Abstract
Myelodysplastic neoplasm (MDS) is a heterogeneous group of myeloid neoplasms affected by germline and somatic genetic alterations. The incidence of MDS increases with age but rarely occurs at a young age. We investigated the germline and somatic genetic alterations of Korean patients with young-onset MDS (<40 years). Among the thirty-one patients, five (16.1%) had causative germline variants predisposing them to myeloid neoplasms (three with GATA2 variants and one each with PGM3 and ETV variants). We found that PGM3 deficiency, a subtype of severe immunodeficiency, predisposes patients to MDS. Somatic mutations were identified in 14 patients (45.2%), with lower rates in patients aged < 20 years (11.1%). Nine (29%) patients had U2AF1 S34F/Y mutations, and patients with U2AF1 mutations showed significantly worse progression-free survival (p < 0.001) and overall survival (p = 0.006) than those without U2AF1 mutations. A UBA1 M41T mutation that causes VEXAS syndrome was identified in a male patient. In conclusion, a germline predisposition to myeloid neoplasms occurred in ~16% of young-onset MDS patients and was largely associated with primary immunodeficiencies, including GATA2 deficiency. Furthermore, the high frequency of somatic U2AF1 mutations in patients with young-onset MDS suggests the presence of a distinct MDS subtype.
1. Introduction
Myelodysplastic neoplasm (MDS) is a heterogeneous group of myeloid neoplasms characterized by ineffective hematopoiesis, dysplastic hematopoietic cells, peripheral blood cytopenia, and a risk of progression to acute myeloid leukemia (AML) [1]. MDS is characterized by distorted hematopoietic stem cell function, inflammatory and innate immune dysregulation, deregulated apoptosis, and multiple genomic events, which result in heterogeneous clinical symptoms and outcomes [2]. The incidence of MDS increases with age, starting to rise rapidly after the age of 50 years, and is most prevalent in those aged 70–80 years [3,4,5]. Conversely, MDS rarely occurs in individuals under the age of 40, with an incidence rate of 0.14 per 100,000 in the United States [3]. In Korea, the rates of 0.48 and 0.39 per 100,000 individuals have been reported for males and females under 35 years of age, respectively [5].
Both germline and somatic genetic alterations contribute to the development of MDS [6]. Furthermore, these genetic alterations in patients with MDS play a crucial role in the clinical phenotypes, prognosis, and responses to treatment, making them crucial for predicting the patient’s clinical course and optimizing treatment and management [7]. Somatic alterations are found in approximately 78–90% of patients with MDS and primarily occur in genes associated with DNA methylation (TET2, DNMT3A, IDH1/IDH2), chromatin modification (ASXL1, EZH2), RNA splicing (SF3B1, SRSF2, U2AF1), and DNA damage pathway (TP53) [2,7,8]. However, due to the low prevalence of MDS in young age groups, most of the patients included in studies on somatic mutations in MDS are older, and it is not well understood whether somatic mutations occurring in young patients differ from those occurring in older patients.
Germline predisposition to myeloid neoplasms has been reported in relation to germline alterations in CEBPA, DDX41, TP53, RUNX1, ANKRD26, ETV6, GATA2, and SAMD9/SAMD9L as well as alterations in genes associated with bone marrow failure syndromes (Fanconi anemia, dyskeratosis congenita and related telomere biology disorders, Diamond–Blackfan anemia, and Shwachman–Diamond syndrome) [9,10,11,12]. The causative genes and their frequencies in these germline predispositions have been reported to vary across different studies, and additional causative genes are still being discovered [13,14,15]. In particular, bone marrow failure syndromes that overlap clinically with MDS share overlapping clinical features with primary immunodeficiencies, and a recent study reported that approximately 17% of patients with bone marrow failure have underlying primary immunodeficiencies [16]. Furthermore, germline predisposition genes exhibit age-related differences, with germline DDX41 variants observed in approximately 2.4–3.8% of patients with myeloid neoplasms and are prevalent in older age [17,18,19]. In contrast, genes other than DDX41 are known to cause myeloid neoplasms in younger age groups, and there may be racial differences (for instance, between European and East Asian) in the distribution and frequency of genes that cause germline predisposition.
In this study, we aimed to investigate the germline and somatic genetic characteristics of young-onset Korean patients with MDS and to examine the clinical and prognostic features associated with genetic factors.
