The Landscape of Lipid Metabolism in Lung Cancer: The Role of Structural Profiling
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Chemicals and Reagents
2.3. Sample Collection and Preparation
2.4. HPLC-MS/MS Analysis
2.5. Data Processing and Statistical Analysis
3. Results
3.1. Characteristics of the Participants
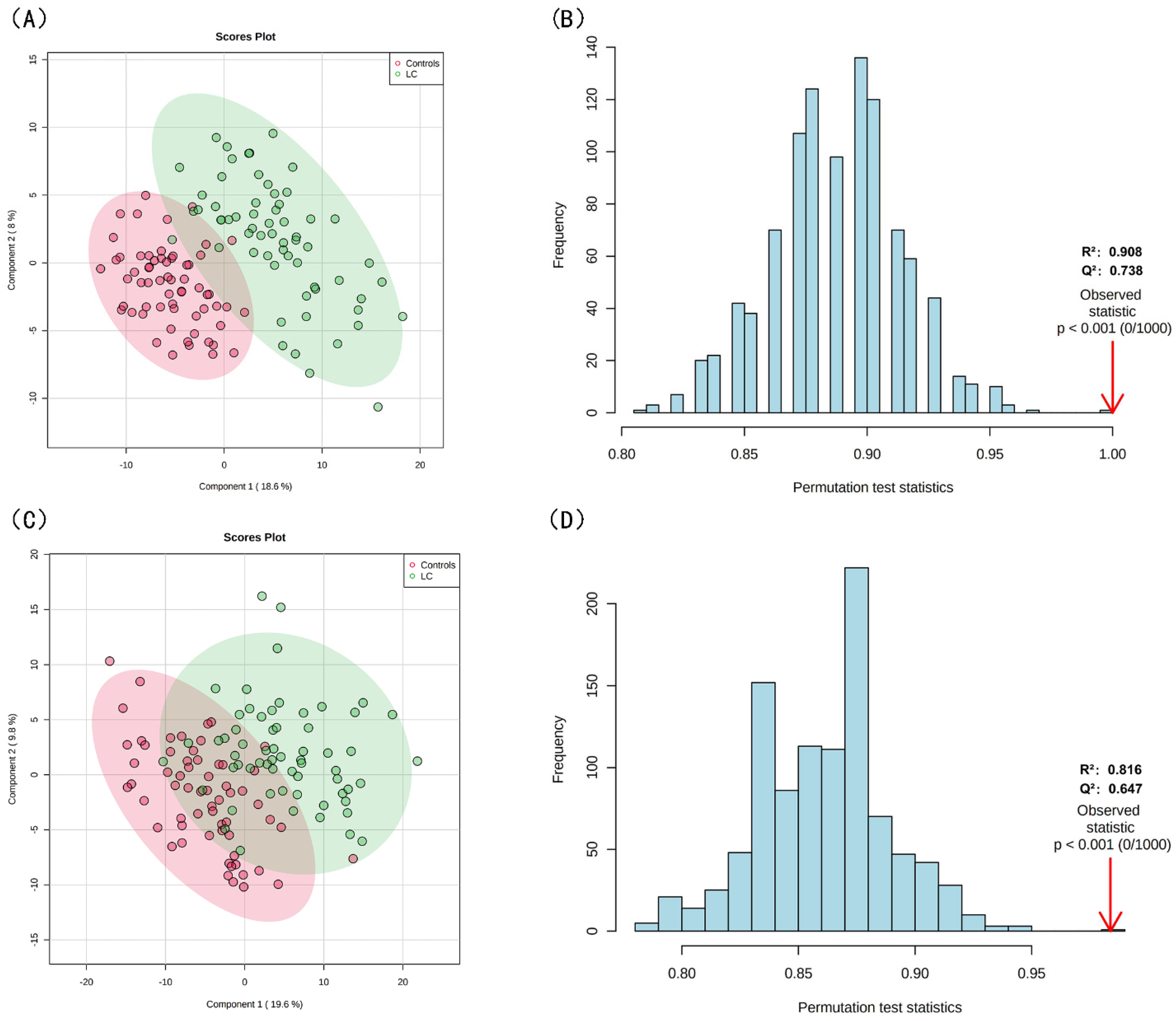
3.2. Lipid Profiling Grouped by Lipid Structure and Risk of LC
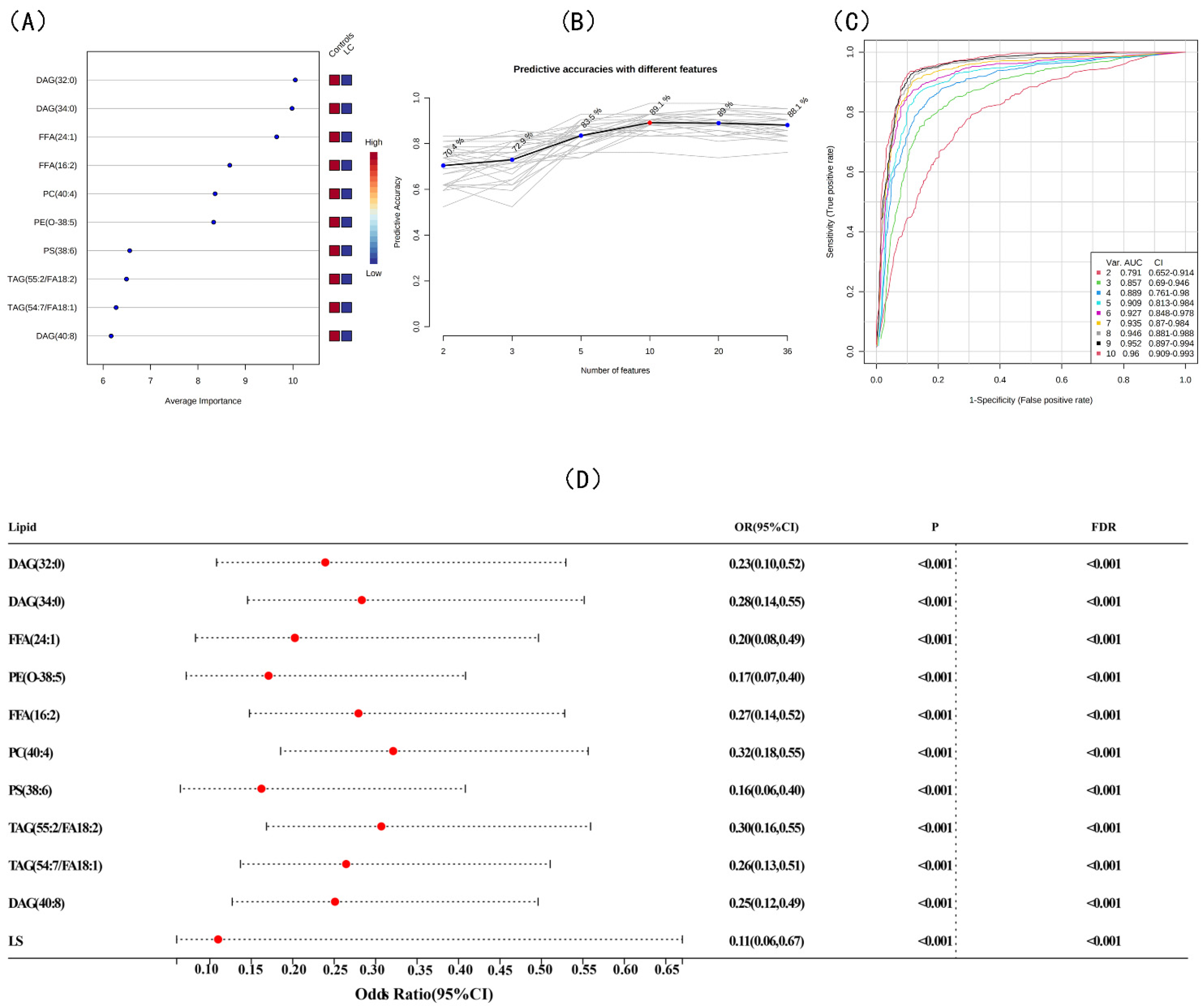
3.3. Screening for Differential Lipids and the Risk of LC
3.4. Mediation Effects for Blood Indexes between Lipid Biomarkers and LC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Wang, G.; Qiu, M.; Xing, X.; Zhou, J.; Yao, H.; Li, M.; Yin, R.; Hou, Y.; Li, Y.; Pan, S.; et al. Lung cancer scRNA-seq and lipidomics reveal aberrant lipid metabolism for early-stage diagnosis. Sci. Transl. Med. 2022, 14, eabk2756. [Google Scholar] [CrossRef] [PubMed]
- Lesko, J.; Triebl, A.; Stacher-Priehse, E.; Fink-Neuböck, N.; Lindenmann, J.; Smolle-Jüttner, F.M.; Köfeler, H.C.; Hrzenjak, A.; Olschewski, H.; Leithner, K. Phospholipid dynamics in ex vivo lung cancer and normal lung explants. Exp. Mol. Med. 2021, 53, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhu, B.; Zeng, Y.; Shen, H.; Zhang, J.; Wang, X. Clinical lipidomics in understanding of lung cancer: Opportunity and challenge. Cancer Lett. 2020, 470, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Hu, X.; Zhao, X.; Kong, X.; Meng, Y.M.; Chen, Y.; Su, L.; Jiang, X.; Qiu, X.; Huang, C.; et al. A circular network of coregulated sphingolipids dictates lung cancer growth and progression. EBioMedicine 2021, 66, 103301. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.E.; Clair, G.; Bandyopadhyay, G.; Misra, R.S.; Zink, E.M.; Bloodsworth, K.J.; Shukla, A.K.; Du, Y.; Lillis, J.; Myers, J.R.; et al. Cell type-resolved human lung lipidome reveals cellular cooperation in lung function. Sci. Rep. 2018, 8, 13455. [Google Scholar] [CrossRef]
- Fan, Y.; Noreldeen, H.A.A.; You, L.; Liu, X.; Pan, X.; Hou, Z.; Li, Q.; Li, X.; Xu, G. Lipid alterations and subtyping maker discovery of lung cancer based on nontargeted tissue lipidomics using liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2020, 190, 113520. [Google Scholar] [CrossRef]
- Cang, S.; Liu, R.; Mu, K.; Tang, Q.; Cui, H.; Bi, K.; Zhang, Y.; Li, Q. Assessment of plasma amino acids, purines, tricarboxylic acid cycle metabolites, and lipids levels in NSCLC patients based on LC-MS/MS quantification. J. Pharm. Biomed. Anal. 2022, 221, 114990. [Google Scholar] [CrossRef]
