Abstract
The emergence of extended-spectrum β-lactamase-producing Klebsiella pneumoniae, including CRKP infections, has resulted in significant morbidity and mortality worldwide. We aimed to explore the presence of bla genes (CTX-M, TEM, and SHV) in CRKP isolates. A total of 24 CRKP isolates were randomly selected from the Salmaniya Medical Complex Microbiology Laboratory. These isolates, which were positive for carbapenemases, were further explored for CTX-M, TEM, and SHV genes using PCR. All the CTX-M PCR amplicons were sent for sequencing. To determine genetic relatedness, molecular typing by ERIC-PCR was performed. The bla gene testing demonstrated that a significant proportion of these isolates harbored SHV, CTX-M, and TEM genes (100%, 91.6%, and 45.8%), respectively. Bioinformatic analyses confirmed CTX-M-15 in these isolates. ERIC-PCR analysis showed three clusters demonstrating genetic relatedness. The study findings reveal the concomitant carriage of the SHV and CTX-M-15 and a comparatively lower carriage of TEM genes in CRKP isolates. Our findings highlight the significance of routinely reporting the presence of antibiotic resistance genes along with regular antibiotic sensitivity reports, as this will aid clinicians in prescribing appropriate antibiotics.
1. Introduction
The public health threat posed by multidrug-resistant (MDR) bacterial infections has grown globally. Klebsiella pneumoniae (KP), one of the most prevalent members of Enterobacterales causing community- and hospital-acquired illnesses, tends to be resistant to multiple antibiotics [1]. The synthesis of beta-lactamases is the most typical mechanism of beta-lactam antibiotic resistance. Extended-spectrum beta-lactamases (ESBLs) are to blame for the rise in resistance in modern medicine. They are plasmid-encoded enzymes that are inhibited by clavulanic acid and have the ability to hydrolyze bonds of β-lactam rings from antibiotics like penicillins, cephalosporins, and aztreonam [2]. Presently, more than 600 ESBL variations have been identified, with the majority being associated with Temoneira (TEM), the Sulfhydryl Variable (SHV), and Cefotaxime hydrolyzing capabilities (CTX-M) (http://www.lahey.org/studies/webt.htm accessed on 15 July 2023). The majority of ESBLs found are either of the SHV or TEM types, and they are linked to nosocomial illnesses brought on by Gram-negative bacteria [3]. The presence of ESBLs carries enormous clinical importance. The ESBLs are frequently plasmid-encoded. Plasmids responsible for ESBL production frequently also carry resistance genes to other antibiotics. Although serious infections caused by ESBL-producing organisms are often treated with carbapenems, carbapenem-resistant isolates have recently been discovered [3]. In particular, the emergence of Enterobacterales resistant to third-generation cephalosporins and aztreonam, which is frequently associated with the expression of ESBLs, has caused antibiotic resistance among Gram-negative bacteria to rise rapidly over the past two decades [4]. According to the Clinical and Laboratory Standards Institute (CLSI) standards, regardless of the minimum inhibitory concentration (MIC) of a given cephalosporin, isolates with a positive phenotypic confirmatory test (combination disc tests/broth microdilution) should be reported as resistant to all cephalosporins (apart from the cephamycins, cefoxitin, and cefotetan) and aztreonam [5]. Furthermore, because the genes for ESBL synthesis are easily transferred by plasmids, many ESBL-producing Enterobacterales are also resistant to other antimicrobial drugs such as aminoglycosides, trimethoprim, and quinolones. This creates a severe dilemma in the treatment of ESBL-producing infections [6]. The incidence of ESBL-producing Enterobacteriaceae in hospitals and the general population varies greatly. Infections caused by Enterobacterales that produce ESBL are now of concern for a variety of reasons, including rising hospital expenditures, length of stay, and mortality rates [5].
Antibiotic resistance in K. pneumoniae is mainly driven by the acquisition of β-lactamases, such as extended-spectrum β-lactamases (ESBLs) and carbapenemases [7]. There is a greater risk of high mortality from infections caused by carbapenem-resistant, ESBL-producing, and multidrug-resistant Enterobacterales [8]. In fact, MDR K. pneumoniae, which produces carbapenemases, is regarded as a severe hazard to global health [1]. Although carbapenem antibiotics are the most effective therapeutic drug, due to their extensive usage, carbapenem-resistant K. pneumoniae (CRKP) is becoming more common throughout the world [9]. Several variables, such as the usage of antibiotics, horizontal gene transfer, and selective pressure in clinical settings, contribute to the emergence and spread of carbapenem resistance in K. pneumoniae. Colistin, in combination with tigecycline, rifampin, or carbapenem; fosfomycin plus colistin or amikacin; and double-carbapenem antibiotics (a combination of doripenem and ertapenem) are a few examples of combination antibiotics that are frequently used to treat CRKP [10]. Even though carbapenem-resistant Enterobacterales (CRE) and ESBL-producing strains frequently have similar genetic backgrounds, carry some of the same antibiotic-resistance determinants, and experience similar antibiotic pressures from beta-lactams, they are not always considered as a group when analyzing the epidemiology of multidrug-resistant Enterobacterales. Further explanation of the possible connections between the resistance profiles of CRE and ESBL-producing strains, as well as the role that these connections play in the spread of resistance, is required [11].
