The Gut Microbiome and Metastatic Renal Cell Carcinoma
Abstract
1. Introduction
2. Gut Microbiome
3. Gut Microbiome and Cancer
4. Gut Microbiome and Renal Cell Carcinoma
5. Challenges and Opportunities
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Key Statistics about Kidney Cancer. Available online: https://www.cancer.org/cancer/kidney-cancer/about/key-statistics.html (accessed on 27 May 2022).
- Motzer, R.J.; Hutson, T.E.; Tomczak, P.; Michaelson, M.D.; Bukowski, R.M.; Rixe, O.; Oudard, S.; Negrier, S.; Szczylik, C.; Kim, S.T.; et al. Sunitinib versus Interferon Alfa in Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 2007, 356, 115–124. [Google Scholar] [CrossRef]
- Sternberg, C.N.; Davis, I.D.; Mardiak, J.; Szczylik, C.; Lee, E.; Wagstaff, J.; Barrios, C.H.; Salman, P.; Gladkov, O.A.; Kavina, A.; et al. Pazopanib in Locally Advanced or Metastatic Renal Cell Carcinoma: Results of a Randomized Phase III Trial. J. Clin. Oncol. 2010, 28, 1061–1068. [Google Scholar] [CrossRef]
- Rini, B.I.; Escudier, B.; Tomczak, P.; Kaprin, A.; Szczylik, C.; Hutson, T.E.; Michaelson, M.D.; Gorbunova, V.A.; Gore, M.E.; Rusakov, I.G.; et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): A randomised phase 3 trial. Lancet 2011, 378, 1931–1939. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Halabi, S.; Sanford, B.L.; Hahn, O.; Michaelson, M.D.; Walsh, M.K.; Feldman, D.; Olencki, T.; Picus, J.; Small, E.J.; et al. Cabozantinib Versus Sunitinib As Initial Targeted Therapy for Patients with Metastatic Renal Cell Carcinoma of Poor or Intermediate Risk: The Alliance A031203 CABOSUN Trial. J. Clin. Oncol. 2017, 35, 591–597. [Google Scholar] [CrossRef]
- Motzer, R.J.; Hutson, T.E.; Glen, H.; Michaelson, M.D.; Molina, A.; Eisen, T.; Jassem, J.; Zolnierek, J.; Maroto, J.P.; Mellado, B.; et al. Lenvatinib, everolimus, and the combination in patients with metastatic renal cell carcinoma: A randomised, phase 2, open-label, multicentre trial. Lancet Oncol. 2015, 16, 1473–1482. [Google Scholar] [CrossRef]
- Hudes, G.; Carducci, M.; Tomczak, P.; Dutcher, J.; Figlin, R.; Kapoor, A.; Staroslawska, E.; Sosman, J.; McDermott, D.; Bodrogi, I.; et al. Temsirolimus, Interferon Alfa, or Both for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2007, 356, 2271–2281. [Google Scholar] [CrossRef]
- Meza, L.; Malhotra, J.; Favorito, C.; Pal, S.K. Cabozantinib plus immunotherapy combinations in metastatic renal cell and urothelial carcinoma. Futur. Oncol. 2022, 18, 21–33. [Google Scholar] [CrossRef]
- SEER. Cancer of the Kidney and Renal Pelvis—Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/kidrp.html (accessed on 4 February 2023).