2. Materials and Methods
2.1. Patients
We retrospectively evaluated 31 patients with young-onset MDS for whom specimens for genetic testing were available, diagnosed between 2007 and 2020 at our institution; their characteristics are summarized in Table 1. Young-onset MDS was defined as an onset age of less than 40 years, and the median age of the patients was 24 years (range, 0–39 years). All patients were diagnosed or revised according to the 2016 and 2022 World Health Organization (WHO) classification of myeloid neoplasms [11,20]. Clinical and laboratory information of the patients, including complete blood cell count (CBC), bone marrow (BM) examination, and cytogenetic and molecular genetic studies, were obtained from electronic medical records. Cytogenetic risk stratification was based on the revised international prognostic scoring system (R-IPSS) [21]. As a control group for somatic mutations detected in patients with young-onset MDS, we used targeted NGS data from 50 patients aged 40 years or older who were diagnosed with MDS and underwent targeted NGS testing for 38 genes (Supplementary Table S1) between November 2018 and October 2021. This study was approved by the Institutional Review Board of the Samsung Medical Center, Seoul, Korea (SMC IRB No. 2021-12-150).

Table 1.
Clinical and laboratory characteristics of 31 patients with young-onset MDS.
2.2. Molecular Genetic Study
Genomic DNA was extracted from bone marrow aspirates at the time of diagnosis using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) or the QIAamp DNA Blood Mini Kit (Qiagen, Venlo, The Netherlands), according to the manufacturer’s instructions. Library preparation was performed using a G-Mendeliom panel (Celemics, Seoul, Republic of Korea) containing 5857 disease-related genes, and sequencing was performed on the DNBSEQ-G400 platform (MGI Tech Co., Ltd., Shenzhen, China), with an average sequencing coverage of 160x. Reads were aligned using the BWA-MEM tool (version 0.7.17) to the human genomic reference sequence GRCh37/hg19, variant calling was performed using the GATK package (v4.1.8), and annotation was performed using VEP101 (VariantEffect Predictor) and dbNSFP v4.1. Each variant was annotated with a population database (Genome Aggregation Database (gnomAD), Exome Sequencing Project (ESP), Korean Reference Genome Database (KRGDB)) and disease databases (ClinVar, Human Gene Mutation Database (HGMD), Online Mendelian Inheritance in Man (OMIM), and Catalogue of Somatic Mutations in Cancer (COSMIC)). In silico analyses were performed using SIFT, PolyPhen-2, MutationTaster, and SpliceAI. To identify germline variants predisposing patients to myeloid neoplasms, we performed a variant analysis of 524 genes that were possibly associated with myeloid neoplasm predisposition (Supplementary Table S2). We considered variants with a variant allele frequency (VAF) of ≥40% as presumed germline variants in these genes, except recurrent variants, reported 10 times or more in COSMIC. We assessed their pathogenicity according to the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) guidelines for sequence variants [22], and referred to pathogenic and likely pathogenic variants as causative variants. Also, we assessed somatic mutations classified as clinically significant (Tier I/II), following the 2017 AMP, American Society of Clinical Oncology (ASCO), and College of American Pathology (CAP) somatic variant guideline [23], in 38 genes where somatic mutations commonly occur in myeloid neoplasms (Supplementary Table S3). All clinically relevant variants were visually inspected using Integrative Genomic Viewer [24].
2.3. Germline Testing
Germline testing was performed for presumed causative germline variants using bone marrow aspirates or peripheral blood in remission. Direct sequencing was performed using primers specific to the target variant on an ABI Prism 3130xl Genetic Analyzer using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA).
2.4. Statistical Analysis
Categorical variables were compared using the chi-square test or Fisher’s exact test, as appropriate, and continuous variables were compared using the Mann–Whitney U test. Progression-free survival (PFS) was determined from the time of initial diagnosis to disease progression or the last follow-up, and overall survival (OS) was determined from the time of initial diagnosis to death or the last follow-up. Survival analysis was performed using Kaplan–Meier plots, and differences in survival were compared using the log-rank test. All statistical analyses were performed using SPSS version 27 software (IBM Corp., Armonk, NY, USA). A p-value < 0.05 was considered statistically significant.