Furthermore, a review article on Gram-negative bacilli from the Gulf Cooperation Council (GCC) that produce β-lactamases revealed that the most prevalent β-lactamases are those with the carbapenemases genes OXA-48, NDM-1, and CTX-M-15 [12]. The rise of these resistant genes in numerous K. pneumoniae isolates in Saudi Arabia hospitals has also been demonstrated in several investigations [13,14,15]. The majority of those isolates were discovered in intensive care units (ICUs) from severely ill patients, and they had high fatality rates [12]. Even though there have not been many studies on CRKP isolates on the Arabian Peninsula, practically all of the GCC nations share similar ESBLs and carbapenemase-producing Enterobacterales, the bulk of which were isolated from nosocomial infections [16]. A study conducted in Saudi Arabia revealed a significant correlation between resistance determinants and the clonal types of K. pneumoniae. Specifically, all ST-152 isolates were found to be positive for NDM-1, while ST-199 isolates exhibited positivity for OXA-48. Additionally, both ST-709 and ST-199 isolates showed positivity for CTX-M-14 [17]. Furthermore, a study conducted in Iran reported the presence of blaSHV, blaTEM, blaCTX-M-15, blaOXA-48, blaKPC, and blaNDM genes in 91.4%, 82.7%, 79.3%, 36.2%, 29.3%, and 6.9% of multidrug-resistant K. pneumoniae isolates, respectively [18]. Mahmoudi et al. demonstrated that out of 30 K. pneumoniae isolates, the frequency of blaSHV, blaCTX-M-15, and blaTEM genes were 83% (n = 25), 70% (n = 21), and 57% (n = 17), respectively [19]. A study conducted in Iran reported that all the CRKP isolates were multidrug resistant. The study uncovered that 96% of CRKP isolates shared four common genes: blaTEM, blaCTX−M-1, blaSHV, and blaCTX−M-15, as reported by Abbasi and Ghaznavi-Rad [20]. Similarly, another study revealed that 93.6% of CRKP isolates shared three genes: blaTEM, blaSHV, and blaCTX−M-15, as reported by Solgi et al. [21]. These resistance genes are typically situated on mobile genetic elements, such as plasmids, which facilitate easy transferability within and between bacterial species. Moreover, studies conducted in the Middle East have demonstrated an increasing frequency of ESBL in K. pneumoniae over the past decade, as highlighted by Beigverdi [22]. Foreign travel was observed in approximately 23.3% of patients infected with CRE in the Gulf region [23]. Among the various travel destinations, India emerged as the primary country, followed by Africa and Pakistan. Notably, travel to and from Middle Eastern countries, including Saudi Arabia, has been identified as a source of OXA-48 carbapenemases in certain reports [23]. The initial case of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States was recognized in a patient who had recently undergone hospitalization in Saudi Arabia [24].
Molecular typing techniques are helpful tools for managing and treating infections brought on by multi-drug-resistance organisms, showing the genetic links in nosocomial infection epidemics, and determining the most likely source of infection. The genetic relatedness of the CRKP isolates using molecular typing techniques such as pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), or whole-genome sequencing (WGS) can help to recognize clonal relationships among the isolates and determine if certain genetic lineages are more prevalent. Molecular tools have limited resolution and a fairly longer turnaround time than whole-genome sequencing in guiding the treatment of MDR, XDR, and PDR infections. A molecular method called the enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) is also used to assess the genetic diversity among members of the Enterobacteriaceae family. The ERIC sequences are non-coding, conserved sections that are 126 bp long. The ERIC method can be applied to analyze genetic differences between bacterial strains. Bacteria contain ERIC sequences in different numbers and distributions. Examining the genetic similarities of bacterial isolates can be done quickly, accurately, and with high reliability using ERIC-PCR [25]. Strategies for control require ongoing monitoring of resistant strains. Understanding antibiotic resistance helps public health organizations, infection control committees, and antimicrobial stewardship programs make informed decisions about how to manage such resistant organisms [26].
Acknowledging the prevalence and dissemination of diverse antibiotic-resistance genes in K. pneumoniae both domestically and globally, as well as the scarcity of data in this specific geographic region, our objective was to examine the occurrence of CTX-M, SHV, and TEM genes in clinical isolates of CRKP from the Kingdom of Bahrain. To accomplish this, we employed molecular methods, including sequencing, for antibiotic resistance genes and also ERIC PCR to unravel the genetic relatedness among these isolates.
2. Materials and Methods
2.1. Bacterial Isolates and Hospital Setting
A total of twenty-four non-duplicate CRKP clinical isolates (December 2020 to June 2021) were included in this study. The bacterial strains were collected from Salmaniya Medical Complex, which is a multispecialty healthcare facility that provides secondary, tertiary, and emergency healthcare services in the Kingdom of Bahrain. It has approximately 1200 beds, with an average of 900 to 1000 people visiting the hospital each day, and it has more than 2000 doctors, nurses, and other workers. The bacterial strains were identified from the urine, blood, and endotracheal aspirate of the patients admitted to Salmaniya tertiary care hospital.
2.2. Bacterial Identification and Antimicrobial Susceptibility Testing
Bacterial species-level identification was established by using a mass spectrometry system (MALDI-TOF Bruker Daltonik GmbH, Bremen, Germany), and antibiotic susceptibility testing of isolates was performed with the VITEK-2 compact automated microbiological system (bioMerieux, Marcy L, Etoile, France). Only the isolates that were carbapenemase producers were included for further molecular analysis. The types of carbapenemases, the antibiotic resistance pattern, the genetic environment of blaNDM, integrons analysis, and molecular characterization of the plasmids of these isolates have already been published in our previous article [27].
2.3. DNA Isolation and Polymerase Chain Reaction-Based Amplification of Antibiotic-Resistant Genes
Fresh colonies from pure culture plates of K. pneumoniae clinical isolates were used to prepare whole-cell DNA. Each colony was suspended in 250 μL of nuclease-free water and incubated in a water bath at 95 °C for 15 min, followed by centrifugation at 12,000 rpm at 4 °C for 8 min. The supernatant was used as a template of DNA for polymerase chain reaction (PCR) on an Applied Biosystems GeneAmp PCR System 9700 (Foster City, CA, USA) with gene-specific primers as described earlier [28] to detect antimicrobial-resistant markers (blaCTX-M, blaSHV, and blaTEM). We have used bacterial strains having these resistant markers as positive control from strains available in Biorepository in the Department of Microbiology, Immunology, and Infectious Diseases, College of Medicine & Medical Sciences, Arabian Gulf University, Kingdom of Bahrain.
2.4. DNA Sequencing
At Geno Screen Lab, PCR-generated CTX-M fragments were sequenced (Campus Institut Pasteur de, La Calmette, France). Using the Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/ accessed on 1 June 2023), the derived protein sequence was aligned with blaCTX-M variants to verify the amino acid substitution in the query sequence for known variants. Additionally, online BLAST (versions 2.2.26) software (http://www.ncbi.nlm.nih.gov/BLAST/ accessed on 1 June 2023) was used to analyze the similarities between the amplified nucleotide sequence and the deduced protein sequences and was confirmed as blaCTX-M-15.