- Dudani, S.; Savard, M.-F.; Heng, D.Y. An Update on Predictive Biomarkers in Metastatic Renal Cell Carcinoma. Eur. Urol. Focus 2020, 6, 34–36. [Google Scholar] [CrossRef]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLOS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Thomas, S.; Izard, J.; Walsh, E.; Batich, K.; Chongsathidkiet, P.; Clarke, G.; Sela, D.A.; Muller, A.J.; Mullin, J.M.; Albert, K.; et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res. 2017, 77, 1783–1812. [Google Scholar] [CrossRef]
- Schluter, J.; Peled, J.; Taylor, B.P.; Markey, K.A.; Smith, J.A.; Taur, Y.; Niehus, R.; Staffas, A.; Dai, A.; Fontana, E.; et al. The gut microbiota is associated with immune cell dynamics in humans. Nature 2020, 588, 303–307. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Gomez de Agüero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef]
- Olin, A.; Henckel, E.; Chen, Y.; Lakshmikanth, T.; Pou, C.; Mikes, J.; Gustafsson, A.; Bernhardsson, A.K.; Zhang, C.; Bohlin, K.; et al. Stereotypic Immune System Development in Newborn Children. Cell 2018, 174, 1277–1292.e14. [Google Scholar] [CrossRef]
- Tan, T.G.; Sefik, E.; Geva-Zatorsky, N.; Kua, L.; Naskar, D.; Teng, F.; Pasman, L.; Ortiz-Lopez, A.; Jupp, R.; Wu, H.-J.J.; et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E8141–E8150. [Google Scholar] [CrossRef]
- Deshmukh, H.S.; Liu, Y.; Menkiti, O.R.; Mei, J.; Dai, N.; O’Leary, C.E.; Oliver, P.M.; Kolls, J.K.; Weiser, J.N.; Worthen, G.S. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 2014, 20, 524–530. [Google Scholar] [CrossRef]
- Ivanov, I.I.; de Llanos Frutos, R.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef]
- Zaneveld, J.; Turnbaugh, P.; Lozupone, C.; Ley, R.; Hamady, M.; Gordon, J.I.; Knight, R. Host-bacterial coevolution and the search for new drug targets. Curr. Opin. Chem. Biol. 2008, 12, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Khan, I.; Usman, M.; Wei, Z.X.; Ping, X.; Khan, S.; Khan, F.; Jianye, Z.; Zhiqiang, L.; Lizhe, A. Circulating microbiota and metabolites: Insights into cardiovascular diseases. J. Clin. Lab. Anal. 2022, 36, e24779. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Azzouz, D.; Omarbekova, A.; Heguy, A.; Schwudke, D.; Gisch, N.; Rovin, B.H.; Caricchio, R.; Buyon, J.P.; Alekseyenko, A.V.; Silverman, G.J. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019, 78, 947–956. [Google Scholar] [CrossRef]
- Ianiro, G.; Maida, M.; Burisch, J.; Simonelli, C.; Hold, G.; Ventimiglia, M.; Gasbarrini, A.; Cammarota, G. Efficacy of different faecal microbiota transplantation protocols for Clostridium difficile infection: A systematic review and meta-analysis. United Eur. Gastroenterol. J. 2018, 6, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Lloyd-Price, J.; Vatanen, T.; Seksik, P.; Beaugerie, L.; Simon, T.; Vlamakis, H.; Sokol, H.; Xavier, R.J. Linking Strain Engraftment in Fecal Microbiota Transplantation with Maintenance of Remission in Crohn’s Disease. Gastroenterology 2020, 159, 2193–2202.e5. [Google Scholar] [CrossRef] [PubMed]