3. Results
3.1. Germline Variants Predisposing to Myeloid Neoplasms in Young-Onset MDS
Among the thirty-one patients, five (16.1%) had causative germline variants predisposing them to myeloid neoplasms in the GATA2, PGM3, and ETV6 genes (Table 2), and all of these identified variants have been previously reported [25,26,27,28,29,30]. GATA2 was the most common causative gene (9.7%), and three distinct GATA2 heterozygous variants were identified in three male patients aged 22–28 years. Among them, one patient (R361H) had warts in the nostrils; another (K390del) had prolonged and severe infections, ultimately leading to uncontrolled pneumonia and death; and the third patient (Y141*) had previously been found to have a hypocellular marrow. Cytogenetic abnormalities, including trisomy 8 or 7q deletions, were found in two patients with GATA2 variant. A homozygous splicing variant (c.871+5G>A) of PGM3 was found in a 3-year-old girl. She had experienced recurrent pneumonia since early childhood. At the time of admission, she presented with anemia and severe neutropenia accompanied by eosinophilia, and BM examination revealed hypocellular marrow along with dysplasia in granulocytes and megakaryocytes. Given that the previous microarray analysis confirmed a heterozygous 6q14.1q14.3 microdeletion encompassing PGM3 in this patient, we concluded that the patient carried compound heterozygous variants consisting of a heterozygous PGM3 deletion and a heterozygous splicing variant. Finally, a heterozygous nonsense variant (R359*) of ETV6 was identified in a 28-year-old male, which was clinically consistent with a previous history of thrombocytopenia.

Table 2.
Patients with MDS harboring causative germline variants predisposing to myeloid neoplasms and their clinical and laboratory information.
3.2. Somatic Mutations and Prognostic Significance of U2AF1 Mutation in Young-Onset MDS
Somatic mutations were identified in 14 patients (45.2%) (Table 3), and patients younger than 20 years had a lower somatic mutation rate than those older than 20 years (11.1% vs. 59.1%; p = 0.015). The presence of somatic mutations was not significantly different between patients with and without an underlying germline predisposition (60% vs. 42.3%, p = 0.467). Notably, nine (29%) patients had U2AF1 mutations. All U2AF1 mutations were missense mutations occurring in codon 34, with a median VAF of 42% (range, 37–52%). Specifically, S34F and S34Y were observed in five (16.1%) and four (12.9%) patients, respectively, and there was no difference in VAF among the mutations (42% vs. 43.2%; p = 0.806). In contrast, mutations in other genes, including ASXL1, DNMT3A, SF3B1, STAG2, and TP53, were observed at lower frequencies, ranging from once to twice.

Table 3.
Somatic mutations identified in patients with young-onset MDS.
When compared with a cohort of 50 patients with late-onset MDS (≥40 years), U2AF1 mutations were significantly more frequent in patients with young-onset MDS (29% vs. 6%; p = 0.008), whereas other somatic mutations were observed at much lower frequencies (Table 4). In the survival analysis stratified by U2AF1 mutation status, patients with U2AF1 mutations showed inferior PFS and OS compared to those without U2AF1 mutations (p < 0.001 and p = 0.006, respectively) (Figure 1). However, no significant differences were observed in the other clinical and laboratory findings based on the presence of U2AF1 mutations (Supplementary Table S4).

Table 4.
Comparison of major somatic mutations according to the onset age in patients with MDS.
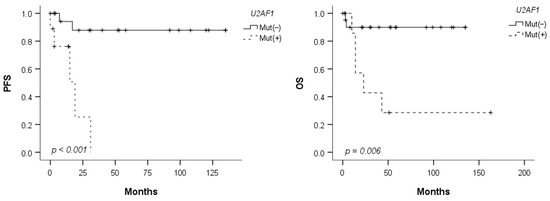
Figure 1.
Progression-free survival (PFS) and overall survival (OS) according to the U2AF1 mutation status.