2.5. Molecular Genotyping
The genetic relatedness of ESBL-producing K. pneumoniae isolates was determined using enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) with specific primers as in our earlier published study [29]. Gel-electrophoresis was used to separate amplified PCR products in a 1.5% agarose gel containing ethidium bromide (0.5 µg/mL) with TAE running buffer. The Azure Biosystem C-200 Gel Documentation System was used to visualize gel images. PyElph version 1.4 software was used to analyze bands and create dendrogram clustering using the unweighted pair group method with arithmetic averages (UPGMA) [30].
3. Results
3.1. Antibiotic Resistance Genes Detection
All 24 K. pneumoniae isolates, which produce Extended Spectrum Beta-Lactamase (ESBL), were subjected to PCR assays to detect antibiotic-resistant determinants, including blaCTX-M, blaSHV, and blaTEM genes. The results revealed a significant presence of these genes within the isolates: All isolates (100%) carried blaSHV (24/24), 91.6% had blaCTX-M (22/24), and 54.8% contained blaTEM genes (13/24) (Table 1).

Table 1.
Distribution of bla genes in Klebsiella pneumonia isolates with their respective accession number.
All 24 K. pneumoniae isolates, were found in various genes combinations, with 45.8% of isolates possessing all three genes (blaSHV + blaCTX-M + blaTEM, n = 11), 45.8% carrying a combination of blaSHV + blaCTX-M (n = 11), and a minority of 8.35% having only blaSHV (n = 2). None of the isolates showed the presence of only (blaCTX-M and blaTEM) genes (Figure 1).
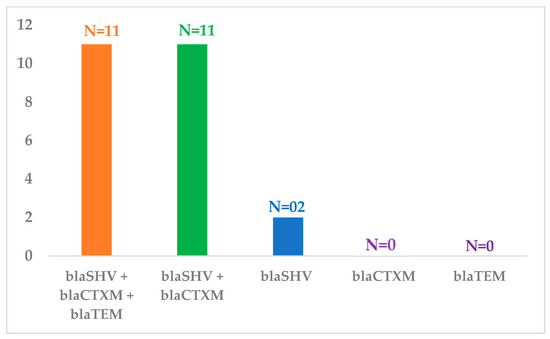
Figure 1.
Frequency of bla genes present either in a single or in a combination in Klebsiella pneumonia isolates.
3.2. Sequencing
Sequencing of the blaCTX-M amplicons revealed the confirmation of blaCTX-M-15. Under the following accession numbers, these sequences have been added to the GenBank nucleotide database: OP807050, OP807051, OP807052, OP807053, OP807054, OP807055, OP807056, OP807057, OP807058, OP807059, OP807060, OP807061, OP807062, OP807063, OP807064, OP807065, OP807066, OP807067, OP807068, OP807069, OP807070, and OP807071 (Table 1), accessible at the National Center of Biotechnology Information website (http://www.ncbi.nlm.nih.gov accessed on 10 July 2023)
3.3. CRKP Isolates Clustering
The ERIC-PCR profiles allowed differentiating 24 isolates into 3 ERIC clusters (A to C), where cluster A was further grouped into subgroup A1 and cluster B into four sub-clusters (B1, B2, B3, and B4) as shown in Figure 2. The isolates grouped within cluster A1 harbored an identical repertoire of resistance genes. Similarly, the isolates in cluster C possessed an identical complement of resistance determinants. Generally, the electrophoretic analysis of the PCR reaction products has revealed that the number of bands in particular electrophoretic paths ranged from 1 to 9. The sizes of the PCR products ranged from 400 bp to about 2000 bp.

Figure 2.
The dendrogram of CRKP isolates clustering based on ERIC patterns (A, B and C) along with the bla genes.
3.4. ERIC Clusters and Their Antibiotic Resistance Pattern
Cluster A with subgroup A1 consisted of five isolates, and all these strains were isolated from blood. The CTX-M-15 and SHV genes were present in all five isolates. All of these isolates had the same pattern of antibiotic resistance, except for MIID-C22, which was resistant to both amikacin and ceftazidime-avibactam (Figure 3).
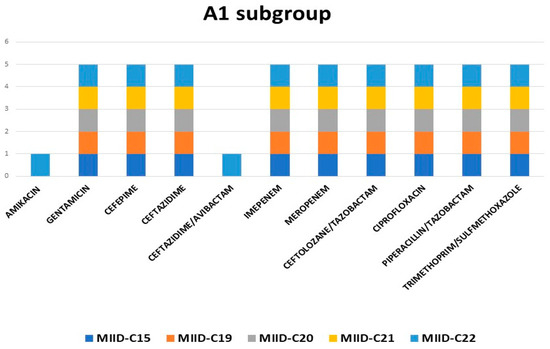
Figure 3.
Antibiotic resistance pattern of subgroup A1.
Cluster B was made up of four subclusters, B1 through B4. The resistance gene profiles within each subcluster varied significantly, according to ERIC analysis. Testing for antibiotic susceptibility also revealed variations in the antibiotic resistance patterns amongst the four subclusters. Cluster C consisted of seven isolates wherein MIID-C2, MIID-C3, MIID-C4 were isolated from blood, MIID-C1, MIID-C5, and MIID-C7 were from urine, and MIID-C12 was isolated from endotracheal aspirate of patients. The CTX-M-15, TEM, and SHV genes were present in all seven isolates. These isolates shared a common pattern of resistance to antibiotics. Except for ceftazidime-avibactam, all of these isolates were resistant to all known antibiotics (Figure 4).

Figure 4.
Antibiotic resistance pattern of cluster C isolates.
