Convalescent Adaptive Immunity Is Highly Heterogenous after SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. PBMC Preparation from Cones
2.3. Anti-SARS-CoV-2 Spike Antibody Measurement
2.4. Identification of HLA-A2-Positive Patients and HLA-A2-Binding Peptides
2.5. Antigen Stimulation Procedures
2.6. FC AIM Assays
2.7. IFN-γ ELISpot Assay
2.8. Cytotoxicity Assay
2.9. Statistical Analysis
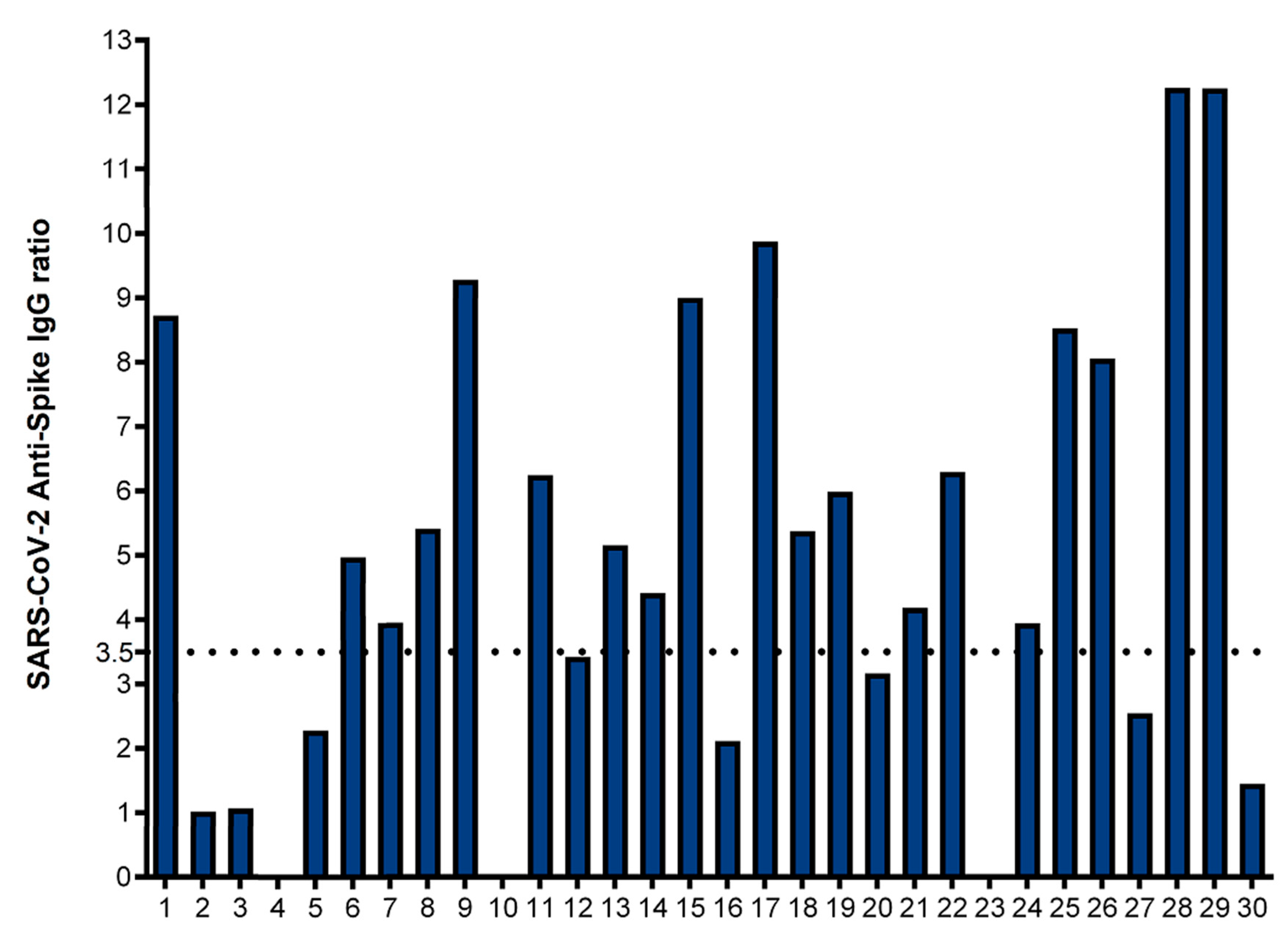
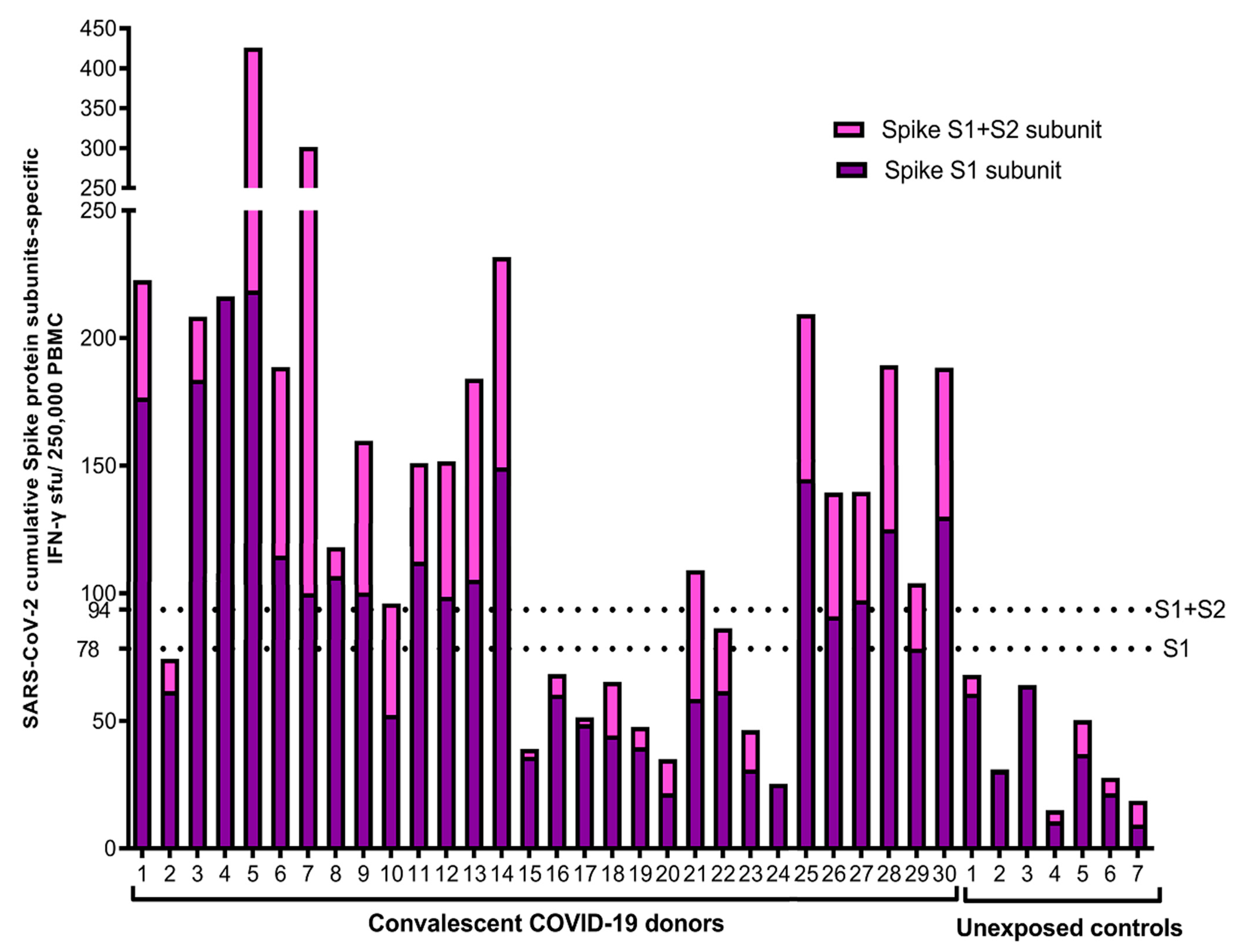
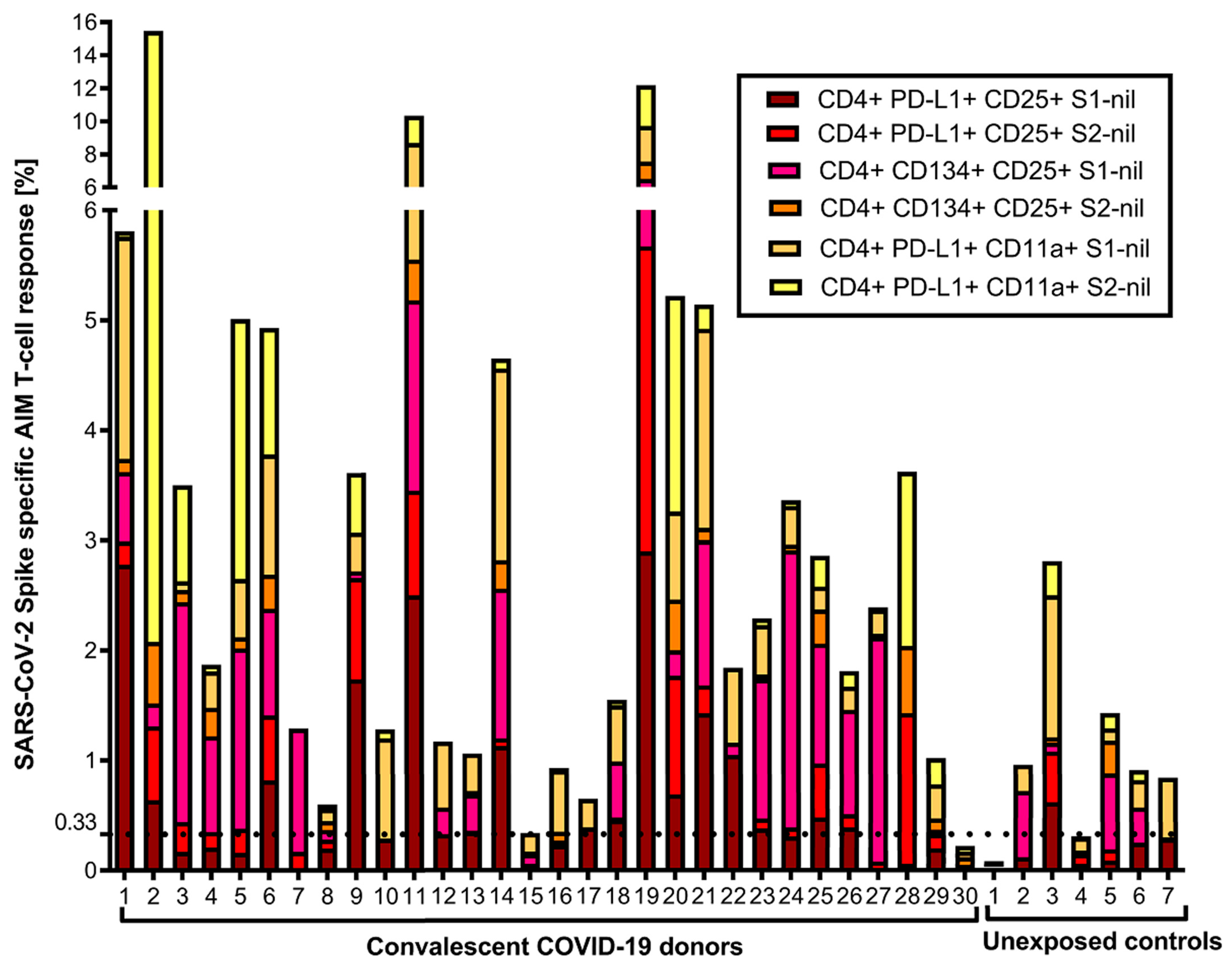
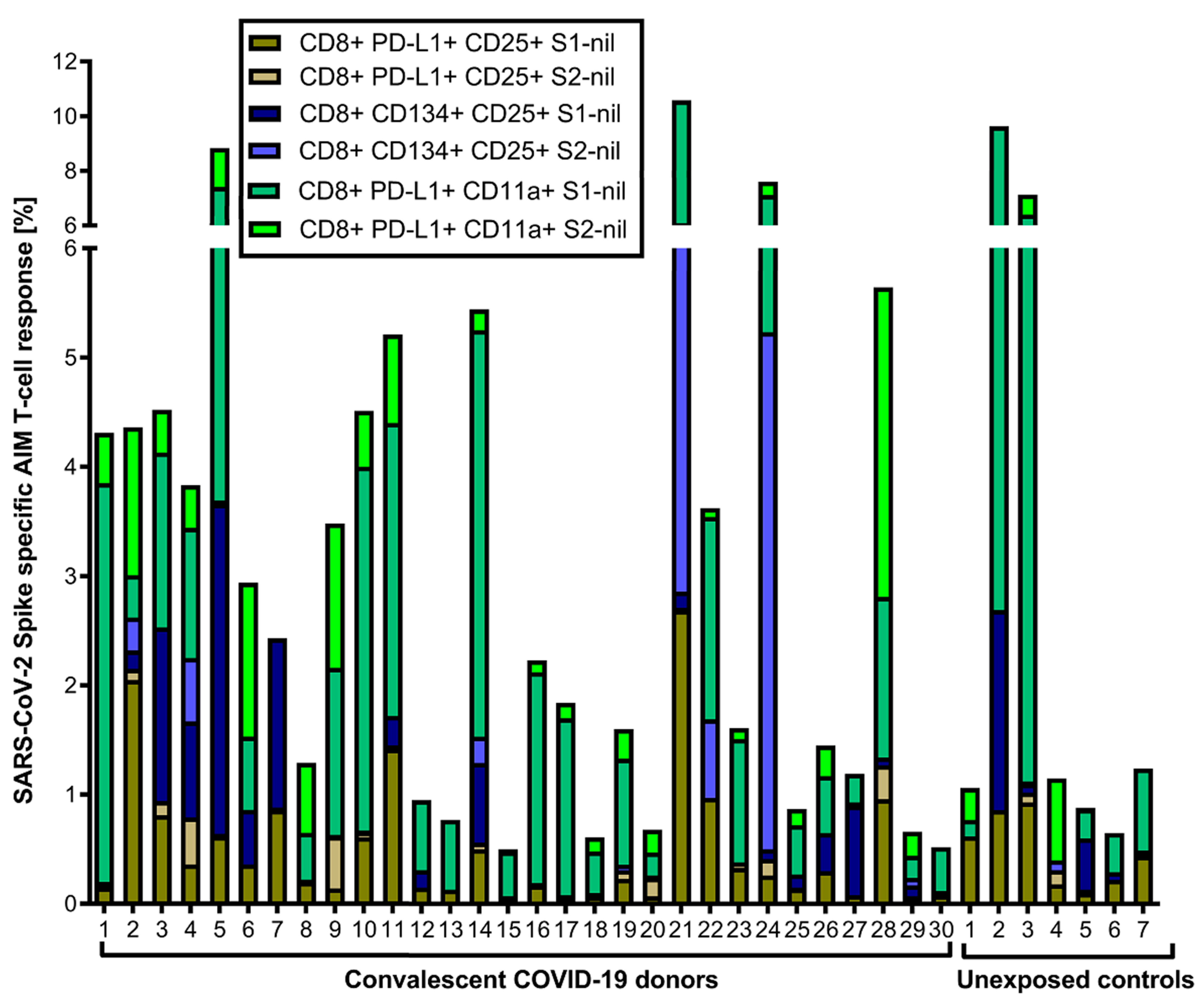
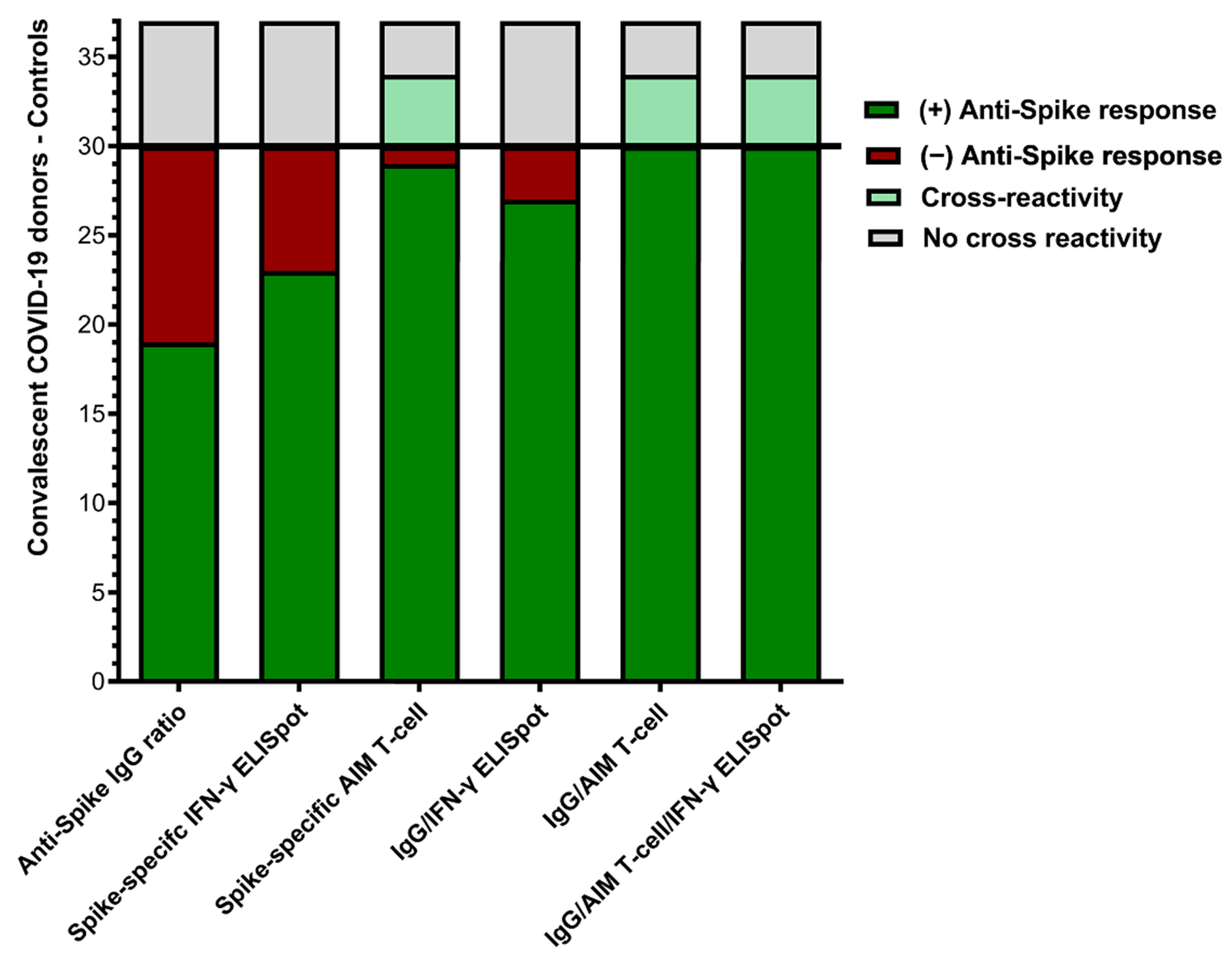
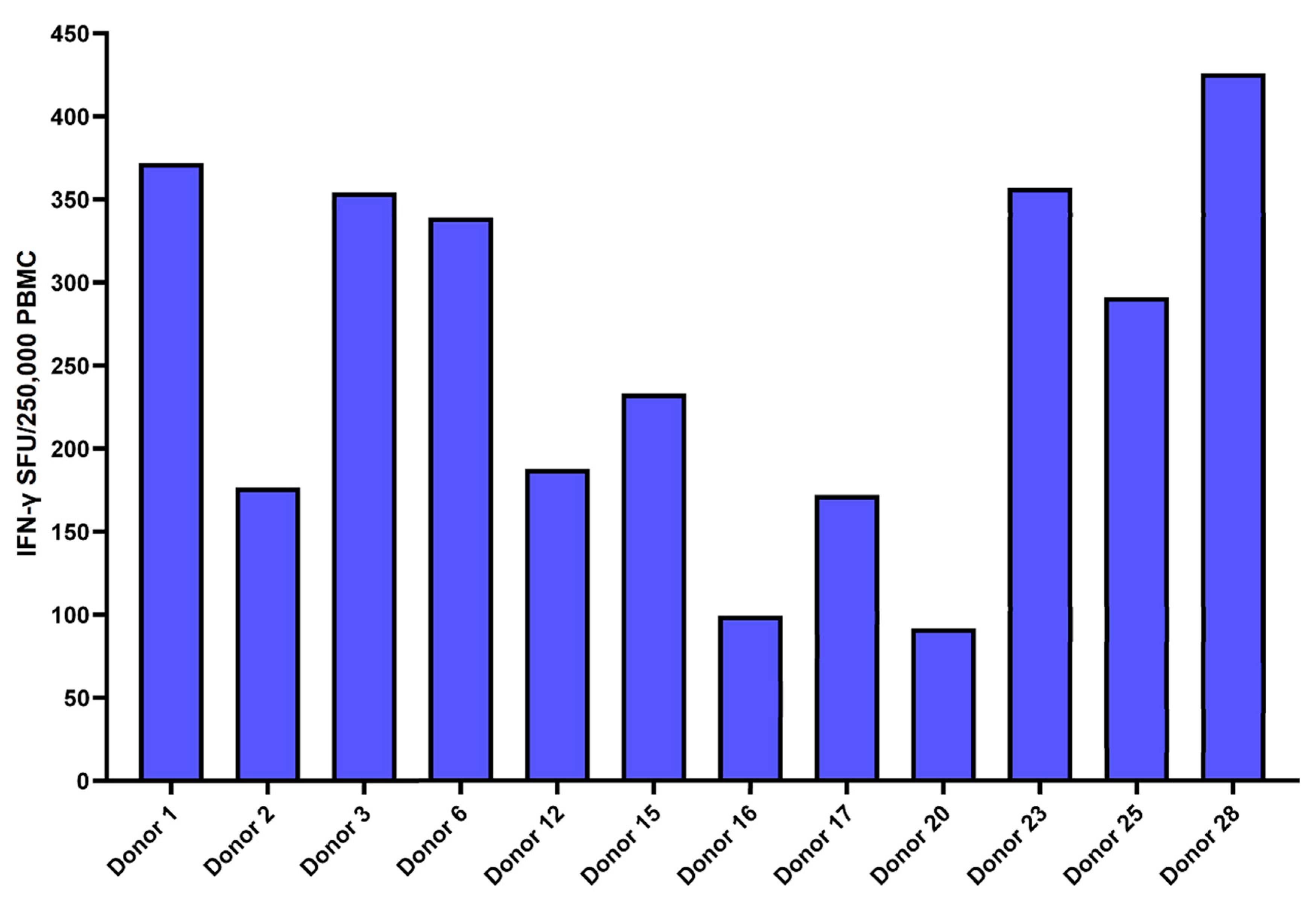
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crotty, S. Hybrid immunity. Science 2021, 372, 1392–1393. [Google Scholar] [CrossRef]
- DiPiazza, A.T.; Graham, B.S.; Ruckwardt, T.J. T cell immunity to SARS-CoV-2 following natural infection and vaccination. Biochem. Biophys. Res. Commun. 2021, 538, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Jarjour, N.N.; Masopust, D.; Jameson, S.C. T Cell Memory: Understanding COVID-19. Immunity 2021, 54, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.-B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e114. [Google Scholar] [CrossRef]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021, 2, 100204. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e1019. [Google Scholar] [CrossRef]
- Nguyen, T.H.O.; Cohen, C.A.; Rowntree, L.C.; Bull, M.B.; Hachim, A.; Kedzierska, K.; Valkenburg, S.A. T Cells Targeting SARS-CoV-2: By Infection, Vaccination, and Against Future Variants. Front. Med. 2021, 8, 793102. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19). JAMA 2020, 324, 782. [Google Scholar] [CrossRef]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef]
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Lu, Z.; Zhang, L.; Fan, T.; Xiong, R.; Shen, X.; Feng, H.; Meng, H.; Lin, W.; Jiang, W.; et al. The clinical course and its correlated immune status in COVID-19 pneumonia. J. Clin. Virol. 2020, 127, 104361. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Björkander, S.; Du, L.; Zuo, F.; Ekström, S.; Wang, Y.; Wan, H.; Sherina, N.; Schoutens, L.; Andréll, J.; Andersson, N.; et al. SARS-CoV-2 specific B- and T-cell immunity in a population-based study of young Swedish adults. J. Allergy Clin. Immunol. 2021, 149, 65–75.e8. [Google Scholar] [CrossRef] [PubMed]