3.3. Clinical Course of a Case with Somatic UBA1 Mutation
Interestingly, the diagnostic marker for VEXAS syndrome, UBA1 M41T mutation (VAF, 83.3%), was identified in a 38-year-old male patient, and we reviewed his clinical disease course. Three years prior to the diagnosis of MDS, he developed a progressive erythematous maculopapular rash accompanied by fever, myalgia, and tenderness, along with auricular chondritis and polyarthritis. At that time, the CBC showed Hb 10.6 g/dL, WBC 3.73 × 109/L, and a platelet count of 167 × 109/L. A BM examination performed due to a history of leukopenia showed normocellular marrow with no evidence of dysplasia but accompanied by granulocytic hyperplasia. Subsequently, the patient was diagnosed with relapsing polychondritis and treated with prednisolone and/or cyclosporine. During follow-up, as cytopenia worsened, the second BM examination was performed three years later. CBC showed Hb 9.5 g/dL, WBC 3.08 × 109/L, and platelets 128 × 109/L. BM examination revealed a hypercellular marrow with trilineage dysplasia, leading to the diagnosis of MDS with multilineage dysplasia according to the 2016 WHO classification. While receiving conservative treatment for MDS, the cytopenia deteriorated significantly. Six years after the diagnosis of MDS, the patient underwent allogeneic hematopoietic stem cell transplantation, and disease-related symptoms improved with complete engraftment. In the pre-transplant bone marrow examination, characteristic findings of VEXAS syndrome, specifically cytoplasmic vacuolation of myeloid precursor cells, were observed (Figure 2).
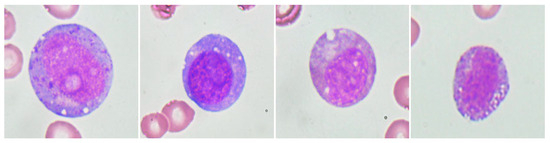
Figure 2.
Cytoplasmic vacuolation of myeloid precursor cells observed in the bone marrow of a patient with somatic UBA1 mutation.
4. Discussion
This study identified diverse genetic characteristics in young-onset MDS patients from germline and somatic perspectives. The identification of germline variants predisposing patients to myeloid neoplasms is critical for making treatment decisions, identifying at-risk family members, and selecting donors for hematopoietic stem cell transplantation [26]. However, the frequency of variants and spectrum of genes involved vary slightly among studies, and racial differences may contribute to these variations. In Western populations, the frequency of germline predisposition in patients with young-onset MDS is approximately 7–17% [13,14,15]. Keel et al. reported that 13.6% of 110 children and young adults (<40 years old) with MDS had a germline predisposition, with germline variants in FANCA, MPL, RTEL4, SBDS, TERT, TINF2, GATA2, RUNX1, and TP53 [13]. Wlodarski et al. conducted a study targeting GATA2 and found germline GATA2 variants in 7% of 426 children and adolescents with de novo MDS, particularly in 15% of the patients with advanced MDS [14]. Moreover, Schwartz et al. found germline SAMD9 or SAMD9L variants in 17% of 46 patients with de novo MDS [15]. In our study, a germline predisposition was identified in 16.1% of the patients with young-onset MDS, and the overall frequency was similar to that reported in previous studies [13,14,15]. However, the patients primarily harbored GATA2 variants, and no patients with classical bone marrow failure syndrome (such as Fanconi anemia) were identified.
GATA2 deficiency, caused by heterozygous germline GATA2 variants, is associated with MDS/AML, monocytopenia and mycobacterial infections (MonoMAC), dendritic cell, monocyte, B and natural killer (NK) lymphoid deficiency (DMLC), and/or lymphedema [9,28]. In a study reporting the natural history of GATA2 deficiency in French and Belgian patients, the median age at the first clinical symptom onset was 19 years [28]. MDS (70%) was the most common hematological malignancy, followed by cutaneous or genital recurrent HPV-related warts (40%) and severe bacterial infections (56%). In our study, GATA2 variants were identified in 9.7% of patients, all of whom were men in their 20s, and the clinical manifestations related to GATA2 deficiency were heterogeneous among the patients. Studies have indicated a higher frequency of leukemia in patients with GATA2 missense variants and an increased risk of lymphedema in those with null variants [28,31]. However, the genotype–phenotype correlation in GATA2 deficiency has not been clearly observed, and phenotypic heterogeneity has been reported even within families with the same variant [32].
PGM3 deficiency, a rare autosomal recessive congenital glycosylation disorder, is a subtype of the severe immunodeficiency caused by variants in PGM3. It is characterized by various clinical phenotypes, including leukopenia, severe neutropenia, T/B lymphopenia, skeletal dysplasia, and progression to bone marrow failure [33]. In this study, we found for the first time that PGM3 deficiency predisposes patients to MDS. In primary immunodeficiencies, the lack of effective immune surveillance against persistent and recurrent infections has been suggested to contribute to an increased risk of tumors and myeloid neoplasms [34,35]. Overall, in our study, primary immunodeficiencies, including GATA2 deficiency, were identified as the predominant factor, accounting for 12.9% of germline predispositions in young-onset MDS patients.