- Kootte, R.S.; Levin, E.; Salojärvi, J.; Smits, L.P.; Hartstra, A.V.; Udayappan, S.D.; Hermes, G.; Bouter, K.E.; Koopen, A.M.; Holst, J.J.; et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab. 2017, 26, 611–619.e6. [Google Scholar] [CrossRef]
- Rous, P. A Sarcoma of the Fowl Transmissible by an Agent Separable from the Tumor Cells. J. Exp. Med. 1911, 13, 397–411. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, S.; Ye, Y.; Jia, L.; Lou, Y. Global research trends on the links between gut microbiota and cancer immunotherapy: A bibliometric analysis (2012–2021). Front. Immunol. 2022, 13, 952546. [Google Scholar] [CrossRef]
- White, M.K.; Pagano, J.S.; Khalili, K. Viruses and Human Cancers: A Long Road of Discovery of Molecular Paradigms. Clin. Microbiol. Rev. 2014, 27, 463–481. [Google Scholar] [CrossRef]
- Knippel, R.J.; Drewes, J.L.; Sears, C.L. The Cancer Microbiome: Recent Highlights and Knowledge Gaps. Cancer Discov. 2021, 11, 2378–2395. [Google Scholar] [CrossRef] [PubMed]
- Park, E.M.; Chelvanambi, M.; Bhutiani, N.; Kroemer, G.; Zitvogel, L.; Wargo, J.A. Targeting the gut and tumor microbiota in cancer. Nat. Med. 2022, 28, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Yonekura, S.; Terrisse, S.; Silva, C.A.C.; Lafarge, A.; Iebba, V.; Ferrere, G.; Goubet, A.-G.; Fahrner, J.-E.; Lahmar, I.; Ueda, K.; et al. Cancer Induces a Stress Ileopathy Depending on β-Adrenergic Receptors and Promoting Dysbiosis that Contributes to Carcinogenesis. Cancer Discov. 2021, 12, 1128–1151. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jobin, C. Novel insights into microbiome in colitis and colorectal cancer. Curr. Opin. Gastroenterol. 2017, 33, 422–427. [Google Scholar] [CrossRef]
- Mei, S.; Deng, Z.; Chen, Y.; Ning, D.; Guo, Y.; Fan, X.; Wang, R.; Meng, Y.; Zhou, Q.; Tian, X. Dysbiosis: The first hit for digestive system cancer. Front. Physiol. 2022, 13, 1040991. [Google Scholar] [CrossRef]
- Mima, K.; Nakagawa, S.; Sawayama, H.; Ishimoto, T.; Imai, K.; Iwatsuki, M.; Hashimoto, D.; Baba, Y.; Yamashita, Y.-I.; Yoshida, N.; et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 2017, 402, 9–15. [Google Scholar] [CrossRef]
- Wu, R.; Yu, I.; Tokumaru, Y.; Asaoka, M.; Oshi, M.; Yan, L.; Okuda, S.; Ishikawa, T.; Takabe, K. Elevated bile acid metabolism and microbiome are associated with suppressed cell proliferation and better survival in breast cancer. Am. J. Cancer Res. 2022, 12, 5271–5285. [Google Scholar]
- Mekadim, C.; Skalnikova, H.K.; Cizkova, J.; Cizkova, V.; Palanova, A.; Horak, V.; Mrazek, J. Dysbiosis of skin microbiome and gut microbiome in melanoma progression. BMC Microbiol. 2022, 22, 63. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science 2019, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.; Oefner, P.J.; Hiergeist, A.; Koestler, J.; Gessner, A.; Weber, M.; Hahn, J.; Wolff, D.; Stämmler, F.; Spang, R.; et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 2015, 126, 1723–1728. [Google Scholar] [CrossRef]