- Boyton, R.J.; Altmann, D.M. The immunology of asymptomatic SARS-CoV-2 infection: What are the key questions? Nat. Rev. Immunol. 2021, 21, 762–768. [Google Scholar] [CrossRef]
- Olea, B.; Albert, E.; Torres, I.; Amat, P.; Remigia, M.J.; Gozalbo-Rovira, R.; Rodríguez-Díaz, J.; Buesa, J.; Blasco, M.L.; Redón, J.; et al. Adaptive immune responses to SARS-CoV-2 in recovered severe COVID-19 patients. J. Clin. Virol. 2021, 142, 104943. [Google Scholar] [CrossRef]
- Bao, Y.; Ling, Y.; Chen, Y.y.; Tian, D.; Zhao, G.p.; Zhang, X.h.; Hang, H.; Li, Y.; Su, B.; Lu, H.z.; et al. Dynamic anti-spike protein antibody profiles in COVID-19 patients. Int. J. Infect. Dis. 2021, 103, 540–548. [Google Scholar] [CrossRef]
- Cox, R.J.; Brokstad, K.A. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat. Rev. Immunol. 2020, 20, 581–582. [Google Scholar] [CrossRef]
- Long, Q.-X.; Tang, X.-J.; Shi, Q.-L.; Li, Q.; Deng, H.-J.; Yuan, J.; Hu, J.-L.; Xu, W.; Zhang, Y.; Lv, F.-J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Müller, M.A.; Li, W.; Wang, C.; Geurtsvankessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; Bruin, E.D.; Chandler, F.D.; et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg. Infect. Dis. 2020, 26, 1478–1488. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef]
- Havervall, S.; Ng, H.; Falk, A.J.; Greilert-Norin, N.; Månberg, A.; Marking, U.; Laurén, I.; Gabrielsson, L.; Salomonsson, A.C.; Aguilera, K.; et al. Robust humoral and cellular immune responses and low risk for reinfection at least eight months following asymptomatic to mild COVID-19. J. Intern. Med. 2021, 291, 72–80. [Google Scholar] [CrossRef]
- Gallais, F.; Gantner, P.; Bruel, T.; Velay, A.; Planas, D.; Wendling, M.-J.; Bayer, S.; Solis, M.; Laugel, E.; Reix, N.; et al. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine 2021, 71, 103561. [Google Scholar] [CrossRef]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.W.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature 2021, 602, 676–681. [Google Scholar] [CrossRef]