Few studies have reported somatic mutations focusing on patients with young-onset MDS [36,37]. In this study, we found U2AF1 mutations in 29% of the patients, suggesting that these mutations are enriched in young-onset MDS. Conversely, mutations in DNMT3A, ASXL1, SF3B1, and TP53, which are typically prevalent in MDS and frequently occur during age-related hematopoiesis [7], were rarely observed in our study. U2AF1 is a small subunit of U2 snRNP auxiliary factor (U2AF) that is involved in pre-mRNA processing. U2AF1 mutations have been reported in 7–11% of patients with MDS, primarily involving codon S34 and Q157 mutations [37,38,39,40]. Although some studies have not shown a significant association between U2AF1 mutations and patient age [6,39,40], a higher prevalence of U2AF1 mutations in young patients has been reported in several studies [37,41,42]. Wu et al. reported that U2AF1 mutations were observed at a frequency of 7.5% in 478 patients with MDS, with a notably higher prevalence of 13.2% in patients under the age of 40 years [37]. In a study by Kim et al. involving 152 Korean patients with MDS, the frequency of U2AF1 mutations was 16%, which was higher than that in Western populations [41]. Furthermore, the S34F mutation was associated with younger onset. Similarly, Li et al. reported the presence of U2AF1 mutations in 17% of 511 Chinese patients with MDS, with higher prevalence in young patients [42]. In particular, in patients aged 40 or younger, the frequency of U2AF1 mutations slightly exceeded 20%. Given that U2AF1 mutations have been suggested to be early initiating genetic events in MDS [39,42], our findings indicate that U2AF1 mutations play a particularly important role in the pathogenesis of MDS, occurring at a younger age. Additionally, while previous studies have suggested a potential association between U2AF1 mutations and trisomy 8 [41,42], we did not observe such an association in our study. Moreover, U2AF1 mutations have been reported to be associated with poor prognosis in MDS [43,44]. In line with these findings, our study revealed a distinct difference in patient PFS and OS based on the presence of U2AF1 mutations, indicating a significant adverse prognostic impact of U2AF1 mutations, particularly in young-onset MDS.
Moreover, we identified a somatic UBA1 mutation, which was the underlying cause of VEXAS syndrome (Vacuoles, E1 enzyme, X-linked, Autoinflammatory, Somatic), in one patient. VEXAS syndrome is a disease characterized by rheumatic and hematological features and was first described in 2020 [45]. The majority of patients exhibit characteristic clinical manifestations of inflammatory conditions, such as relapsing polychondritis, Sweet syndrome, and nodular vasculitis, with 25–50% of patients developing MDS [45,46]. It primarily occurs in males, and M41T, which was observed in our patient, is the most common mutation, followed by M41V/L. To date, limited data have been published on the treatment of VEXAS, particularly in cases related to MDS [47]. In this study, we observed an improvement in relapsing polychondritis and MDS-related findings following allogeneic hematopoietic stem cell transplantation.
Our study has several limitations. Because BM samples were used, there is a possibility that germline variants predisposing to myeloid neoplasms may have been masked by somatic genetic rescue, albeit rare. Furthermore, the sensitivity of the NGS testing performed in patients with young-onset MDS was lower than that of the testing used in the control group. Thus, we cannot rule out the possibility that low-burden somatic mutations might not have been detected in patients with young-onset MDS, resulting in a low somatic mutation rate.
5. Conclusions
To the best of our knowledge, this is the first comprehensive investigation of germline predisposition gene variants in Korean patients with MDS. The prevalence of germline predisposition to myeloid neoplasm in young-onset MDS was approximately 16%, largely associated with primary immunodeficiencies, and we have demonstrated the PGM3 variants as the cause of germline predisposition for the first time. Furthermore, somatic U2AF1 mutations associated with poor prognosis were observed in approximately one third of the patients, suggesting the possibility of a distinct subtype in young-onset MDS patients. However, additional validation studies in a multi-center cohort are necessary to confirm these findings.