- Jenq, R.R.; Ubeda, C.; Taur, Y.; Menezes, C.C.; Khanin, R.; Dudakov, J.A.; Liu, C.; West, M.L.; Singer, N.V.; Equinda, M.J.; et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med. 2012, 209, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.C.; Shanahan, E.R.; Batten, M.; Reijers, I.L.M.; Read, M.; Silva, I.P.; Versluis, J.M.; Ribeiro, R.; Angelatos, A.S.; Tan, J.; et al. Diet-driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome. Nat. Med. 2022, 28, 2344–2352. [Google Scholar] [CrossRef]
- Liu, W.; Ma, F.; Sun, B.; Liu, Y.; Tang, H.; Luo, J.; Chen, H.; Luo, Z. Intestinal Microbiome Associated with Immune-Related Adverse Events for Patients Treated with Anti-PD-1 Inhibitors, a Real-World Study. Front. Immunol. 2021, 12, 5334. Available online: https://www.frontiersin.org/articles/10.3389/fimmu.2021.756872 (accessed on 5 February 2023). [CrossRef]
- Andrews, M.C.; Duong, C.P.M.; Gopalakrishnan, V.; Iebba, V.; Chen, W.-S.; Derosa, L.; Khan, A.W.; Cogdill, A.P.; White, M.G.; Wong, M.C.; et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat. Med. 2021, 27, 1432–1441. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wiesnoski, D.H.; Helmink, B.A.; Gopalakrishnan, V.; Choi, K.; DuPont, H.L.; Jiang, Z.-D.; Abu-Sbeih, H.; Sanchez, C.A.; Chang, C.-C.; et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 2018, 24, 1804–1808. [Google Scholar] [CrossRef]
- Routy, B.; le Chatelier, E.; DeRosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Aggarwal, P.; Costa, R.G.F.; Cole, A.M.; Trinchieri, G. Targeting the gut microbiota for cancer therapy. Nat. Rev. Cancer 2022, 22, 703–722. [Google Scholar] [CrossRef] [PubMed]
- Tomita, Y.; Ikeda, T.; Sakata, S.; Saruwatari, K.; Sato, R.; Iyama, S.; Jodai, T.; Akaike, K.; Ishizuka, S.; Saeki, S.; et al. Association of Probiotic Clostridium butyricum Therapy with Survival and Response to Immune Checkpoint Blockade in Patients with Lung Cancer. Cancer Immunol. Res. 2020, 8, 1236–1242. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.-M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti–PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2020, 371, 602–609. [Google Scholar] [CrossRef]
- Spencer, C.N.; McQuade, J.L.; Gopalakrishnan, V.; McCulloch, J.A.; Vetizou, M.; Cogdill, A.P.; Khan, A.W.; Zhang, X.; White, M.G.; Peterson, C.B.; et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021, 374, 1632–1640. [Google Scholar] [CrossRef]
- Lythgoe, M.P.; Mullish, B.H.; Frampton, A.E.; Krell, J. Polymorphic microbes: A new emerging hallmark of cancer. Trends Microbiol. 2022, 30, 1131–1134. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; Van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Cho, C.H.; Yun, M.S.; Jang, S.J.; You, H.J.; Kim, J.-H.; Han, D.; Cha, K.H.; Moon, S.H.; Lee, K.; et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat. Microbiol. 2021, 6, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Derosa, L.; Routy, B.; Desilets, A.; Daillère, R.; Terrisse, S.; Kroemer, G.; Zitvogel, L. Microbiota-Centered Interventions: The Next Breakthrough in Immuno-Oncology? Cancer Discov. 2021, 11, 2396–2412. [Google Scholar] [CrossRef] [PubMed]