- Carreño, J.M.; Alshammary, H.; Tcheou, J.; Singh, G.; Raskin, A.; Kawabata, H.; Sominsky, L.; Clark, J.; Adelsberg, D.C.; Bielak, D.; et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 2021, 602, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.-H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2021, 602, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.-K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2021, 602, 487–695. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2021, 602, 654–656. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2021, 602, 657–663. [Google Scholar] [CrossRef]
- Ramezani, A.; Sorouri, R.; Haji Maghsoudi, S.; Dahmardeh, S.; Doroud, D.; Sadat Larijani, M.; Eybpoosh, S.; Mostafavi, E.; Olyaeemanesh, A.; Salehi-Vaziri, M.; et al. PastoCovac and PastoCovac Plus as protein subunit COVID-19 vaccines led to great humoral immune responses in BBIP-CorV immunized individuals. Sci. Rep. 2023, 13, 8065. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977.e973. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Vogler, I.; Derhovanessian, E.; Kranz, L.M.; Vormehr, M.; Quandt, J.; Bidmon, N.; Ulges, A.; Baum, A.; et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021, 595, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Andreatta, M.; Nielsen, M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef]
- Escalante, P.; Peikert, T.; Van Keulen, V.P.; Erskine, C.L.; Bornhorst, C.L.; Andrist, B.R.; McCoy, K.; Pease, L.R.; Abraham, R.S.; Knutson, K.L.; et al. Combinatorial immunoprofiling in latent tuberculosis infection: Toward better risk stratification. Am. J. Respir. Crit. Care Med. 2015, 192, 605–617. [Google Scholar] [CrossRef]
- Bowyer, G.; Rampling, T.; Powlson, J.; Morter, R.; Wright, D.; Hill, A.V.S.; Ewer, K.J. Activation-induce markers detect vaccine-specific CD4+ T cell responses not measured by assays conventionally used in clinical trials. Vaccines 2018, 6, 50. [Google Scholar] [CrossRef]