Supplementary Materials
The online version contains supplementary material available at https://www.mdpi.com/article/10.3390/jcm12247651/s1, Table S1. The gene list of the targeted NGS panel conducted in the control group (N = 38); Table S2: Target gene list for detecting germline variant predisposing to myeloid neoplasm in patients with young-onset MDS (N = 524); Table S3: Target gene list for somatic mutation detection in patients with young-onset MDS (N = 38); Table S4: Comparison of clinical and laboratory characteristics of 31 patients with young-onset MDS according to somatic U2AF1 mutation status.
Author Contributions
H.-Y.K. designed the study, analyzed the data, and wrote the manuscript. K.H.Y. and C.W.J. collected patient data. H.-J.K. and S.-H.K. interpreted the data and contributed to manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by a grant from the National Research Foundation of Korea (grant number 2022R1F1A1073963).
Institutional Review Board Statement
The study was reviewed and approved by the Institutional Review Board of Samsung Medical Center, Seoul, Korea (SMC IRB No. 2021-12-150).
Informed Consent Statement
This study complied with the Declaration of Helsinki. All BM samples were collected after obtaining informed consent.
Data Availability Statement
All data relevant to the study have been included in the article or uploaded as supplementary information. The additional data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tanaka, T.N.; Bejar, R. MDS overlap disorders and diagnostic boundaries. Blood 2019, 133, 1086–1095. [Google Scholar] [CrossRef]
- Garcia-Manero, G. Myelodysplastic syndromes: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2023, 98, 1307–1325. [Google Scholar] [CrossRef] [PubMed]
- Rollison, D.E.; Howlader, N.; Smith, M.T.; Strom, S.S.; Merritt, W.D.; Ries, L.A.; Edwards, B.K.; List, A.F. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States, 2001–2004, using data from the NAACCR and SEER programs. Blood 2008, 112, 45–52. [Google Scholar] [CrossRef]
- Goldberg, S.L.; Chen, E.; Corral, M.; Guo, A.; Mody-Patel, N.; Pecora, A.L.; Laouri, M. Incidence and clinical complications of myelodysplastic syndromes among United States Medicare beneficiaries. J. Clin. Oncol. 2010, 28, 2847–2852. [Google Scholar] [CrossRef] [PubMed]
- Park, E.H.; Lee, H.; Won, Y.J.; Ju, H.Y.; Oh, C.M.; Ingabire, C.; Kong, H.J.; Park, B.K.; Yoon, J.Y.; Eom, H.S.; et al. Nationwide statistical analysis of myeloid malignancies in Korea: Incidence and survival rate from 1999 to 2012. Blood Res. 2015, 50, 204–217. [Google Scholar] [CrossRef]
- Saygin, C.; Godley, L.A. Genetics of Myelodysplastic Syndromes. Cancers 2021, 13, 3380. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S. Genetics of MDS. Blood 2019, 133, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Klco, J.M.; Mullighan, C.G. Advances in germline predisposition to acute leukaemias and myeloid neoplasms. Nat. Rev. Cancer 2021, 21, 122–137. [Google Scholar] [CrossRef]
- University of Chicago Hematopoietic Malignancies Cancer Risk Team. How I diagnose and manage individuals at risk for inherited myeloid malignancies. Blood 2016, 128, 1800–1813. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef] [PubMed]
- Keel, S.B.; Scott, A.; Sanchez-Bonilla, M.; Ho, P.A.; Gulsuner, S.; Pritchard, C.C.; Abkowitz, J.L.; King, M.C.; Walsh, T.; Shimamura, A. Genetic features of myelodysplastic syndrome and aplastic anemia in pediatric and young adult patients. Haematologica 2016, 101, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Wlodarski, M.W.; Hirabayashi, S.; Pastor, V.; Starý, J.; Hasle, H.; Masetti, R.; Dworzak, M.; Schmugge, M.; van den Heuvel-Eibrink, M.; Ussowicz, M.; et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood 2016, 127, 1387–1397; quiz 1518. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.R.; Ma, J.; Lamprecht, T.; Walsh, M.; Wang, S.; Bryant, V.; Song, G.; Wu, G.; Easton, J.; Kesserwan, C.; et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat. Commun. 2017, 8, 1557. [Google Scholar] [CrossRef] [PubMed]