4. Discussion
Klebsiella pneumoniae is generally resistant to multiple antibiotics and is considered a reservoir of antimicrobial resistance markers, and these can horizontally transfer to other Gram-negative pathogens [31]. The primary reason for therapy failure is the spread of CRKP, which also raises morbidity and mortality rates among hospital patients [32]. To effectively stop the spread of CRKP, accurate and rapid detection using molecular techniques, including PCR and/or whole genome sequencing, is crucial [16]. The primary factor causing carbapenem resistance in clinical isolates of K. pneumoniae is the emergence of several types of β-lactamases. It has been proven that K. pneumoniae’s resistance to broad-spectrum β-lactams and carbapenems is influenced by the production of ESBLs [33].
The simultaneous existence and evolution of antibiotic-resistant markers are considered to have the most worrying potential, as they may lead to the emergence of uncurable, disruptive K. pneumoniae infections [34]. In our study, the production of blaSHV, blaCTX-M-15, and blaTEM combinations were found as two or three resistance genes in a single CRKP isolate. Moreover, it was observed that all the CRKP isolates demonstrated a significant presence of the SHV, CTX-M-15, and TEM genes (100%, 91.6%, and 45.8%), respectively. The SHV-type ESBL gene was the most prevalent among the isolates, followed by the CTX-M-type, which agrees with two previous reports [35,36]. Tijet et al. carried out a molecular characterization of KPC-producing Enterobacteriaceae submitted to the provincial reference laboratory in Ontario, Canada, due to the paucity of comprehensive reports of Klebsiella pneumoniae carbapenemase (KPC)-producing enterobacteria in that province. Their isolates harbored blaKPC-2 or blaKPC-3. The commonly detected gene was blaTEM-1, and occasionally blaOXA-1 and blaCTX-M-15 were detected. All K. pneumoniae isolates carried blaSHV-11 in the study [35]. Uz Zaman et al. reported the presence of CTX-M and SHV genes in all their isolates, with CTX-M-15 and SHV-1 types predominating among these extended-spectrum beta-lactamases (ESBLs). All isolates but one contained TEM-1 [36].
In Saudi Arabia, though carbapenem resistance remained rare among Enterobacterales, the first outbreak of CRKP was reported in 2010 [37]. In their study, a cluster of eight CRKPs were found in March 2010 in the adult intensive care unit. Two more CRKPs were discovered after a study of K. pneumoniae isolates from the previous six months. On performing PFGE, the bulk of the strains throughout the outbreak period were genetically similar or indistinguishable [37]. Later, the molecular basis of this resistance was found to be due to the involvement of carbapenemase enzyme (OXA-48) in combination with CTX-M-15 genes [36]. A study from India was conducted to ascertain the frequency and genetic makeup of K. pneumoniae strains that produce ESBL and carbapenemase isolated from intensive care units of a tertiary care hospital. Here, they reported the coexistence of the bla gene along with carbapenemase producers to be 69% and 50% of the SHV and TEM, respectively [38]. Previous Indian studies have documented the coexistence of carbapenemase producers (NDM) with the bla genes [39,40]. Similar international studies have revealed the presence of bla genes along with carbapenemase [41,42]. Carbapenemases-producing KP strains appear to frequently co-produce ESBL; as a result, they are resistant to both cephalosporins and carbapenems. Numerous researchers have noted this tendency, which shows that isolates that are resistant to multiple drugs frequently produce carbapenemase, which is also noticed in our study [42]. A study from China investigated the occurrence and spread of carbapenemase and extended-spectrum β-lactamase encoding genes co-existence in sporadic K. pneumoniae ST307 in pediatric patients from the Shenzhen Children’s Hospital, China, and reported the presence of SHV (92%) CTX-M (53%) among 36 CRKP isolates from pediatric patients [43]. A study from India reported blaCTX-M, blaTEM, and blaSHV in coexistence with blaNDM-1 [44].
Globally, the CTX-M gene is linked to antibiotic resistance [36]. CTX-M sequencing of our isolates confirmed CTX-M-15 in our region. The same variant has been reported by Zaman et al. from Saudi Arabia as the most abundant ESBL gene detected in 47/71 (66.2%) CRKP isolates [17]. This is consistent with earlier studies [17,45,46] and suggests that it is endemic to this region. A study from China observed that among blaCTX-M producers, blaCTX-M-2 was the most prevalent genotype (44/45, 97.8%), followed by blaCTX-M-3 (20/45, 44.4%), blaCTX-M-1 (18/45, 40%), blaCTX-M-9 (17/45, 37.8%), and blaCTX-M-15 (5/45, 11.1%) [47]. According to a study from Tanzania, ESBL genes were distributed as follows: 29/32 (90.6%) had blaCTX-M-15, two had blaSHV-12, and one had both blaCTX-M-15 and blaSHV-12. The percentage of hospital and community isolates with blaCTX-M-15 was 69% (20/29) and 31% (9/29), respectively. Only infections acquired in hospitals were found to have blaSHV-12 genotypes [48]. The rise of CRKP isolates, as well as co-existing K. pneumoniae strains of blaCTX-M-1 and blaSHV in Lagos hospitals, were documented by a study from Nigeria [49]. In contrast to our findings, a study from Syria in 2015 reported the presence of blaCTX-M-1 (100%) as the most prevalent ESBL gene in K. pneumoniae [50]. Another study from Saudi Arabia reported blaCTX-M-1 (60%) and blaCTX-M-9-like genes (40%) among K. pneumoniae [51].
Types of CTX-M ESBLs, especially CTX-M-15, are well known for spreading quickly among Enterobacteriaceae members all over the world [52]. A study by Blanco et al. reported that complicated UTI was strongly associated with ESBL-producing E. coli infections. In their study, CTX-M-15-producing E. coli showed ten different plusotypes; 65% were PT1 or PT4 and corresponded to ST131 [52]. Additionally, it has been proposed that the widespread use of ceftriaxone and cefotaxime may have contributed to the establishment and spread of CTX-M enzymes [53]. The emergence of carbapenemase-producing Gram-negatives is a worry because it is frequently linked with an outbreak of MDR isolates, which leads to restrictions on alternative treatments [54]. Although the carbapenemases had previously been reported in Saudi Arabia and Kuwait, no isolate was discovered to produce KPC, VIM, or IMP beta-lactamases. Since no carbapenemase activity or genes were found in the study isolates, there was a possibility that the resistance mechanism is unrelated to carbapenemase. These isolates generated genes of the CTX-M-15 type and most likely produced extended-spectrum beta-lactamases in connection with lower outer membrane permeability [54]. In the present study, findings of antibiotic susceptibility of K. pneumoniae isolates are in keeping with the reported MDR phenotype associated with detected antibiotic resistance markers of blaSHV, blaCTX-M-15, and blaTEM. However, the K. pneumoniae isolates revealed high resistance against multiple antibiotic groups, especially the carbapenem class. Resistance to carbapenem can be associated with other mechanisms, such as the production of AmpC and ESBL or modifications to the structure of the outer membrane [36,55].