- Godoy-Corchuelo, J.M.; Fernández-Beltrán, L.C.; Ali, Z.; Gil-Moreno, M.J.; López-Carbonero, J.I.; Guerrero-Sola, A.; Larrad-Sainz, A.; Matias-Guiu, J.; Matias-Guiu, J.A.; Cunningham, T.J.; et al. Lipid Metabolic Alterations in the ALS&ndash. FTD Spectr. Disord. 2022, 10, 1105. [Google Scholar]
- Corn, K.C.; Windham, M.A.; Rafat, M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020, 80, 101055. [Google Scholar] [CrossRef] [PubMed]
- Juloski, J.; Popović, T.B.; Debeljak-Martačić, J.; Ćuk, V.V.; Milić Perović, M.S.; Stanković, M.S.; Trbovich, A.M.; De Luka, S.R. Fatty acid in colorectal cancer in adult and aged patients of both sexes. J. BUON 2021, 26, 1898–1907. [Google Scholar]
- Noreldeen, H.A.A.; Du, L.; Li, W.; Liu, X.; Wang, Y.; Xu, G. Serum lipidomic biomarkers for non-small cell lung cancer in nonsmoking female patients. J. Pharm. Biomed. Anal. 2020, 185, 113220. [Google Scholar] [CrossRef]
- Liu, W.; Deng, Y.; Li, Z.; Chen, Y.; Zhu, X.; Tan, X.; Cao, G. Cancer Evo-Dev: A Theory of Inflammation-Induced Oncogenesis. Front. Immunol. 2021, 12, 768098. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Song, H.; Chen, Y.; Han, F.; Wang, Q.; Cui, Y. Neutrophil-to-Lymphocyte Ratio and Platelet-to-Lymphocyte Ratio in Blood to Distinguish Lung Cancer Patients from Healthy Subjects. Dis. Markers 2020, 2020, 8844698. [Google Scholar] [CrossRef]
- Chen, J.L.; Wu, J.N.; Lv, X.D.; Yang, Q.C.; Chen, J.R.; Zhang, D.M. The value of red blood cell distribution width, neutrophil-to-lymphocyte ratio, and hemoglobin-to-red blood cell distribution width ratio in the progression of non-small cell lung cancer. PLoS ONE 2020, 15, e0237947. [Google Scholar] [CrossRef]
- Lu, J.; Lam, S.; Wan, Q.; Shi, L.; Huo, Y.; Chen, L.; Tang, X.; Li, B.; Wu, X.; Peng, K.; et al. High-Coverage Targeted Lipidomics Reveals Novel Serum Lipid Predictors and Lipid Pathway Dysregulation Antecedent to Type 2 Diabetes Onset in Normoglycemic Chinese Adults. Diabetes Care 2019, 42, 2117–2126. [Google Scholar] [CrossRef]
- Steen, J.; Loeys, T.; Moerkerke, B.; Vansteelandt, S. Medflex: An R Package for Flexible Mediation Analysis using Natural Effect Models. J. Stat. Softw. 2017, 76, 1–46. [Google Scholar] [CrossRef]
- Liu, Z.; Gomez, C.R.; Espinoza, I.; Le, T.P.T.; Shenoy, V.; Zhou, X. Correlation of cholesteryl ester metabolism to pathogenesis, progression and disparities in colorectal Cancer. Lipids Health Dis. 2022, 21, 22. [Google Scholar] [CrossRef]