- Agilent. xCELLigence RTCA S16-Pilot Scale RUO. Available online: https://www.agilent.com/en/product/cell-analysis/real-time-cell-analysis/rtca-analyzers/xcelligence-rtca-s16-pilot-scale-741231 (accessed on 4 October 2023).
- Azqueta, A.; Stopper, H.; Zegura, B.; Dusinska, M.; Møller, P. Do cytotoxicity and cell death cause false positive results in the in vitro comet assay? Mutat. Res. Genet. Toxicol. Environ. Mutagen 2022, 881, 503520. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Begom, S.; Hoschler, K.; Bermingham, A.; Adamson, W.; Carman, W.; Riley, S.; Lalvani, A. Longevity and Determinants of Protective Humoral Immunity after Pandemic Influenza Infection. Am. J. Respir. Crit. Care Med. 2015, 191, 325–332. [Google Scholar] [CrossRef]
- Mantei, A.; Meyer, T.; Schürmann, M.; Beßler, C.; Bias, H.; Krieger, D.; Bauer, T.; Bacher, P.; Helmuth, J.; Volk, H.-D.; et al. Mycobacterium tuberculosis -specific CD4 T-cell scoring discriminates tuberculosis infection from disease. Eur. Respir. J. 2022, 60, 2101780. [Google Scholar] [CrossRef] [PubMed]
- Niessl, J.; Sekine, T.; Buggert, M. T cell immunity to SARS-CoV-2. Semin. Immunol. 2021, 55, 101505. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.T.d.; Ortega, M.M.; Tiyo, B.T.; Viana, I.F.T.; Lima, T.E.d.; Tozetto-Mendoza, T.R.; Oliveira, L.M.d.S.; Teixeira, F.M.E.; Lins, R.D.; Almeida, A.d.; et al. SARS-CoV-2 recombinant proteins stimulate distinct cellular and humoral immune response profiles in samples from COVID-19 convalescent patients. Clinics 2021, 76, e3548. [Google Scholar] [CrossRef] [PubMed]
- Valletta, J.J.; Recker, M. Identification of immune signatures predictive of clinical protection from malaria. PLoS Comput. Biol. 2017, 13, e1005812. [Google Scholar] [CrossRef]
- Dan, J.M.; Lindestam Arlehamn, C.S.; Weiskopf, D.; da Silva Antunes, R.; Havenar-Daughton, C.; Reiss, S.M.; Brigger, M.; Bothwell, M.; Sette, A.; Crotty, S. A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4 + T Cells in Blood. J. Immunol. 2016, 197, 983–993. [Google Scholar] [CrossRef]