- Okumura, S.; Konishi, Y.; Narukawa, M.; Sugiura, Y.; Yoshimoto, S.; Arai, Y.; Sato, S.; Yoshida, Y.; Tsuji, S.; Uemura, K.; et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat. Commun. 2021, 12, 5674. [Google Scholar] [CrossRef]
- Rossi, T.; Vergara, D.; Fanini, F.; Maffia, M.; Bravaccini, S.; Pirini, F. Microbiota-Derived Metabolites in Tumor Progression and Metastasis. Int. J. Mol. Sci. 2020, 21, 5786. [Google Scholar] [CrossRef] [PubMed]
- Blake, S.J.; James, J.; Ryan, F.J.; Caparros-Martin, J.; Eden, G.L.; Tee, Y.C.; Salamon, J.R.; Benson, S.C.; Tumes, D.J.; Sribnaia, A.; et al. The immunotoxicity, but not anti-tumor efficacy, of anti-CD40 and anti-CD137 immunotherapies is dependent on the gut microbiota. Cell Rep. Med. 2021, 2, 100464. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Kespohl, M.; Vachharajani, N.; Luu, M.; Harb, H.; Pautz, S.; Wolff, S.; Sillner, N.; Walker, A.; Schmitt-Kopplin, P.; Boettger, T.; et al. The Microbial Metabolite Butyrate Induces Expression of Th1-Associated Factors in CD4+ T Cells. Front. Immunol. 2017, 8, 1036. [Google Scholar] [CrossRef]
- Luu, M.; Weigand, K.; Wedi, F.; Breidenbend, C.; Leister, H.; Pautz, S.; Adhikary, T.; Visekruna, A. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 2018, 8, 14430. [Google Scholar] [CrossRef]
- Salvi, P.S.; Cowles, R.A. Butyrate and the Intestinal Epithelium: Modulation of Proliferation and Inflammation in Homeostasis and Disease. Cells 2021, 10, 1775. [Google Scholar] [CrossRef]
- Bordonaro, M.; Lazarova, D.L.; Augenlicht, L.H.; Sartorelli, A.C. Cell type- and promoter-dependent modulation of the Wnt signaling pathway by sodium butyrate. Int. J. Cancer 2001, 97, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Bordonaro, M.; Lazarova, D.L.; Sartorelli, A.C. Role of Tcf-DNA binding and the chromatin remodeling factor Brg-1 in the modulation of Wnt activity by butyrate. Cell Cycle 2008, 7, 3472–3473. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Olson, D.J.; Sharma, T.; Vadlamudi, R.K.; Kumar, R. Butyric acid induces apoptosis by up-regulating Bax expression via stimulation of the c-Jun N-terminal kinase/activation protein-1 pathway in human colon cancer cells. Gastroenterology 2001, 120, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. GPR109A Is a G-protein–Coupled Receptor for the Bacterial Fermentation Product Butyrate and Functions as a Tumor Suppressor in Colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2019, 469, 456–467. [Google Scholar] [CrossRef]
- Miao, D.; Margolis, C.A.; Vokes, N.I.; Liu, D.; Taylor-Weiner, A.; Wankowicz, S.M.; Adeegbe, D.; Keliher, D.; Schilling, B.; Tracy, A.; et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018, 50, 1271–1281. [Google Scholar] [CrossRef]
