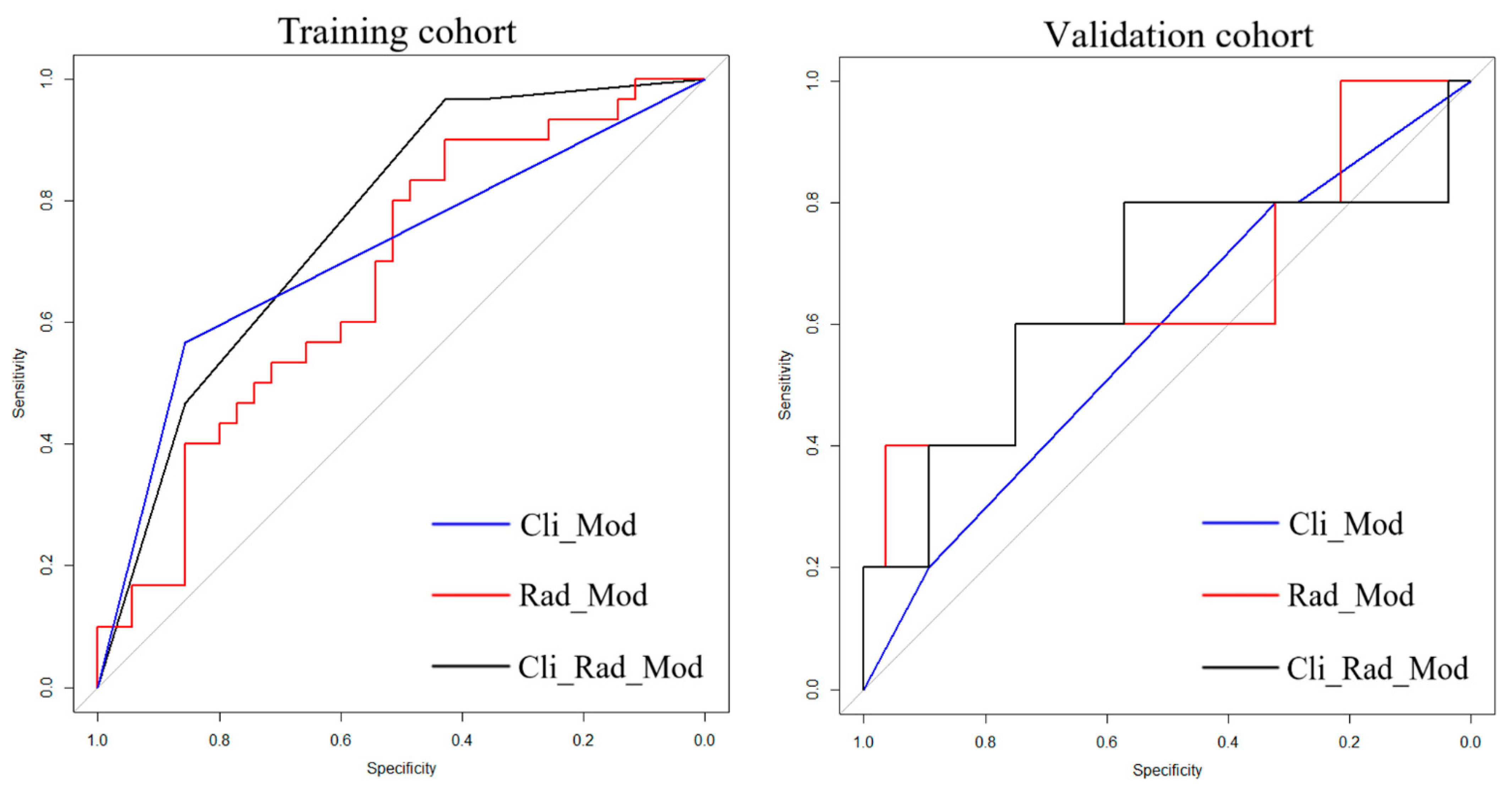
| Cli_Mod | 0.692 (0.542, 0.804) | 0.667 | 0.600 | Reference | 0.582 (0.305, 0.860) | 0.600 | 0.821 | Reference |
| Rad_Mod | 0.673 (0.645, 0.877) | 0.500 | 0.886 | 0.328 | 0.650 (0.350, 0.950) | 0.600 | 0.786 | 0.058 |
| Cli_Rad_Mod | 0.761 (0.698, 0.906) | 0.567 | 0.857 | 0.042 * | 0.650 (0.339, 0.961) | 0.800 | 0.786 | 0.036 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, B.; Huang, G.; Huang, H.; Wang, T.; Han, X.; Shen, L.; Chen, Y.; Hou, J. Machine Learning Model Based on Optimized Radiomics Feature from 18F-FDG-PET/CT and Clinical Characteristics Predicts Prognosis of Multiple Myeloma: A Preliminary Study. J. Clin. Med. 2023, 12, 2280. https://doi.org/10.3390/jcm12062280
Ni B, Huang G, Huang H, Wang T, Han X, Shen L, Chen Y, Hou J. Machine Learning Model Based on Optimized Radiomics Feature from 18F-FDG-PET/CT and Clinical Characteristics Predicts Prognosis of Multiple Myeloma: A Preliminary Study. Journal of Clinical Medicine. 2023; 12(6):2280. https://doi.org/10.3390/jcm12062280
Chicago/Turabian StyleNi, Beiwen, Gan Huang, Honghui Huang, Ting Wang, Xiaofeng Han, Lijing Shen, Yumei Chen, and Jian Hou. 2023. "Machine Learning Model Based on Optimized Radiomics Feature from 18F-FDG-PET/CT and Clinical Characteristics Predicts Prognosis of Multiple Myeloma: A Preliminary Study" Journal of Clinical Medicine 12, no. 6: 2280. https://doi.org/10.3390/jcm12062280
APA StyleNi, B., Huang, G., Huang, H., Wang, T., Han, X., Shen, L., Chen, Y., & Hou, J. (2023). Machine Learning Model Based on Optimized Radiomics Feature from 18F-FDG-PET/CT and Clinical Characteristics Predicts Prognosis of Multiple Myeloma: A Preliminary Study. Journal of Clinical Medicine, 12(6), 2280. https://doi.org/10.3390/jcm12062280