A wide range of bacteria can be successfully typed using enterobacterial repetitive intergenic consensus (ERIC-PCR) sequences [56,57]. In our study, the ERIC-PCR profiles allowed differentiating CRKP isolates into 3 ERIC clusters. In a Danish investigation, a semi-automated rep-PCR typing method was utilized to identify the relationship between the strains associated with the outbreak and the ESBLs generated by local K. pneumoniae strains [58]. According to Kholy and Manakhly from Egypt, only 5 KPC-positive and two producers were genetically related when 27 CRKP isolates were analyzed using ERIC-PCR; the majority of the isolates were polyclonal [58]. ERIC-PCR can be used to look into the epidemiological relationships between MDR K. pneumoniae isolates and to determine whether any plausible outbreaks might exist. According to research by Kundu et al., no outbreaks or nosocomial clustering occurred throughout the planned sampling period based on ERIC-PCR typing results and their relationship to hospital wards and units of data [59].
In this study, the isolates of ERIC types A and C had similar antibiotic resistance patterns. The cluster A and C isolates were resistant to all the antibiotics except amikacin and ceftazidime/avibactam. Among this resistance pattern, an exception was MIID-C22, which was resistant to both amikacin and ceftazidime-avibactam. This could be due to the presence of other antibiotic resistance mechanisms that were not studied here. A study on CRKP colonization in an intensive care unit by Madni et al. identified five primary ERIC clusters. Except for ERIC cluster 2, where isolates showed two antibiograms, isolates from the same ERIC cluster typically had identical antibiograms [60]. According to research by Shen et al., CRKP isolates had lower resistance to the antibiotics trimethoprim/sulfamethoxazole (24.5%), ciprofloxacin (23.4%), gentamicin (22.3%), levofloxacin (17.0%), tobramycin (16.0%), and amikacin (14.9%). According to their findings, 76.6% of the isolates were resistant to ceftazidime/avibactam, while 21.3% were resistant to tigecycline [61].
The small sample size is the main limitation of this study. Due to this, it is difficult to generalize the findings to other carbapenemase-producing CRKP strains that harbor these bla genes. Hence, an extensive multi-center investigation is needed. Another limitation is that this study was restricted to carbapenemases in CRKP isolates, so the details of other molecular mechanisms were not known. Additionally, the inability to perform whole-genome sequencing and analysis would have revealed more about the genomic determinants of antibiotic resistance and virulence in these isolates.
To the best of our knowledge, this is the first publication that describes the bla genes and molecular typing of CRKP isolates from this region.
5. Conclusions
The study findings reveal the concomitant carriage of the SHV, CTX-M, and comparatively lower carriage of TEM genes in CRKP isolates. Sequencing of the CTX-M positive isolates confirmed CTX-M-15 in our region. CRKP infection is a significant clinical concern with a high mortality rate due to the difficult nature of treating CRKP and the lack of an optimum medication regimen. Our findings highlight the significance of routinely using molecular characterization assays in hospital laboratories for the accurate detection of antibiotic-resistance gene-carrying bacteria. It is crucial to report the presence of antibiotic-resistance genes along with regular antibiotic sensitivity reports, as this will aid the clinician in prescribing the appropriate antibiotics. This study has contributed to the understanding of antibiotic resistance genes and has provided valuable data on bla gene carriage in CRKP isolates. Studies with a larger sample size are required to objectively assess the trends and comprehend the dynamics of spread and efficient control strategies.
Author Contributions
M.S. (Mohammad Shahid) conceived the project proposal. N.K.S. provided clinical CRKP isolates, identified them, and performed antibiotic susceptibility on automated systems. N.A., M.S. (Mohd Shadab) and A.A.-M. performed molecular experimentations. M.S. (Mohammad Shahid), F.K.D., N.K.S., K.M.B., A.Y.I. and K.S.T. evaluated the data and provided expertise and feedback. R.M.J., N.A. and M.S. (Mohammad Shahid) wrote the preliminary draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Internal funding for the research grant (G06/AGU-11/19) from the Arabian Gulf University.
Institutional Review Board Statement
This study protocol was approved by the Research and Ethics Committee, College of Medicine and Medical Sciences, Arabian Gulf University (E012-PI-11/19), and the Ministry of Health (AURS/305/2020).
Informed Consent Statement
We have taken informed consent from the patients in this study.
Data Availability Statement
All data used for analysis are presented as figures in this article.
