A Microplate-Based Approach to Map Interactions between TDP-43 and α-Synuclein
Abstract
1. Introduction
2. Materials and Methods
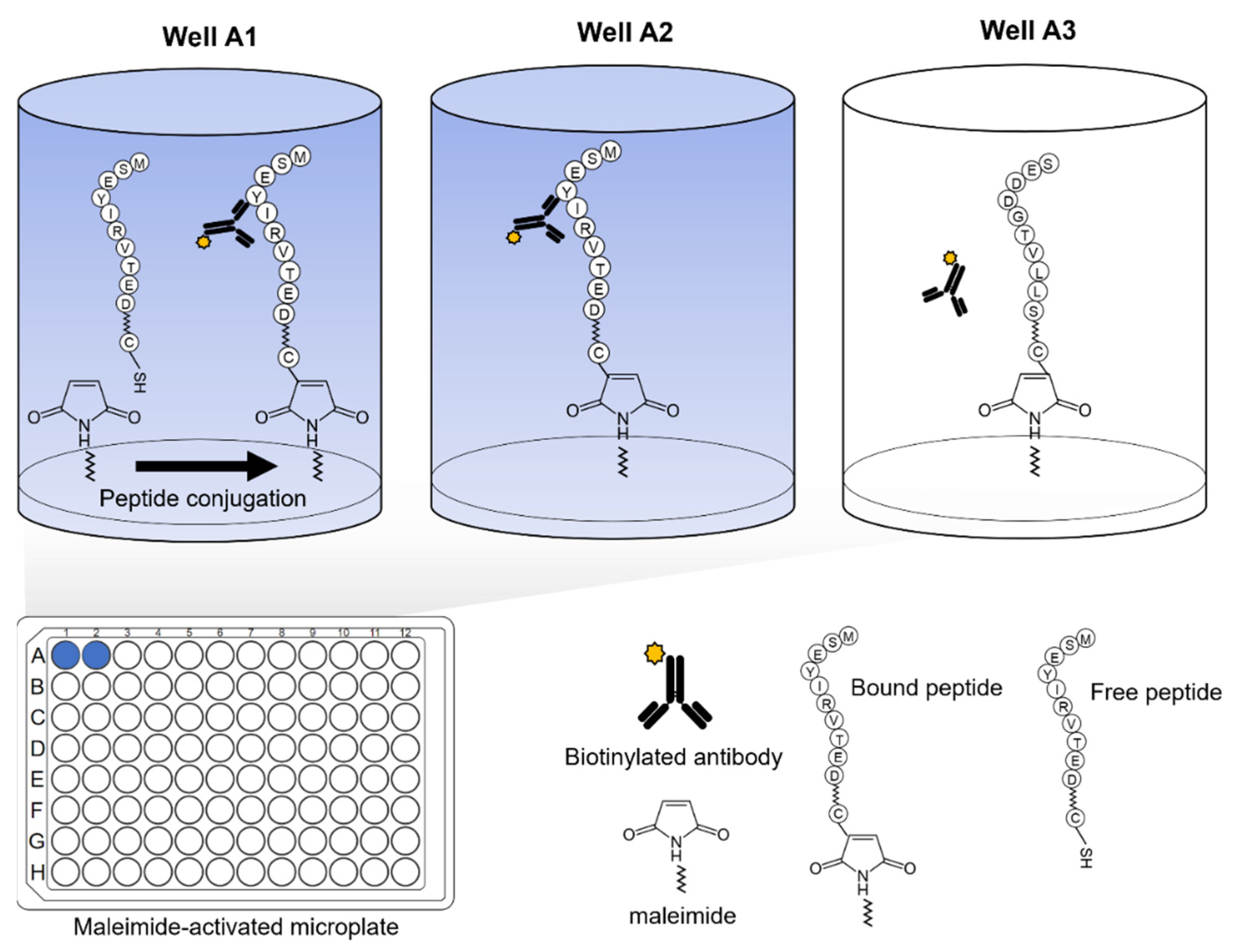
2.1. Peptide Synthesis and Preparation