- Miano, M.; Grossi, A.; Dell’Orso, G.; Lanciotti, M.; Fioredda, F.; Palmisani, E.; Lanza, T.; Guardo, D.; Beccaria, A.; Ravera, S.; et al. Genetic screening of children with marrow failure. The role of primary Immunodeficiencies. Am. J. Hematol. 2021, 96, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.E.; Routbort, M.J.; DiNardo, C.D.; Bueso-Ramos, C.E.; Kanagal-Shamanna, R.; Khoury, J.D.; Thakral, B.; Zuo, Z.; Yin, C.C.; Loghavi, S.; et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am. J. Hematol. 2019, 94, 757–766. [Google Scholar] [CrossRef]
- Sébert, M.; Passet, M.; Raimbault, A.; Rahmé, R.; Raffoux, E.; Sicre de Fontbrune, F.; Cerrano, M.; Quentin, S.; Vasquez, N.; Da Costa, M.; et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood 2019, 134, 1441–1444. [Google Scholar] [CrossRef]
- Makishima, H.; Saiki, R.; Nannya, Y.; Korotev, S.; Gurnari, C.; Takeda, J.; Momozawa, Y.; Best, S.; Krishnamurthy, P.; Yoshizato, T.; et al. Germ line DDX41 mutations define a unique subtype of myeloid neoplasms. Blood 2023, 141, 534–549. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, R.E.; Milne, P.; Jardine, L.; Zandi, S.; Swierczek, S.I.; McGovern, N.; Cookson, S.; Ferozepurwalla, Z.; Langridge, A.; Pagan, S.; et al. The evolution of cellular deficiency in GATA2 mutation. Blood 2014, 123, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Keel, S.B.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Watts, A.C.; Pritchard, C.C.; Salipante, S.J.; Jeng, M.R.; Hofmann, I.; et al. Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity. Haematologica 2015, 100, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Bluteau, O.; Sebert, M.; Leblanc, T.; Peffault de Latour, R.; Quentin, S.; Lainey, E.; Hernandez, L.; Dalle, J.H.; Sicre de Fontbrune, F.; Lengline, E.; et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood 2018, 131, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Donadieu, J.; Lamant, M.; Fieschi, C.; de Fontbrune, F.S.; Caye, A.; Ouachee, M.; Beaupain, B.; Bustamante, J.; Poirel, H.A.; Isidor, B.; et al. Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica 2018, 103, 1278–1287. [Google Scholar] [CrossRef]
- Moriyama, T.; Metzger, M.L.; Wu, G.; Nishii, R.; Qian, M.; Devidas, M.; Yang, W.; Cheng, C.; Cao, X.; Quinn, E.; et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: A systematic genetic study. Lancet Oncol. 2015, 16, 1659–1666. [Google Scholar] [CrossRef]
- Okano, T.; Imai, K.; Naruto, T.; Okada, S.; Yamashita, M.; Yeh, T.W.; Ono, S.; Tanaka, K.; Okamoto, K.; Tanita, K.; et al. Whole-Exome Sequencing-Based Approach for Germline Mutations in Patients with Inborn Errors of Immunity. J. Clin. Immunol. 2020, 40, 729–740. [Google Scholar] [CrossRef]
- Spinner, M.A.; Sanchez, L.A.; Hsu, A.P.; Shaw, P.A.; Zerbe, C.S.; Calvo, K.R.; Arthur, D.C.; Gu, W.; Gould, C.M.; Brewer, C.C.; et al. GATA2 deficiency: A protean disorder of hematopoiesis, lymphatics, and immunity. Blood 2014, 123, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.F.; Buechner, J.; Myhre, A.E.; Galteland, E.; Spetalen, S.; Kulseth, M.A.; Sorte, H.S.; Holla, O.L.; Lundman, E.; Alme, C.; et al. A Nationwide Study of GATA2 Deficiency in Norway-the Majority of Patients Have Undergone Allo-HSCT. J. Clin. Immunol. 2022, 42, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Stray-Pedersen, A.; Backe, P.H.; Sorte, H.S.; Mørkrid, L.; Chokshi, N.Y.; Erichsen, H.C.; Gambin, T.; Elgstøen, K.B.; Bjørås, M.; Wlodarski, M.W.; et al. PGM3 mutations cause a congenital disorder of glycosylation with severe immunodeficiency and skeletal dysplasia. Am. J. Hum. Genet. 2014, 95, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Barcellini, W.; Fattizzo, B. Immune Phenomena in Myeloid Neoplasms: An “Egg or Chicken” Question. Front. Immunol. 2021, 12, 751630. [Google Scholar] [CrossRef]