- Ueno, G.; Iwagami, Y.; Kobayashi, S.; Mitsufuji, S.; Yamada, D.; Tomimaru, Y.; Akita, H.; Asaoka, T.; Noda, T.; Gotoh, K.; et al. ACAT-1-Regulated Cholesteryl Ester Accumulation Modulates Gemcitabine Resistance in Biliary Tract Cancer. Ann. Surg. Oncol. 2022, 29, 2899–2909. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Zou, X.; Yan, X.; Cong, P.; Li, H.; Wang, H.; Xue, C.; Xu, J. Deep mining and quantification of oxidized cholesteryl esters discovers potential biomarkers involved in breast cancer by liquid chromatography-mass spectrometry. J. chromatogr. A 2022, 1663, 462764. [Google Scholar] [CrossRef] [PubMed]
- Hall, Z.; Wilson, C.H.; Burkhart, D.L.; Ashmore, T.; Evan, G.I.; Griffin, J.L. Myc linked to dysregulation of cholesterol transport and storage in nonsmall cell lung cancer. J. Lipid Res. 2020, 61, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, H.; Zhu, Y.; Ai, J.; Li, Y.; Gu, W.; Borgia, J.A.; Zhang, J.; Jiang, B.; Chen, W.; et al. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: A pilot study. Oncol. Lett. 2018, 16, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Hoppstädter, J.; Dembek, A.; Höring, M.; Schymik, H.S.; Dahlem, C.; Sultan, A.; Wirth, N.; Al-Fityan, S.; Diesel, B.; Gasparoni, G.; et al. Dysregulation of cholesterol homeostasis in human lung cancer tissue and tumour-associated macrophages. EBioMedicine 2021, 72, 103578. [Google Scholar] [CrossRef]
- Khan, S.A.; Goliwas, K.F.; Deshane, J.S. Sphingolipids in Lung Pathology in the Coronavirus Disease Era: A Review of Sphingolipid Involvement in the Pathogenesis of Lung Damage. Front. Physiol. 2021, 12, 760638. [Google Scholar] [CrossRef]
- Canals, D. Peeking inside the sphingolipid network in lung cancer. EBioMedicine 2021, 67, 103340. [Google Scholar] [CrossRef]
- Baxter, A.A.; Hulett, M.D.; Poon, I.K. The phospholipid code: A key component of dying cell recognition, tumor progression and host-microbe interactions. Cell Death Differ. 2015, 22, 1893–1905. [Google Scholar] [CrossRef]
- Stoica, C.; Ferreira, A.K.; Hannan, K.; Bakovic, M. Bilayer Forming Phospholipids as Targets for Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 5266. [Google Scholar] [CrossRef]
- Bandu, R.; Mok, H.J.; Kim, K.P. Phospholipids as cancer biomarkers: Mass spectrometry-based analysis. Mass Spectrum. Rev. 2018, 37, 107–138. [Google Scholar] [CrossRef]