- McDermott, D.F.; Huseni, M.A.; Atkins, M.B.; Motzer, R.J.; Rini, B.I.; Escudier, B.; Fong, L.; Joseph, R.W.; Pal, S.K.; Reeves, J.A.; et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018, 24, 749–757. [Google Scholar] [CrossRef]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the Treatment of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Albiges, L.; Tannir, N.M.; Burotto, M.; McDermott, D.; Plimack, E.R.; Barthélémy, P.; Porta, C.; Powles, T.; Donskov, F.; George, S.; et al. Nivolumab plus ipilimumab versus sunitinib for first-line treatment of advanced renal cell carcinoma: Extended 4-year follow-up of the phase III CheckMate 214 trial. ESMO Open 2020, 5, e001079. [Google Scholar] [CrossRef]
- Derosa, L.; Hellmann, M.D.; Spaziano, M.; Halpenny, D.; Fidelle, M.; Rizvi, H.; Long, N.; Plodkowski, A.J.; Arbour, K.C.; Chaft, J.E.; et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018, 29, 1437–1444. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Fidelle, M.; Iebba, V.; Alla, L.; Pasolli, E.; Segata, N.; Desnoyer, A.; Pietrantonio, F.; Ferrere, G.; et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur. Urol. 2020, 78, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Salgia, N.J.; Bergerot, P.G.; Maia, M.C.; Dizman, N.; Hsu, J.; Gillece, J.D.; Folkerts, M.; Reining, L.; Trent, J.; Highlander, S.K.; et al. Stool Microbiome Profiling of Patients with Metastatic Renal Cell Carcinoma Receiving Anti–PD-1 Immune Checkpoint Inhibitors. Eur. Urol. 2020, 78, 498–502. [Google Scholar] [CrossRef]
- Silva, C.A.C. Longitudinal analysis reveals gut microbiota shift during standard therapies in metastatic renal cell carcinoma (mRCC). 2022, 33 (Suppl. 7), pp. S660–S680. Available online: https://oncologypro.esmo.org/meeting-resources/esmo-congress/longitudinal-analysis-reveals-gut-microbiota-shift-during-standard-therapies-in-metastatic-renal-cell-carcinoma-mrcc (accessed on 10 January 2023).
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Kuroki, Y.; Ariyoshi, T.; Higashi, S.; Fukuda, K.; Yamashita, R.; Matsumoto, A.; Mori, T.; Mimura, K.; Yamaguchi, N.; et al. Clostridium butyricum Modulates the Microbiome to Protect Intestinal Barrier Function in Mice with Antibiotic-Induced Dysbiosis. Iscience 2019, 23, 100772. [Google Scholar] [CrossRef] [PubMed]
- Dizman, N.; Hsu, J.; Bergerot, P.G.; Gillece, J.D.; Folkerts, M.; Reining, L.; Trent, J.; Highlander, S.K.; Pal, S.K. Randomized trial assessing impact of probiotic supplementation on gut microbiome and clinical outcome from targeted therapy in metastatic renal cell carcinoma. Cancer Med. 2020, 10, 79–86. [Google Scholar] [CrossRef]
- Dizman, N.; Meza, L.; Bergerot, P.; Alcantara, M.; Dorff, T.; Lyou, Y.; Frankel, P.; Cui, Y.; Mira, V.; Llamas, M.; et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: A randomized phase 1 trial. Nat. Med. 2022, 28, 704–712. [Google Scholar] [CrossRef]
- Balan, M.; Teran, E.M.Y.; Waaga-Gasser, A.M.; Gasser, M.; Choueiri, T.K.; Freeman, G.; Pal, S. Novel Roles of c-Met in the Survival of Renal Cancer Cells through the Regulation of HO-1 and PD-L1 Expression. J. Biol. Chem. 2015, 290, 8110–8120. [Google Scholar] [CrossRef]