- Dijkman, K.; Aguilo, N.; Boot, C.; Hofman, S.O.; Sombroek, C.C.; Vervenne, R.A.W.; Kocken, C.H.M.; Marinova, D.; Thole, J.; Rodríguez, E.; et al. Pulmonary MTBVAC vaccination induces immune signatures previously correlated with prevention of tuberculosis infection. Cell Rep. Med. 2021, 2, 100187. [Google Scholar] [CrossRef]
- Lewinsohn, D.M.; Lewinsohn, D.A. The Missing Link in Correlates of Protective Tuberculosis Immunity: Recognizing the Infected Cell. Front. Immunol. 2022, 13, 869057. [Google Scholar] [CrossRef]
- Swadling, L.; Maini, M.K. T cells in COVID-19—United in diversity. Nat. Immunol. 2020, 21, 1307–1308. [Google Scholar] [CrossRef]
- Boonyaratanakornkit, J.; Morishima, C.; Selke, S.; Zamora, D.; McGuffin, S.; Shapiro, A.E.; Campbell, V.L.; McClurkan, C.L.; Jing, L.; Gross, R.; et al. Clinical, laboratory, and temporal predictors of neutralizing antibodies against SARS-CoV-2 among COVID-19 convalescent plasma donor candidates. J. Clin. Investig. 2021, 131, e144930. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Laing, E.D.; Pena DaMata, J.; Pohida, K.; Tso, M.S.; Samuels, E.C.; Epsi, N.J.; Dorjbal, B.; Lake, C.; Richard, S.A.; et al. Durability of SARS-CoV-2-specific T cell responses at 12-months post-infection. J. Infect. Dis. 2021, 224, 2010–2019. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Shi, J.; Fan, Q.; Wang, Y.; Huang, H.; Chen, F.; Tang, G.; Li, Y.; Li, P.; Li, J.; et al. Protective humoral and cellular immune responses to SARS-CoV-2 persist up to 1 year after recovery. Nat. Commun. 2021, 12, 4984. [Google Scholar] [CrossRef] [PubMed]
- Lau, E.H.; Hui, D.S.; Tsang, O.T.; Chan, W.H.; Kwan, M.Y.; Chiu, S.S.; Cheng, S.M.; Ko, R.L.; Li, J.K.; Chaothai, S.; et al. Long-term persistence of SARS-CoV-2 neutralizing antibody responses after infection and estimates of the duration of protection. EClinicalMedicine 2021, 41, 101174. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, J.; Mangalam, A.K.; Channappanavar, R.; Fett, C.; Meyerholz, D.K.; Agnihothram, S.; Baric, R.S.; David, C.S.; Perlman, S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 2016, 44, 1379–1391. [Google Scholar] [CrossRef]