- Guo, Y.; Ren, J.; Li, X.; Liu, X.; Liu, N.; Wang, Y.; Li, Z. Simultaneous Quantification of Serum Multi-Phospholipids as Potential Biomarkers for Differentiating Different Pathophysiological states of lung, stomach, intestine, and pancreas. J. Cancer 2017, 8, 2191–2204. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Grapov, D.; DeFelice, B.C.; Taylor, S.; Kim, K.; Kelly, K.; Wikoff, W.R.; Pass, H.; Rom, W.N.; Fiehn, O.; et al. Serum phosphatidylethanolamine levels distinguish benign from malignant solitary pulmonary nodules and represent a potential diagnostic biomarker for lung cancer. Cancer Biomark. Sec. A Dis. Mark. 2016, 16, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Cao, Q.; Guan, X.; Meng, H.; Feng, Y.; Wang, C.; Fu, M.; Hong, S.; Zhou, Y.; Yuan, F.; et al. Metabolic linkages between zinc exposure and lung cancer risk: A nested case-control study. Sci. Total Environ. 2022, 837, 155796. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.; Zhou, X.; Casado-Medrano, V.; Lopez-Haber, C.; Baker, M.J.; Garg, R.; Ann, J.; Lee, J.; Blumberg, P.M.; Kazanietz, M.G. Characterization of AJH-836, a diacylglycerol-lactone with selectivity for novel PKC isozymes. J. Biol. Chem. 2018, 293, 8330–8341. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.; Casado-Medrano, V.; Ann, J.; Lee, J.; Blumberg, P.M.; Abba, M.C.; Kazanietz, M.G. Differential Regulation of Gene Expression in Lung Cancer Cells by Diacyglycerol-Lactones and a Phorbol Ester Via Selective Activation of Protein Kinase C Isozymes. Sci. Rep. 2019, 9, 6041. [Google Scholar] [CrossRef]
- Zhang, L.; Han, L.; He, J.; Lv, J.; Pan, R.; Lv, T. A high serum-free fatty acid level is associated with cancer. J. Cancer Res. Clin. Oncol. 2020, 146, 705–710. [Google Scholar] [CrossRef]
- Liu, X.; Peng, Y.; Tao, R.; Meng, L.; Li, X. Mendelian Randomization Study of Causal Relationship between Omega-3 Fatty Acids and Risk of Lung Cancer. BioMed Res. Int. 2022, 2022, 2786567. [Google Scholar] [CrossRef]
- Luu, H.N.; Cai, H.; Murff, H.J.; Xiang, Y.B.; Cai, Q.; Li, H.; Gao, J.; Yang, G.; Lan, Q.; Gao, Y.T.; et al. A prospective study of dietary polyunsaturated fatty acids intake and lung cancer risk. Int. J. Cancer 2018, 143, 2225–2237. [Google Scholar] [CrossRef]
- Biswas, P.; Datta, C.; Rathi, P.; Bhattacharjee, A. Fatty acids and their lipid mediators in the induction of cellular apoptosis in cancer cells. Prostaglandins Other Lipid Mediat. 2022, 160, 106637. [Google Scholar] [CrossRef]