Acknowledgments
All authors acknowledge and thank AGU for providing a research grant. Nayeem Ahmad acknowledges and gratefully thanks AGU for providing a Postdoc Research Fellowship. The authors also acknowledge Genoscreen Lab, France, for providing sequence facility support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mil-Homens, D.; Martins, M.; Barbosa, J.; Serafim, G.; Sarmento, M.J.; Pires, R.F.; Rodrigues, V.; Bonifácio, V.D.B.; Pinto, S.N. Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates: In Vivo Virulence Assessment in Galleria mellonella and Potential Therapeutics by Polycationic Oligoethyleneimine. Antibiotics 2021, 10, 56. [Google Scholar]
- Eliopoulos, G.M.; Bush, K. New β-lactamases in Gram-negative bacteria: Diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 2001, 32, 1085–1089. [Google Scholar]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [PubMed]
- Giriyapur, R.S.; Nandihal, N.W.; Patil, A.B. Comparison of disc diffusion methods for the detection of extended-spectrum beta lactamase-producing Enterobacteriaceae. J. Lab. Physicians 2011, 3, 033–036. [Google Scholar] [CrossRef] [PubMed]
- Leverstein-van Hall, M.A.; Fluit, A.C.; Paauw, A.; Box, A.T.; Brisse, S.; Verhoef, J. Evaluation of the Etest ESBL and the BD Phoenix, VITEK 1, and VITEK 2 automated instruments for detection of extended-spectrum beta-lactamases in multiresistant Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 2002, 40, 3703–3711. [Google Scholar] [CrossRef] [PubMed]
- Tada, D.G.; Gandhi, P.J.; Patel, K.N. A study on antibiotic related resistance in UTI patients: A comparison between community acquired and hospital acquired E. coli. Natl. J. Community Med. 2012, 3, 255–258. [Google Scholar]
- Ramadan, R.A.; Bedawy, A.M.; Negm, E.M.; Hassan, T.H.; Ibrahim, D.A.; ElSheikh, S.M.; Amer, R.M. Carbapenem-resistant Klebsiella pneumoniae among patients with ventilator-associated pneumonia: Evaluation of antibiotic combinations and susceptibility to new antibiotics. Infect. Drug Resist. 2022, 15, 3537–3548. [Google Scholar] [CrossRef]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar]
- Zhang, R.; Chan, E.W.; Zhou, H.; Chen, S. Prevalence and genetic characteristics of carbapenem-resistant Enterobacteriaceae strains in China. Lancet. Infect. Dis. 2017, 17, 256–257. [Google Scholar]
- Jafari, Z.; Harati, A.A.; Haeili, M.; Kardan-Yamchi, J.; Jafari, S.; Jabalameli, F.; Meysamie, A.; Abdollahi, A.; Feizabadi, M.M. Molecular Epidemiology and Drug Resistance Pattern of Carbapenem-Resistant Klebsiella pneumoniae Isolates from Iran. Microb. Drug Resist. 2019, 25, 336–343. [Google Scholar]
- Tian, X.; Sun, S.; Jia, X.; Zou, H.; Li, S.; Zhang, L. Epidemiology of and risk factors for infection with extended-spectrum β-lactamase-producing carbapenem-resistant Enterobacteriaceae: Results of a double case–control study. Infect. Drug Resist. 2018, 11, 1339. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M.; Balkhy, H.H.; Walsh, T.R.; Paterson, D.L. β-Lactamase production in key gram-negative pathogen isolates from the Arabian Peninsula. Clin. Microbiol. Rev. 2013, 26, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, F. Carbapenem-Resistant Enterobacteriaceae: An update narrative review from Saudi Arabia. J. Infect. Public Health 2019, 12, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdely, H.; AlHababi, R.; Dada, H.M.; Roushdy, H.; Alanazi, M.M.; Alessa, A.A.; Gad, N.M.; Alasmari, A.M.; Radwan, E.E.; Al-Dughmani, H.; et al. Molecular characterization of carbapenem-resistant Enterobacterales in thirteen tertiary care hospitals in Saudi Arabia. Ann. Saudi Med. 2021, 41, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Alghoribi, M.F.; Binkhamis, K.; Alswaji, A.A.; Alhijji, A.; Alsharidi, A.; Balkhy, H.H.; Doumith, M.; Somily, A. Genomic analysis of the first KPC-producing Klebsiella pneumoniae isolated from a patient in Riyadh: A new public health concern in Saudi Arabia. J. Infect. Public Health 2020, 13, 647–650. [Google Scholar] [CrossRef]
- Al-Zahrani, I.A.; Alasiri, B.A. The emergence of carbapenem-resistant Klebsiella pneumoniae isolates producing OXA-48 and NDM in the Southern (Asir) province, Saudi Arabia. Saudi Med. J. 2018, 39, 23. [Google Scholar] [CrossRef]
- Alrodayyan, M.; Albladi, M.; Aldrees, M.; Siddique, M.I.; Aljohani, S.; Balkhy, H.H. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arab. BMC Infect. Dis. 2018, 18, 205. [Google Scholar]
- Farhadi, M.; Ahanjan, M.; Goli, H.R.; Haghshenas, M.R.; Gholami, M. High frequency of multidrug-resistant (MDR) Klebsiella pneumoniae harboring several β-lactamase and integron genes collected from several hospitals in the north of Iran. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 70. [Google Scholar] [CrossRef]
- Mahmoudi, S.; Pourakbari, B.; Rahbarimanesh, A.; Abdosalehi, M.R.; Ghadiri, K.; Mamishi, S. An outbreak of ESBL-producing Klebsiella pneumoniae in an Iranian referral hospital: Epidemiology and molecular typing. Infect. Disord.-Drug Targets 2019, 19, 46–54. [Google Scholar] [CrossRef]
- Abbasi, E.; Ghaznavi-Rad, E. High frequency of NDM-1 and OXA-48 carbapenemase genes among Klebsiella pneumoniae isolates in central Iran. BMC Microbiol. 2023, 23, 98. [Google Scholar] [CrossRef]
- Solgi, H.; Badmasti, F.; Giske, C.G.; Aghamohammad, S.; Shahcheraghi, F. Molecular epidemiology of NDM-1-and OXA-48-producing Klebsiella pneumoniae in an iranian hospital: Clonal dissemination of ST11 and ST893. J. Antimicrob. Chemother. 2018, 73, 151. [Google Scholar] [CrossRef] [PubMed]