- Paolino, M.; Penninger, J.M. The Role of TAM Family Receptors in Immune Cell Function: Implications for Cancer Therapy. Cancers 2016, 8, 97. [Google Scholar] [CrossRef]
- Voron, T.; Marcheteau, E.; Pernot, S.; Colussi, O.; Tartour, E.; Taieb, J.; Terme, M. Control of the Immune Response by Pro-Angiogenic Factors. Front. Oncol. 2014, 4, 70. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.E.; Caenepeel, S.; Wu, L.C. Targeted Therapy and Checkpoint Immunotherapy Combinations for the Treatment of Cancer. Trends Immunol. 2016, 37, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Meza, L.A.; Malhotra, J.; Zengin, Z.B.; Dizman, N.; Hsu, J.; Chawla, N.S.; Chehrazi-Raffle, A.; Muddasani, R.; Govindarajan, A.; Castro, D.V.; et al. A phase I trial to evaluate the biologic effect of CBM588 (Clostridium butyricum) in combination with cabozantinib plus nivolumab for patients with metastatic renal cell carcinoma (mRCC). J. Clin. Oncol. 2022, 40, TPS4606. Available online: https://ascopubs.org/doi/abs/10.1200/JCO.2022.40.16_suppl.TPS4606 (accessed on 24 August 2022). [CrossRef]
- Meza, L.; Govindarajan, A.; Feng, M.; Pal, S.K. Live bacterial supplementation for improving treatment response in metastatic renal cell carcinoma. Clin. Transl. Med. 2022, 12, e948. [Google Scholar] [CrossRef] [PubMed]
- Ninkov, M.; Schmerk, C.L.; Moradizadeh, M.; Parvathy, S.N.; Figueredo, R.; Burton, J.P.; Silverman, M.S.; Fernandes, R.; Vareki, S.M.; Haeryfar, S.M.M. Improved MAIT cell functions following fecal microbiota transplantation for metastatic renal cell carcinoma. Cancer Immunol. Immunother. 2022, 1–14. [Google Scholar] [CrossRef]
- Ianiro, G.; Rossi, E.; Thomas, A.M.; Schinzari, G.; Masucci, L.; Quaranta, G.; Settanni, C.R.; Lopetuso, L.R.; Armanini, F.; Blanco-Miguez, A.; et al. Faecal microbiota transplantation for the treatment of diarrhoea induced by tyrosine-kinase inhibitors in patients with metastatic renal cell carcinoma. Nat. Commun. 2020, 11, 4333. [Google Scholar] [CrossRef]
- Lawson Health Research Institute. Preventing Immune-Related Adverse Events in Renal Cell Carcinoma Patients Treated with Combination Immunotherapy Using Fecal Microbiota Transplantation. 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT04163289 (accessed on 19 December 2022).
- GIANLUCA, I. Targeting Gut Microbiota to Improve Efficacy of Immune Checkpoint Inhibitors in Patients with Advanced Renal Cell Carcinoma. 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT04758507 (accessed on 19 December 2022).
- Kyte, J.A. MITRIC: Microbiota Transplant to Cancer Patients Who Have Failed Immunotherapy Using Faeces from Clincal Responders. 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT05286294 (accessed on 19 December 2022).