- Chen, J.Y.-F.; Hwang, C.-C.; Chen, W.-Y.; Lee, J.-C.; Fu, T.-F.; Fang, K.; Chu, Y.; Huang, Y.-L.; Lin, J.; Tsai, W.H.; et al. Additive effects of C(2)-ceramide on paclitaxel-induced premature senescence of human lung cancer cells. Life Sci. 2010, 87, 350–357. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, Z.; Shen, X.; Li, L.; Zhong, J.; Min, L.S.; Xu, L.; Li, H.; Zhang, J.; Dai, L. Serum Lipidomics Profiling to Identify Biomarkers for Non-Small Cell Lung Cancer. BioMed Res. Int. 2018, 2018, 5276240. [Google Scholar] [CrossRef]
- Reczyńska, K.; Khanal, D.; Pielichowska, K.; Pamuła, E.; Chrzanowski, W. Distinct Influence of Saturated Fatty Acids on Malignant and Nonmalignant Human Lung Epithelial Cells. Lipids 2020, 55, 117–126. [Google Scholar] [CrossRef]
- Liu, Y.; Tian, Y.; Guo, Y.; Yan, Z.; Xue, C.; Wang, J. DHA-enriched phosphatidylcholine suppressed angiogenesis by activating PPARγ and modulating the VEGFR2/Ras/ERK pathway in human umbilical vein endothelial cells. Food Sci. Biotechnol. 2021, 30, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Montecillo-Aguado, M.; Tirado-Rodriguez, B.; Antonio-Andres, G.; Morales-Martinez, M.; Tong, Z.; Yang, J.; Hammock, B.D.; Hernandez-Pando, R.; Huerta-Yepez, S. Omega-6 Polyunsaturated Fatty Acids Enhance Tumor Aggressiveness in Experimental Lung Cancer Model: Important Role of Oxylipins. Int. J. Mol. Sci. 2022, 23, 6179. [Google Scholar] [CrossRef] [PubMed]
- Pietzner, M.; Kaul, A.; Henning, A.K.; Kastenmüller, G.; Artati, A.; Lerch, M.M.; Adamski, J.; Nauck, M.; Friedrich, N. Comprehensive metabolic profiling of chronic low-grade inflammation among generally healthy individuals. BMC Med. 2017, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.; Li, H.; Yu, D.; Cai, H.; Gao, J.; Gao, Y.; Luu, H.; Tran, H.; Xiang, Y.; Zheng, W.; et al. Dietary fatty acids and colorectal cancer risk in men: A report from the Shanghai Men’s Health Study and a meta-analysis. Int. J. Cancer 2021, 148, 77–89. [Google Scholar] [CrossRef]
- Zhu, Q.; Wu, Y.; Mai, J.; Guo, G.; Meng, J.; Fang, X.; Chen, X.; Liu, C.; Zhong, S. Comprehensive Metabolic Profiling of Inflammation Indicated Key Roles of Glycerophospholipid and Arginine Metabolism in Coronary Artery Disease. Front. Immunol. 2022, 13, 829425. [Google Scholar] [CrossRef]
- Wallner, S.; Orsó, E.; Grandl, M.; Konovalova, T.; Liebisch, G.; Schmitz, G. Phosphatidylcholine and phosphatidylethanolamine plasmalogens in lipid loaded human macrophages. PLoS ONE 2018, 13, e0205706. [Google Scholar] [CrossRef]