- Beigverdi, R.; Jabalameli, L.; Jabalameli, F.; Emaneini, M. Prevalence of extended-spectrum β-lactamase-producing Klebsiella pneumoniae: First systematic review and meta-analysis from Iran. J. Glob. Antimicrob. Resist. 2019, 18, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Sonnevend, A.; Ghazawi, A.A.; Hashmey, R.; Jamal, W.; Rotimi, V.O.; Shibl, A.M.; Al-Jardani, A.; Al-Abri, S.S.; Tariq, W.U.; Weber, S.; et al. Characterization of carbapenem-resistant Enterobacteriaceae with high rate of autochthonous transmission in the Arabian Peninsula. PLoS ONE 2015, 10, e0131372. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Hazen, K.C.; Carroll, J.; Yeh, A.J.; Cox, H.L.; Bonomo, R.A.; Sifri, C.D. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: The “menace” arrives in the new world. J. Clin. Microbiol. 2013, 51, 680–683. [Google Scholar] [CrossRef]
- Abdelhamid, S.M.; Abd-Elaal, H.M.; Matareed, M.O.; Baraka, K. Genotyping and Virulence Analysis of Drug Resistant Clinical Klebsiella pneumoniae isolates in Egypt. J. Pure Appl. Microbiol. 2020, 14, 1967–1976. [Google Scholar] [CrossRef]
- Di Tella, D.; Tamburro, M.; Guerrizio, G.; Fanelli, I.; Sammarco, M.L.; Ripabelli, G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infect. Drug Resist. 2019, 12, 3783–3795. [Google Scholar] [CrossRef]
- Shahid, M.; Ahmad, N.; Saeed, N.K.; Shadab, M.; Joji, R.M.; Al-Mahmeed, A.; Bindayna, K.M.; Tabbara, K.S.; Dar, F.K. Clinical carbapenem-resistant Klebsiella pneumoniae isolates simultaneously harboring blaNDM-1, blaOXA types and qnrS genes from the Kingdom of Bahrain: Resistance profile and genetic environment. Front. Cell. Infect. Microbiol. 2022, 12, 1033305. [Google Scholar] [CrossRef]
- Shahid, M. Citrobacter spp. simultaneously harboring blaCTX-M, blaTEM, blaSHV, blaampC, and insertion sequences IS26 and orf513: An evolutionary phenomenon of recent concern for antibiotic resistance. J. Clin. Microbiol. 2010, 48, 1833–1838. [Google Scholar] [CrossRef]
- Shahid, M.; Malik, A.; Akram, M.; Agrawal, L.M.; Khan, A.U.; Agrawal, M. Prevalent phenotypes and antibiotic resistance in Escherichia coli and Klebsiella pneumoniae at an Indian tertiary care hospital: Plasmid-mediated cefoxitin resistance. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2008, 12, 256–264. [Google Scholar] [CrossRef][Green Version]
- Pavel, A.B.; Vasile, C.I. PyElph—A software tool for gel images analysis and phylogenetics. BMC Bioinform. 2012, 13, 9. [Google Scholar] [CrossRef]
- Chung, H.; Karkey, A.; Pham Thanh, D.; Boinett, C.J.; Cain, A.K.; Ellington, M.; Baker, K.S.; Dongol, S.; Thompson, C.; Harris, S.R.; et al. A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol. Med. 2015, 7, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Pavelkovich, A.; Balode, A.; Edquist, P.; Egorova, S.; Ivanova, M.; Kaftyreva, L.; Konovalenko, I.; Kõljalg, S.; Lillo, J.; Lipskaya, L. Detection of carbapenemase-producing enterobacteriaceae in the baltic countries and st. Petersburg area. BioMed Res. Int. 2014, 2014, 548960. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Cuzon, G.; Naas, T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet. Infect. Dis. 2009, 9, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Alraddadi, B.M.; Heaphy, E.L.G.; Aljishi, Y.; Ahmed, W.; Eljaaly, K.; Al-Turkistani, H.H.; Alshukairi, A.N.; Qutub, M.O.; Alodini, K.; Alosaimi, R.; et al. Molecular Epidemiology and Outcome of Carbapenem-Resistant Enterobacterales in Saudi Arabia. BMC Infect. Dis. 2022, 22, 254. [Google Scholar] [CrossRef] [PubMed]
- Tijet, N.; Sheth, P.M.; Lastovetska, O.; Chung, C.; Patel, S.N.; Melano, R.G. Molecular characterization of Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae in Ontario, Canada, 2008–2011. PLoS ONE 2014, 9, e116421. [Google Scholar] [CrossRef]
- Uz Zaman, T.; Aldrees, M.; Al Johani, S.M.; Alrodayyan, M.; Aldughashem, F.A.; Balkhy, H.H. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2014, 28, 186–192. [Google Scholar] [CrossRef]
- Balkhy, H.H.; El-Saed, A.; Al Johani, S.M.; Francis, C.; Al-Qahtani, A.A.; Al-Ahdal, M.N.; Altayeb, H.T.; Arabi, Y.; Alothman, A.; Sallah, M. The epidemiology of the first described carbapenem-resistant Klebsiella pneumoniae outbreak in a tertiary care hospital in Saudi Arabia: How far do we go? Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2012, 31, 1901–1909. [Google Scholar] [CrossRef]
- Bhaskar, B.H.; Mulki, S.S.; Joshi, S.; Adhikary, R.; Venkatesh, B.M. Molecular Characterization of Extended Spectrum β-lactamase and Carbapenemase Producing Klebsiella pneumoniae from a Tertiary Care Hospital. Indian. J. Crit. Care Med. Peer-Rev. Off. Publ. Indian. Soc. Crit. Care Med. 2019, 23, 61–66. [Google Scholar] [CrossRef]
- Ahmad, N.; Khalid, S.; Ali, S.M.; Khan, A.U. Occurrence of bla(NDM) Variants among Enterobacteriaceae from a Neonatal Intensive Care Unit in a Northern India Hospital. Front. Microbiol. 2018, 9, 407. [Google Scholar] [CrossRef]
- Ahmad, N.; Ali, S.M.; Khan, A.U. Co-existence of blaNDM-1 and blaVIM-1 producing Moellerella wisconsensis in NICU of North Indian Hospital. J. Infect. Dev. Ctries. 2020, 14, 228–231. [Google Scholar] [CrossRef]
- Jeremiah, S.S.; Balaji, V.; Anandan, S.; Sahni, R.D. A possible alternative to the error prone modified Hodge test to correctly identify the carbapenemase producing Gram-negative bacteria. Indian. J. Med. Microbiol. 2014, 32, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, G.; Wu, X.; Wang, L.; Cai, J.; Chan, E.W.; Chen, S.; Zhang, R. Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the blaNDM-1 element and clonal spread of progenitor resistant strains. Front. Microbiol. 2015, 6, 595. [Google Scholar] [CrossRef]