- Brunk, F.; Moritz, A.; Nelde, A.; Bilich, T.; Casadei, N.; Fraschka, S.A.K.; Heitmann, J.S.; Hörber, S.; Peter, A.; Rammensee, H.G.; et al. SARS-CoV-2-reactive T-cell receptors isolated from convalescent COVID-19 patients confer potent T-cell effector function. Eur. J. Immunol. 2021, 51, 2651–2664. [Google Scholar] [CrossRef]
- Almendro-Vázquez, P.; Laguna-Goya, R.; Ruiz-Ruigomez, M.; Utrero-Rico, A.; Lalueza, A.; Maestro de la Calle, G.; Delgado, P.; Perez-Ordoño, L.; Muro, E.; Vila, J.; et al. Longitudinal dynamics of SARS-CoV-2-specific cellular and humoral immunity after natural infection or BNT162b2 vaccination. PLoS Pathog. 2021, 17, e1010211. [Google Scholar] [CrossRef]
- Jacot, D.; Moraz, M.; Coste, A.T.; Aubry, C.; Sacks, J.A.; Greub, G.; Croxatto, A. Evaluation of sixteen ELISA SARS-CoV-2 serological tests. J. Clin. Virol. 2021, 142, 104931. [Google Scholar] [CrossRef]
- Zhou, J.; Singanayagam, A.; Goonawardane, N.; Moshe, M.; Sweeney, F.P.; Sukhova, K.; Killingley, B.; Kalinova, M.; Mann, A.J.; Catchpole, A.P.; et al. Viral emissions into the air and environment after SARS-CoV-2 human challenge: A phase 1, open label, first-in-human study. Lancet Microbe 2023, 4, e579–e590. [Google Scholar] [CrossRef]
- Elyanow, R.; Snyder, T.M.; Dalai, S.C.; Gittelman, R.M.; Boonyaratanakornkit, J.; Wald, A.; Selke, S.; Wener, M.H.; Morishima, C.; Greninger, A.L.; et al. T cell receptor sequencing identifies prior SARS-CoV-2 infection and correlates with neutralizing antibodies and disease severity. JCI Insight 2022, 7, e150070. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020, 587, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Marty, P.K.; Van Keulen, V.P.; Erskine, C.L.; Shah, M.; Hummel, A.; Stachowitz, M.; Fatis, S.; Granger, D.; Block, M.S.; Duarte-García, A.; et al. Antigen Specific Humoral and Cellular Immunity Following SARS-CoV-2 Vaccination in ANCA-Associated Vasculitis Patients Receiving B-Cell Depleting Therapy. Front. Immunol. 2022, 13, 834981. [Google Scholar] [CrossRef] [PubMed]