- Xing, F.; Liu, J.; Mo, Y.; Liu, Z.; Qin, Q.; Wang, J.; Fan, Z.; Long, Y.; Liu, N.; Zhao, K.; et al. Lysophosphatidylcholine up-regulates human endothelial nitric oxide synthase gene transactivity by c-Jun N-terminal kinase signalling pathway. J. Cell. Mol. Med. 2009, 13, 1136–1148. [Google Scholar] [CrossRef]
- Zhang, R.; Bai, N.; So, J.; Laher, I.; MacLeod, K.M.; Rodrigues, B. The ischemic metabolite lysophosphatidylcholine increases rat coronary arterial tone by endothelium-dependent mechanisms. J. Mol. Cell. Cardiol. 2009, 47, 112–120. [Google Scholar] [CrossRef]
- Şahin, F.; Aslan, A.F. Relationship between Inflammatory and Biological Markers and Lung Cancer. J. Clin. Med. 2018, 7, 160. [Google Scholar] [CrossRef]
- Wang, K.; Zhao, Q.; Yan, T.; Guo, D.; Liu, J.; Wang, G.; Du, J. The Prognostic Value of Multiple Systemic Inflammatory Biomarkers in Preoperative Patients With Non-small Cell Lung Cancer. Front. Surg. 2022, 9, 830642. [Google Scholar] [CrossRef] [PubMed]
- Stares, M.; Ding, T.E.; Stratton, C.; Thomson, F.; Baxter, M.; Cagney, H.; Cumming, K.; Swan, A.; Ross, F.; Barrie, C.; et al. Biomarkers of systemic inflammation predict survival with first-line immune checkpoint inhibitors in non-small-cell lung cancer. ESMO Open 2022, 7, 100445. [Google Scholar] [CrossRef] [PubMed]
- Nøst, T.H.; Alcala, K.; Urbarova, I.; Byrne, K.S.; Guida, F.; Sandanger, T.M.; Johansson, M. Systemic inflammation markers and cancer incidence in the UK Biobank. Eur. J. Epidemiol. 2021, 36, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Gao, L.; Wang, Y.; Xu, B.; Maswikiti, E.P.; Li, H.; Zheng, P.; Tao, P.; Xiang, L.; Gu, B.; et al. A Forgotten Corner in Cancer Immunotherapy: The Role of Lipids. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Roudi, R.; Dai, T.; Chen, S.; Fan, B.; Li, H.; Zhou, Y.; Zhou, M.; Zhu, B.; Yin, C.; et al. Immune-related adverse events associated with programmed cell death protein-1 and programmed cell death ligand 1 inhibitors for non-small cell lung cancer: A PRISMA systematic review and meta-analysis. BMC Cancer 2019, 19, 558. [Google Scholar] [CrossRef]
- Mabrouk, N.; Lecoeur, B.; Bettaieb, A.; Paul, C.; Végran, F. Impact of Lipid Metabolism on Antitumor Immune Response. Cancers 2022, 14, 1850. [Google Scholar] [CrossRef]

| Variables | N | LC | Controls | p2 |
|---|---|---|---|---|
| Sex | ||||
| Male | 80 | 40 (64.5%) | 40 (64.5%) | |
| Female | 44 | 22 (35.5%) | 22 (35.5%) | |
| Age (years) | ||||
| ≤54 | 64 | 30 (48.4%) | 34 (54.8%) | |
| >54 | 60 | 32 (51.6%) | 28 (45.2%) | |