- Patil, S.; Chen, H.; Guo, C.; Zhang, X.; Ren, P.G.; Francisco, N.M.; Wen, F. Emergence of Klebsiella pneumoniae ST307 Co-Producing CTX-M with SHV and KPC from Paediatric Patients at Shenzhen Children’s Hospital, China. Infect. Drug Resist. 2021, 14, 3581–3588. [Google Scholar] [CrossRef]
- Ahmad, N.; Ali, S.M.; Khan, A.U. First reported New Delhi metallo-β-lactamase-1-producing Cedecea lapagei. Int. J. Antimicrob. Agents 2017, 49, 118–119. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Ko, K.S.; Kang, C.-I.; Chung, D.R.; Peck, K.R.; Song, J.-H. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: Diverse clones and clonal dissemination. Int. J. Antimicrob. Agents 2011, 38, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Al-Marzooq, F.; Mohd Yusof, M.Y.; Tay, S.T. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug resistant Klebsiella pneumoniae. PLoS ONE 2015, 10, e0133654. [Google Scholar] [CrossRef]
- Wang, S.; Dong, H.; Wang, M.; Ma, W.; Cheng, Y.; Zhou, J.; Cheng, Y.; Xu, H.; Yu, X. Molecular Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae in a Tertiary Hospital in Northern China. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 2615753. [Google Scholar] [CrossRef]
- Manyahi, J.; Moyo, S.J.; Tellevik, M.G.; Ndugulile, F.; Urassa, W.; Blomberg, B.; Langeland, N. Detection of CTX-M-15 beta-lactamases in Enterobacteriaceae causing hospital- and community-acquired urinary tract infections as early as 2004, in Dar es Salaam, Tanzania. BMC Infect. Dis. 2017, 17, 282. [Google Scholar] [CrossRef]
- Akinyemi, K.O.; Abegunrin, R.O.; Iwalokun, B.A.; Fakorede, C.O.; Makarewicz, O.; Neubauer, H.; Pletz, M.W.; Wareth, G. The Emergence of Klebsiella pneumoniae with Reduced Susceptibility against Third Generation Cephalosporins and Carbapenems in Lagos Hospitals, Nigeria. Antibiotics 2021, 10, 142. [Google Scholar] [CrossRef]
- AL-Subol, I.; Youssef, N. Prevalence of CTX-M, TEM and SHV Beta-lactamases in Clinical Isolates of Escherichia coli and Klebsiella pneumoniae Isolated from Aleppo University Hospitals, Aleppo, Syria. Arch. Clin. Infect. Dis. 2015, 10, e22540. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.M.; Shibl, A.M.; Tawfik, A.F. Prevalence and molecular characterization of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in Riyadh, Saudi Arabia. Ann. Saudi Med. 2009, 29, 253–257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Blanco, V.M.; Maya, J.J.; Correa, A.; Perenguez, M.; Munoz, J.S.; Motoa, G.; Pallares, C.J.; Rosso, F.; Matta, L.; Celis, Y. Prevalence and risk factors for extended-spectrum β-lactamase-producing Escherichia coli causing community-onset urinary tract infections in Colombia. Enfermedades Infecc. Y Microbiol. Clin. 2016, 34, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kelkar, S.; Wu, W.; Chen, M.; Quinn, J.P. Clinical isolates of Enterobacteriaceae producing extended-spectrum β-lactamases: Prevalence of CTX-M-3 at a hospital in China. Antimicrob. Agents Chemother. 2003, 47, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M.; Sartor, A.L.; Balkhy, H.H.; Walsh, T.R.; Al Johani, S.M.; AlJindan, R.Y.; Alfaresi, M.; Ibrahim, E.; Al-Jardani, A.; Al-Abri, S.; et al. Molecular Characterization of Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae in the Countries of the Gulf Cooperation Council: Dominance of OXA-48 and NDM Producers. Antimicrob. Agents Chemother. 2014, 58, 3085–3090. [Google Scholar] [CrossRef]
- Memish, Z.A.; Assiri, A.; Almasri, M.; Roshdy, H.; Hathout, H.; Kaase, M.; Gatermann, S.G.; Yezli, S. Molecular Characterization of Carbapenemase Production among Gram-Negative Bacteria in Saudi Arabia. Microb. Drug Resist. 2015, 21, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.B.; Skov, M.N.; Jørgensen, R.L.; Heltberg, O.; Hansen, D.S.; Schønning, K. Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 773–778. [Google Scholar] [CrossRef]
- Ahmad, N.; Ali, S.M.; Khan, A.U. Detection of New Delhi Metallo-β-Lactamase Variants NDM-4, NDM-5, and NDM-7 in Enterobacter aerogenes Isolated from a Neonatal Intensive Care Unit of a North India Hospital: A First Report. Microb. Drug Resist. 2018, 24, 161–165. [Google Scholar] [CrossRef]
- Kholy, A.E.; Manakhly, A.E. 2300. Molecular Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae (CRKP) Causing Central Line Associated Blood Stream Infections (CLABSI) in Three ICU Units in Egypt. Open Forum Infect. Dis. 2018, 5 (Suppl. 1), S682. [Google Scholar] [CrossRef]
- Kundu, J.; Kansal, S.; Rathore, S.; Kaundal, M.; Angrup, A.; Biswal, M.; Walia, K.; Ray, P. Evaluation of ERIC-PCR and MALDI-TOF as typing tools for multidrug resistant Klebsiella pneumoniae clinical isolates from a tertiary care center in India. PLoS ONE 2022, 17, e0271652. [Google Scholar] [CrossRef]
- Madni, O.; Amoako, D.G.; Abia, A.L.K.; Rout, J.; Essack, S.Y. Genomic Investigation of Carbapenem-Resistant Klebsiella pneumonia Colonization in an Intensive Care Unit in South Africa. Genes 2021, 12, 951. [Google Scholar] [CrossRef]
- Shen, M.; Chen, X.; He, J.; Xiong, L.; Tian, R.; Yang, G.; Zha, H.; Wu, K. Antimicrobial Resistance Patterns, Sequence Types, Virulence and Carbapenemase Genes of Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates from a Tertiary Care Teaching Hospital in Zunyi, China. Infect. Drug Resist. 2023, 16, 637–649. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).