- Roussy, G.; Campus, C.; Paris, G. A Pilot Study Evaluating a Ketogenic Diet Concomitant to Nivolumab and Ipilimumab in Patients with Metastatic Renal Cell Carcinoma. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT05119010 (accessed on 19 December 2022).
- Ferrere, G.; Alou, M.T.; Liu, P.; Goubet, A.-G.; Fidelle, M.; Kepp, O.; Durand, S.; Iebba, V.; Fluckiger, A.; Daillère, R.; et al. Ketogenic diet and ketone bodies enhance the anticancer effects of PD-1 blockade. J. Clin. Investig. 2021, 6, e145207. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef]
- Cogdill, A.P.; Gaudreau, P.O.; Arora, R.; Gopalakrishnan, V.; Wargo, J.A. The Impact of Intratumoral and Gastrointestinal Microbiota on Systemic Cancer Therapy. Trends Immunol. 2018, 39, 900–920. [Google Scholar] [CrossRef]
- Brooks, A.W.; Priya, S.; Blekhman, R.; Bordenstein, S.R. Gut microbiota diversity across ethnicities in the United States. PLOS Biol. 2018, 16, e2006842. [Google Scholar] [CrossRef]
- The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Guo, G.; Han, J.; Yu, J. Gut microbiome in modulating immune checkpoint inhibitors. Ebiomedicine 2022, 82, 104163. [Google Scholar] [CrossRef]
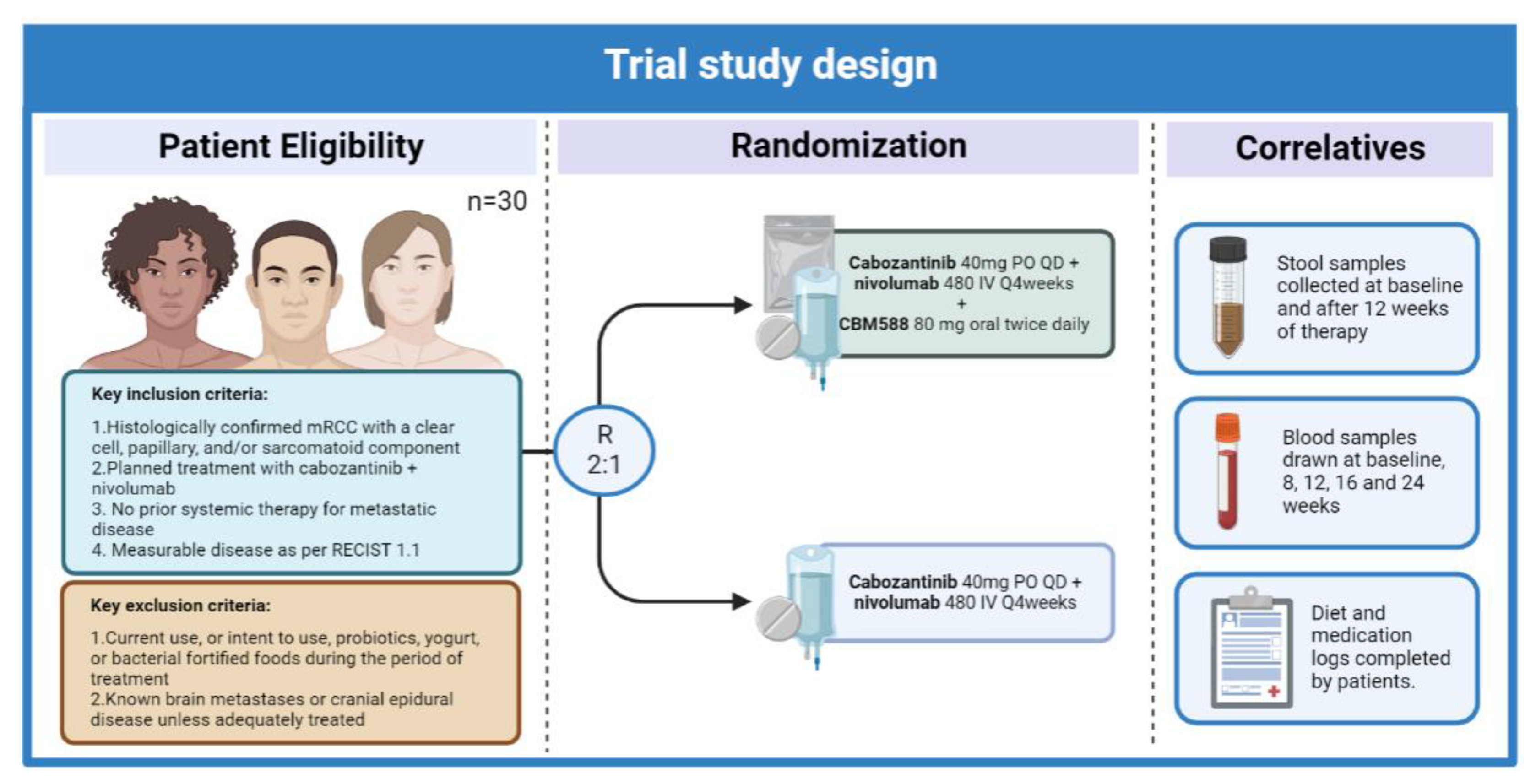
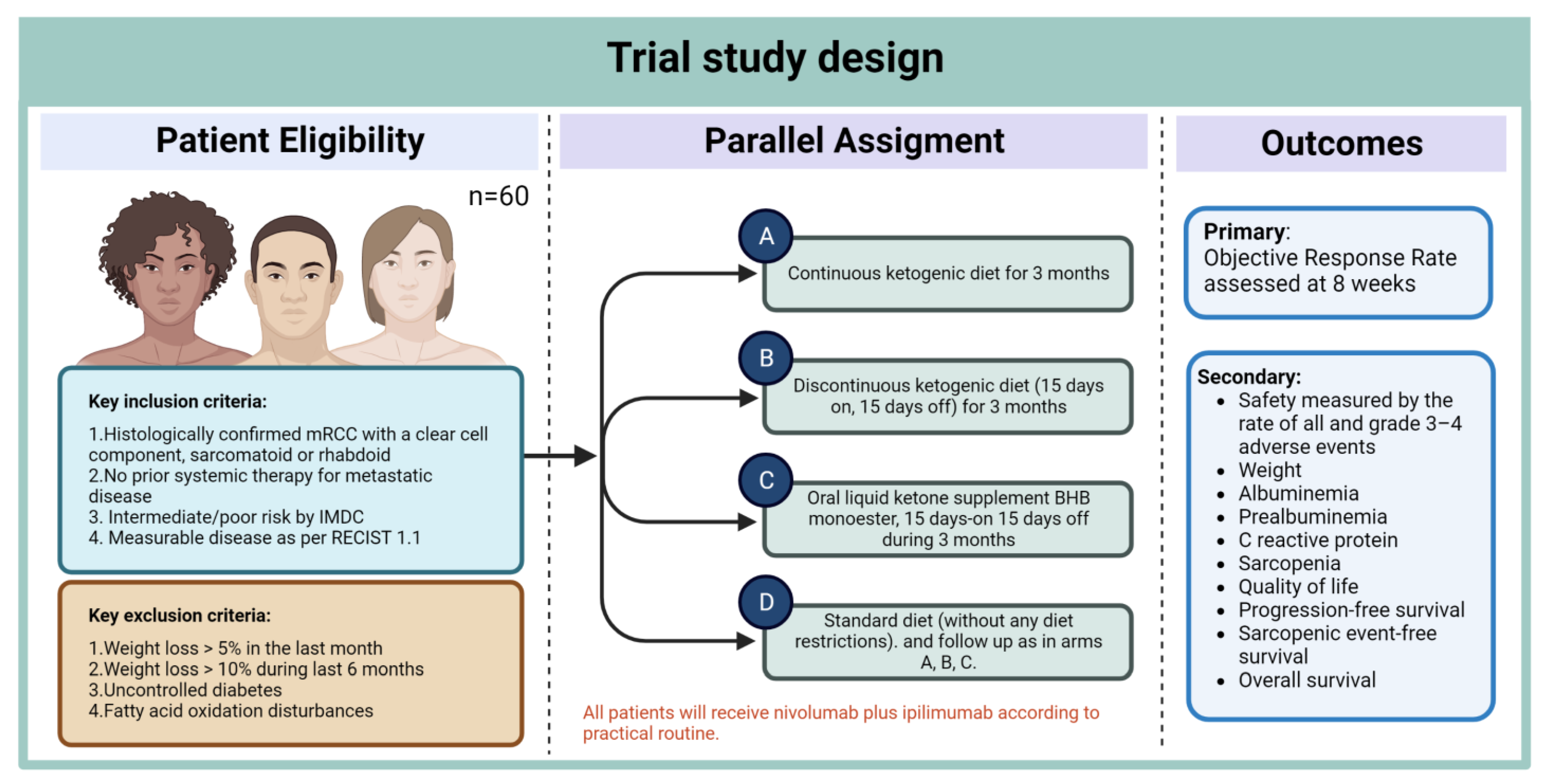
| Study | Patient Population | Microorganism Associated with Response/Clinical Benefit | Treatment |
|---|---|---|---|
| Routy et al, Science (2018) [50] | Patients with metastatic RCC or NSCLC | Akkermansia muciniphila, Ruminococcus, Alistipes, and Eubacterium | Anti PD-L1 |
| Derosa et al, European Urology, (2020) [85] | Patients with advanced RCC | Akkermansia muciniphila, Bacteroides salyersiae, and Eubacterium siraeum | Nivolumab |
| Salgia European Urology (2020) [86] | Patients with metastatic RCC | Akkermansia municiphila, Prevotella copri, Feacalibacterium rumino Bifidobacterium adolescentis, and Barnesiella intestinihominis | Nivolumab or nivolumab with ipilimumab |
| Dizman et al, Cancer Medicine (2021) [90] | Patients with metastatic RCC | Akkermansia muciniphila, Barnesiella intestinihominis and Bacteroides caccae | VEGF-TKI therapy |
| Dizman et al, Nature Medicine (2022) [91] | Patients with metastatic RCC | Bifidobacterium spp. | Nivolumab with ipilimumab +/− CBM588 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meza, L.; Feng, M.; Lee, K.; Sperandio, R.; Pal, S.K. The Gut Microbiome and Metastatic Renal Cell Carcinoma. J. Clin. Med. 2023, 12, 1502. https://doi.org/10.3390/jcm12041502
Meza L, Feng M, Lee K, Sperandio R, Pal SK. The Gut Microbiome and Metastatic Renal Cell Carcinoma. Journal of Clinical Medicine. 2023; 12(4):1502. https://doi.org/10.3390/jcm12041502
Chicago/Turabian StyleMeza, Luis, Matthew Feng, Kyle Lee, Rubens Sperandio, and Sumanta Kumar Pal. 2023. "The Gut Microbiome and Metastatic Renal Cell Carcinoma" Journal of Clinical Medicine 12, no. 4: 1502. https://doi.org/10.3390/jcm12041502
APA StyleMeza, L., Feng, M., Lee, K., Sperandio, R., & Pal, S. K. (2023). The Gut Microbiome and Metastatic Renal Cell Carcinoma. Journal of Clinical Medicine, 12(4), 1502. https://doi.org/10.3390/jcm12041502