| BMI(kg/m²) | 0.853 | |||
| ≤24 | 93 | 46 (74.2%) | 47 (75.8%) | |
| >24 | 31 | 16 (25.8%) | 15 (24.2%) | |
| WBC (×109/L) | 124 | 8.88 (5.43,12.33) | 6.32 (5.97,6.68) | 0.018 |
| NEUT (×109/L) | 124 | 4.73 (4.21,5.24) | 3.54 (3.28,3.80) | <0.001 |
| LYMPH (×109/L) | 124 | 1.67 (1.51,1.84) | 2.17 (2.03,2.31) | <0.001 |
| MONO (×109/L) | 124 | 0.59 (0.44,0.74) | 0.39 (0.37,0.43) | 0.002 |
| PLT (×109/L) | 124 | 284 (257,310) | 239 (227,251) | 0.010 |
| EOS (×109/L) | 124 | 0.21 (0.13,0.29) | 0.17 (0.14,0.20) | 0.384 |
| NLR | 124 | 3.26 (2.76,3.76) | 1.69 (1.55,1.82) | <0.001 |
| PLR | 124 | 199 (166,233) | 116 (107,125) | <0.001 |
| NMR | 124 | 9.86 (8.73,10.99) | 9.21 (8.56,9.87) | <0.001 |
| LMR | 124 | 3.65 (3.13,4.18) | 5.67 (5.33,6.02) | <0.001 |
| OR(95%CI) * | p | FDR | |
|---|---|---|---|
| n-6 PUFA(all) | 1.00 (0.99,1.02) | 0.260 | 0.484 |
| n-3 PUFA(all) | 0.97 (0.96,0.98) | <0.001 | 0.001 |
| n6/n3 ratio(all) | 0.96 (0.90,1.04) | 0.382 | 0.574 |
| CE | |||
| n-6 PUFA | 1.27 (1.03,1.56) | 0.024 | 0.072 |
| n-3 PUFA | 1.09 (0.98,1.22) | 0.099 | 0.215 |
| n6/n3 ratio | 1.06 (0.88,1.29) | 0.494 | 0.600 |
| DAG | |||
| n-6 PUFA | 1.13 (0.99,1.30) | 0.065 | 0.170 |
| n-3 PUFA | 0.53 (0.39,0.73) | <0.001 | 0.001 |
| n6/n3 ratio | 0.98 (0.94,1.01) | 0.334 | 0.556 |
| FFA | |||
| n-6 PUFA | 2.11 (1.45,3.09) | <0.001 | 0.001 |
| n-3 PUFA | 0.95 (0.83,1.07) | 0.438 | 0.600 |
| n6/n3 ratio | 0.99 (0.97,1.01) | 0.573 | 0.638 |
| LPC | |||
| n-6 PUFA | 1.69 (1.28,2.25) | <0.001 | 0.001 |
| n-3 PUFA | 1.12 (1.00,1.25) | 0.043 | 0.121 |
| n6/n3 ratio | 0.99 (0.97,1.01) | 0.562 | 0.638 |
| LPE | |||
| n-6 PUFA | 1.67 (1.23,2.26) | <0.001 | 0.003 |
| n-3 PUFA | 1.13 (0.80,1.61) | 0.466 | 0.600 |
| n6/n3 ratio | 1.00 (0.98,1.01) | 0.945 | 0.945 |
| PA | |||
| n-6 PUFA | 0.81 (0.72,0.91) | <0.001 | 0.003 |
| n-3 PUFA | 0.55 (0.39,0.77) | <0.001 | 0.002 |
| n6/n3 ratio | 1.05 (0.93,1.19) | 0.360 | 0.561 |
| PC | |||
| n-6 PUFA | 1.03 (0.99,1.07) | 0.085 | 0.196 |
| n-3 PUFA | 1.01 (0.97,1.05) | 0.508 | 0.600 |
| n6/n3 ratio | 0.88 (0.77,1.02) | 0.114 | 0.234 |
| PE | |||
| n-6 PUFA | 0.96 (0.93,1.00) | 0.079 | 0.194 |
| n-3 PUFA | 0.87 (0.81,0.93) | <0.001 | 0.001 |
| n6/n3 ratio | 0.99 (0.96,1.01) | 0.494 | 0.600 |
| PG | |||
| n-6 PUFA | 0.95 (0.87,1.04) | 0.281 | 0.498 |
| PI | |||
| n-6 PUFA | 0.99 (0.92,1.06) | 0.831 | 0.853 |
| n-3 PUFA | 0.93 (0.84,1.03) | 0.177 | 0.346 |
| n6/n3 ratio | 0.99 (0.98,1.00) | 0.476 | 0.600 |
| PS | |||
| n-6 PUFA | 0.85 (0.78,0.92) | <0.001 | 0.001 |