- Hueso, T.; Pouderoux, C.; Péré, H.; Beaumont, A.-L.; Raillon, L.-A.; Ader, F.; Chatenoud, L.; Eshagh, D.; Szwebel, T.-A.; Martinot, M.; et al. Convalescent plasma therapy for B-cell-depleted patients with protracted COVID-19. Blood 2020, 136, 2290–2295. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Chia, W.N.; Wan, W.Y.; Teo, A.K.J.; Chong, S.Z.-R.; Tan, N.; Tan, D.S.C.; Chia, A.; Tan, I.B.; Kunasegaran, K.; et al. Widely heterogeneous humoral and cellular immunity after mild SARS-CoV-2 infection in a homogeneous population of healthy young men. Emerg. Microbes Infect. 2021, 10, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Mukund, K.; Nayak, P.; Ashokkumar, C.; Rao, S.; Almeda, J.; Betancourt-Garcia, M.M.; Sindhi, R.; Subramaniam, S. Immune Response in Severe and Non-Severe Coronavirus Disease 2019 (COVID-19) Infection: A Mechanistic Landscape. Front. Immunol. 2021, 12, 738073. [Google Scholar] [CrossRef]
- García-González, P.; Tempio, F.; Fuentes, C.; Merino, C.; Vargas, L.; Simon, V.; Ramirez-Pereira, M.; Rojas, V.; Tobar, E.; Landskron, G.; et al. Dysregulated Immune Responses in COVID-19 Patients Correlating With Disease Severity and Invasive Oxygen Requirements. Front. Immunol. 2021, 12, 769059. [Google Scholar] [CrossRef]

| N-Terminal Amino Acid | Peptide Sequence | Predicted Affinity for HLA-A2 |
|---|---|---|
| 268 | YLQPRTFLL | 5.4 |
| 132 | FQFCNDPFL | 9.2 |
| 690 | SIIAYTMSL | 13.5 |
| 385 | KLNDLCFTNV | 15.3 |
| 514 | FELLHAPATV | 21 |
| 3 | FLVLLPLV | 28.2 |
| 416 | KIADYNYKL | 36.1 |
| 1 | FVFLVLLPLV | 32.6 |
| 267 | GYLQPRTFLL | 36.1 |
| Demographic | Subjects, No. (%) | ||
|---|---|---|---|
| Convalescent Donors (n = 30) | Unexposed Controls (n = 7) | ||
| Sex | p = 0.8110 | ||
| Male | 14 (46.7) | 3 (57.1) | |
| Female | 16 (53.3) | 4 (42.9) | |
| Age (years) | p = 0.0397 | ||
| Mean ± SD | 44 ± 15.4 | 61 ± 16.8 | |
| Range | 21–67 | 35–80 | |
| HLA-A2 + | 12 (40) | N/A | |
| SPIKE HLA-A2 Peptides | Pt 1 | Pt 2 | Pt 3 | Pt 6 | Pt 12 | Pt 15 | Pt 16 | Pt 17 | Pt 20 | Pt 23 | Pt 25 | Pt 28 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cov1 | 21.60 | 6.92 | 43.89 | 6.82 | 47.53 | 27.06 | 46.33 | 61.67 | 22.63 | 35.27 | 30.85 | 58.08 |
| Cov3 | 31.24 | 8.13 | 36.33 | 21.94 | 32.58 | 30.84 | 27.34 | 33.96 | 19.92 | 31.33 | 33.85 | 39.17 |