- Verhoeven, D.; Stoppelenburg, A.J.; Meyer-Wentrup, F.; Boes, M. Increased risk of hematologic malignancies in primary immunodeficiency disorders: Opportunities for immunotherapy. Clin. Immunol. 2018, 190, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Kozyra, E.J.; Hirabayashi, S.; Pastor Loyola, V.B.; Przychodzen, B.; Karow, A.; Catala, A.; De Moerloose, B.; Dworzak, M.; Hasle, H.; Masetti, R.; et al. Clonal Mutational Landscape of Childhood Myelodysplastic Syndromes. Blood 2015, 126, 1662. [Google Scholar] [CrossRef]
- Wu, S.J.; Tang, J.L.; Lin, C.T.; Kuo, Y.Y.; Li, L.Y.; Tseng, M.H.; Huang, C.F.; Lai, Y.J.; Lee, F.Y.; Liu, M.C.; et al. Clinical implications of U2AF1 mutation in patients with myelodysplastic syndrome and its stability during disease progression. Am. J. Hematol. 2013, 88, E277–E282. [Google Scholar] [CrossRef]
- Visconte, V.; Nakashima, M.O.; Rogers, H.J. Mutations in Splicing Factor Genes in Myeloid Malignancies: Significance and Impact on Clinical Features. Cancers 2019, 11, 1844. [Google Scholar] [CrossRef]
- Graubert, T.A.; Shen, D.; Ding, L.; Okeyo-Owuor, T.; Lunn, C.L.; Shao, J.; Krysiak, K.; Harris, C.C.; Koboldt, D.C.; Larson, D.E.; et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2011, 44, 53–57. [Google Scholar] [CrossRef]
- Thol, F.; Kade, S.; Schlarmann, C.; Löffeld, P.; Morgan, M.; Krauter, J.; Wlodarski, M.W.; Kölking, B.; Wichmann, M.; Görlich, K.; et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 2012, 119, 3578–3584. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, K.; Hwang, B.; Im, K.; Park, S.N.; Kim, J.A.; Hwang, S.M.; Bang, D.; Lee, D.S. The high frequency of the U2AF1 S34Y mutation and its association with isolated trisomy 8 in myelodysplastic syndrome in Asians, but not in Caucasians. Leuk. Res. 2017, 61, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, J.; Jia, Y.; Wang, J.; Xu, Z.; Qin, T.; Shi, Z.; Song, Z.; Peng, S.; Huang, H.; et al. Clinical features and biological implications of different U2AF1 mutation types in myelodysplastic syndromes. Genes Chromosomes Cancer 2018, 57, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, N.; Wu, X.; Zheng, X.; Ling, Y.; Gong, Y. Prognostic value of U2AF1 mutant in patients with de novo myelodysplastic syndromes: A meta-analysis. Ann. Hematol. 2019, 98, 2629–2639. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Arango Ossa, J.E.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evid. 2022, 1, EVIDoa2200008. [Google Scholar] [CrossRef]
- Beck, D.B.; Ferrada, M.A.; Sikora, K.A.; Ombrello, A.K.; Collins, J.C.; Pei, W.; Balanda, N.; Ross, D.L.; Ospina Cardona, D.; Wu, Z.; et al. Somatic Mutations in UBA1 and Severe Adult-Onset Autoinflammatory Disease. N. Engl. J. Med. 2020, 383, 2628–2638. [Google Scholar] [CrossRef]
- Templé, M.; Kosmider, O. VEXAS Syndrome: A Novelty in MDS Landscape. Diagnostics 2022, 12, 1590. [Google Scholar] [CrossRef]
- Comont, T.; Heiblig, M.; Rivière, E.; Terriou, L.; Rossignol, J.; Bouscary, D.; Rieu, V.; Le Guenno, G.; Mathian, A.; Aouba, A.; et al. Azacitidine for patients with Vacuoles, E1 Enzyme, X-linked, Autoinflammatory, Somatic syndrome (VEXAS) and myelodysplastic syndrome: Data from the French VEXAS registry Spectrum of clonal hematopoiesis in VEXAS syndrome. Br. J. Haematol. 2022, 196, 969–974. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).