| n-3 PUFA | 0.70 (0.58,0.84) | <0.001 | 0.001 |
| n6/n3 ratio | 1.02 (0.97,1.06) | 0.342 | 0.556 |
| TAG | |||
| n-6 PUFA | 1.09 (1.03,1.14) | <0.001 | 0.002 |
| n-3 PUFA | 0.94 (0.92,0.97) | <0.001 | 0.001 |
| n6/n3 ratio | 0.99 (0.95,1.02) | 0.620 | 0.672 |
| Effects | Β (95%CI) | OR (95%CI) | p |
|---|---|---|---|
| Mediation factor = LMR | |||
| Direct effect(LS→LC) | −2.09 (−3.34,−0.85) | 0.12 (0.03,0.43) | <0.001 |
| Indirect effect(LMR→LS→LC) | −0.24 (−0.41,−0.07) | 0.79 (0.66,0.93) | 0.005 |
| Proportion mediated = 2.87% | |||
| Mediation factor = LYMPH | |||
| Direct effect(LS→LC) | −2.05 (−2.95,−1.13) | 0.13 (0.05,0.32) | <0.001 |
| Indirect effect(LYMPH→LS→LC) | −0.14 (−0.26,−0.03) | 0.87 (0.77,0.97) | 0.014 |
| Proportion mediated = 1.89% | |||
| Mediation factor = PLR | |||
| Direct effect(LS→LC) | −1.99 (−2.94,−1.05) | 0.14 (0.05,0.35) | <0.001 |
| Indirect effect(PLR→LS→LC) | −0.14 (−3.09,−1.18) | 0.87 (0.78,0.97) | 0.013 |
| Proportion mediated = 2.03% | |||
| Mediation factor = NLR | |||
| Direct effect(LS→LC) | −1.68 (−2.62,−0.74) | 0.19 (0.07,0.47) | <0.001 |
| Indirect effect(NLR→LS→LC) | −0.26 (−2.85,−1.04) | 0.77 (0.64,0.91) | 0.003 |
| Proportion mediated = 5.04% | |||
| Mediation factor = PLT | |||
| Direct effect(LS→LC) | −1.90 (−2.83,−0.97) | 0.15 (0.06,0.37) | <0.001 |
| Indirect effect(PLT→LS→LC) | −0.19 (−0.32,−0.07) | 0.83 (0.73,0.93) | 0.002 |
| Proportion mediated = 2.95% | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, C.; Chen, L.; Fan, Y.; Lin, Z.; Tang, X.; Xu, Y.; Zeng, Y.; Hu, Z. The Landscape of Lipid Metabolism in Lung Cancer: The Role of Structural Profiling. J. Clin. Med. 2023, 12, 1736. https://doi.org/10.3390/jcm12051736
Hu C, Chen L, Fan Y, Lin Z, Tang X, Xu Y, Zeng Y, Hu Z. The Landscape of Lipid Metabolism in Lung Cancer: The Role of Structural Profiling. Journal of Clinical Medicine. 2023; 12(5):1736. https://doi.org/10.3390/jcm12051736
Chicago/Turabian StyleHu, Chanchan, Luyang Chen, Yi Fan, Zhifeng Lin, Xuwei Tang, Yuan Xu, Yiming Zeng, and Zhijian Hu. 2023. "The Landscape of Lipid Metabolism in Lung Cancer: The Role of Structural Profiling" Journal of Clinical Medicine 12, no. 5: 1736. https://doi.org/10.3390/jcm12051736
APA StyleHu, C., Chen, L., Fan, Y., Lin, Z., Tang, X., Xu, Y., Zeng, Y., & Hu, Z. (2023). The Landscape of Lipid Metabolism in Lung Cancer: The Role of Structural Profiling. Journal of Clinical Medicine, 12(5), 1736. https://doi.org/10.3390/jcm12051736