| Cov132 | 24.11 | 5.68 | 31.66 | 22.37 | 31.45 | 33.23 | 22.76 | 29.04 | 23.89 | 23.41 | 36.42 | 54.43 |
| Cov267 | 25.93 | 3.21 | 26.52 | 25.95 | 40.69 | 33.02 | 27.05 | 42.35 | 20.17 | 44.20 | 48.82 | 48.60 |
| Cov268 | 20.16 | 1.51 | 22.12 | 17.30 | 42.98 | 31.81 | 29.87 | 37.64 | 19.28 | 17.59 | 25.61 | 29.71 |
| Cov385 | 5.48 | 8.82 | 21.48 | 17.10 | 31.63 | 22.94 | 21.91 | 21.10 | 5.57 | 11.01 | 18.32 | 24.79 |
| Cov416 | 7.80 | 2.85 | 28.81 | 16.19 | 36.67 | 26.22 | 22.47 | 21.69 | 13.24 | 9.66 | 24.08 | 22.31 |
| Cov514 | 10.30 | 4.94 | 24.79 | 13.79 | 10.79 | 13.92 | 14.88 | 14.80 | 12.96 | 22.89 | 5.10 | 18.34 |
| Cov690 | 16.63 | 9.89 | 35.59 | 31.67 | 39.34 | 23.56 | 18.03 | 62.20 | 16.29 | 25.33 | 26.40 | 38.35 |
| # Peptides | 1 | 0 | 4 | 1 | 8 | 4 | 1 | 5 | 0 | 3 | 4 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pathakumari, B.; Marty, P.K.; Shah, M.; Van Keulen, V.P.; Erskine, C.L.; Block, M.S.; Arias-Sanchez, P.; Escalante, P.; Peikert, T. Convalescent Adaptive Immunity Is Highly Heterogenous after SARS-CoV-2 Infection. J. Clin. Med. 2023, 12, 7136. https://doi.org/10.3390/jcm12227136
Pathakumari B, Marty PK, Shah M, Van Keulen VP, Erskine CL, Block MS, Arias-Sanchez P, Escalante P, Peikert T. Convalescent Adaptive Immunity Is Highly Heterogenous after SARS-CoV-2 Infection. Journal of Clinical Medicine. 2023; 12(22):7136. https://doi.org/10.3390/jcm12227136
Chicago/Turabian StylePathakumari, Balaji, Paige K. Marty, Maleeha Shah, Virginia P. Van Keulen, Courtney L. Erskine, Matthew S. Block, Pedro Arias-Sanchez, Patricio Escalante, and Tobias Peikert. 2023. "Convalescent Adaptive Immunity Is Highly Heterogenous after SARS-CoV-2 Infection" Journal of Clinical Medicine 12, no. 22: 7136. https://doi.org/10.3390/jcm12227136
APA StylePathakumari, B., Marty, P. K., Shah, M., Van Keulen, V. P., Erskine, C. L., Block, M. S., Arias-Sanchez, P., Escalante, P., & Peikert, T. (2023). Convalescent Adaptive Immunity Is Highly Heterogenous after SARS-CoV-2 Infection. Journal of Clinical Medicine, 12(22), 7136. https://doi.org/10.3390/jcm12227136