2.2. Microplate-Based Epitope Mapping of Antibodies and Recombinant Proteins
2.3. Interaction of α-Synuclein and TDP-43
2.4. In Silico Sequence-Based Prediction
2.5. Statistical Analysis
3. Results and Discussion
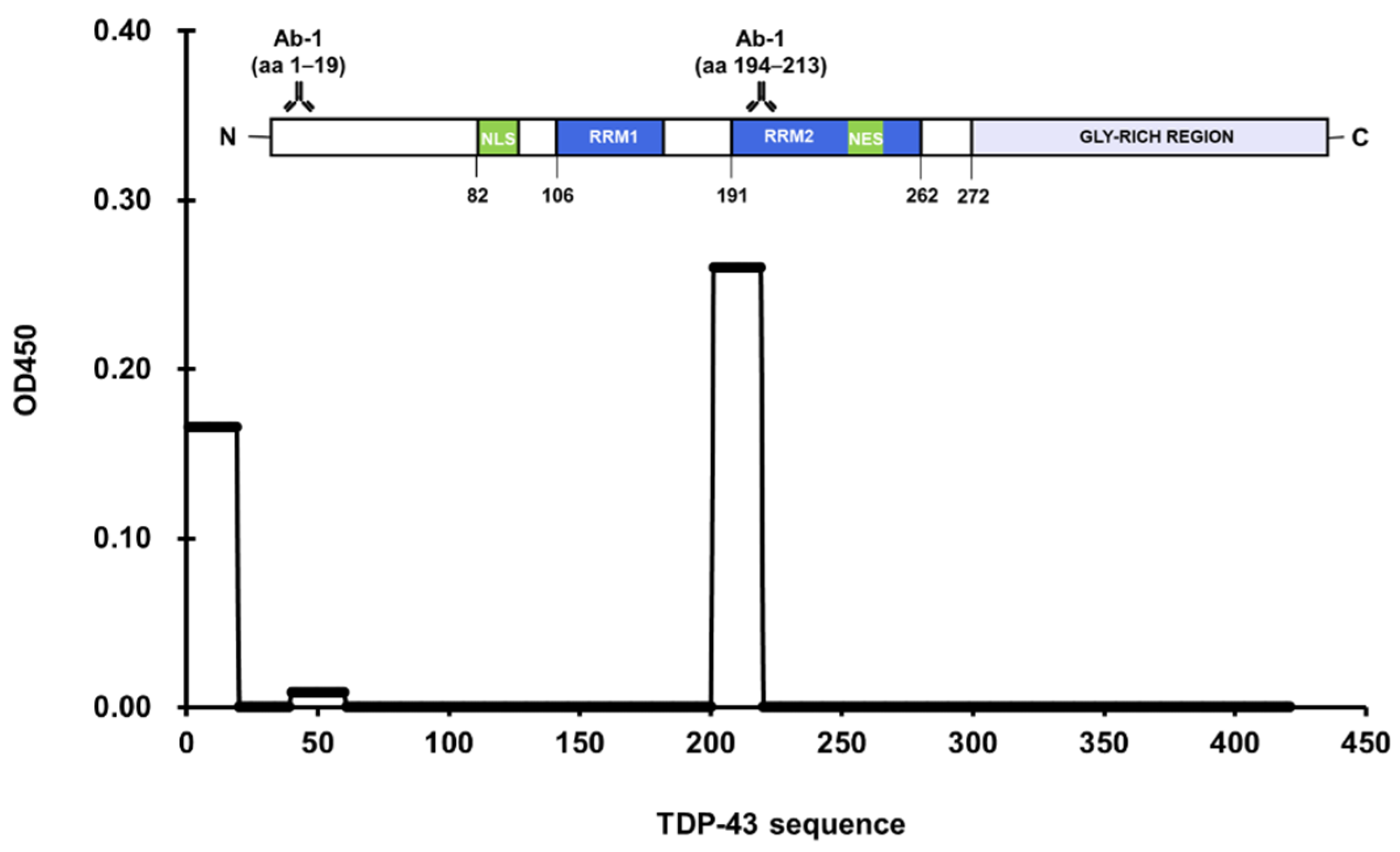
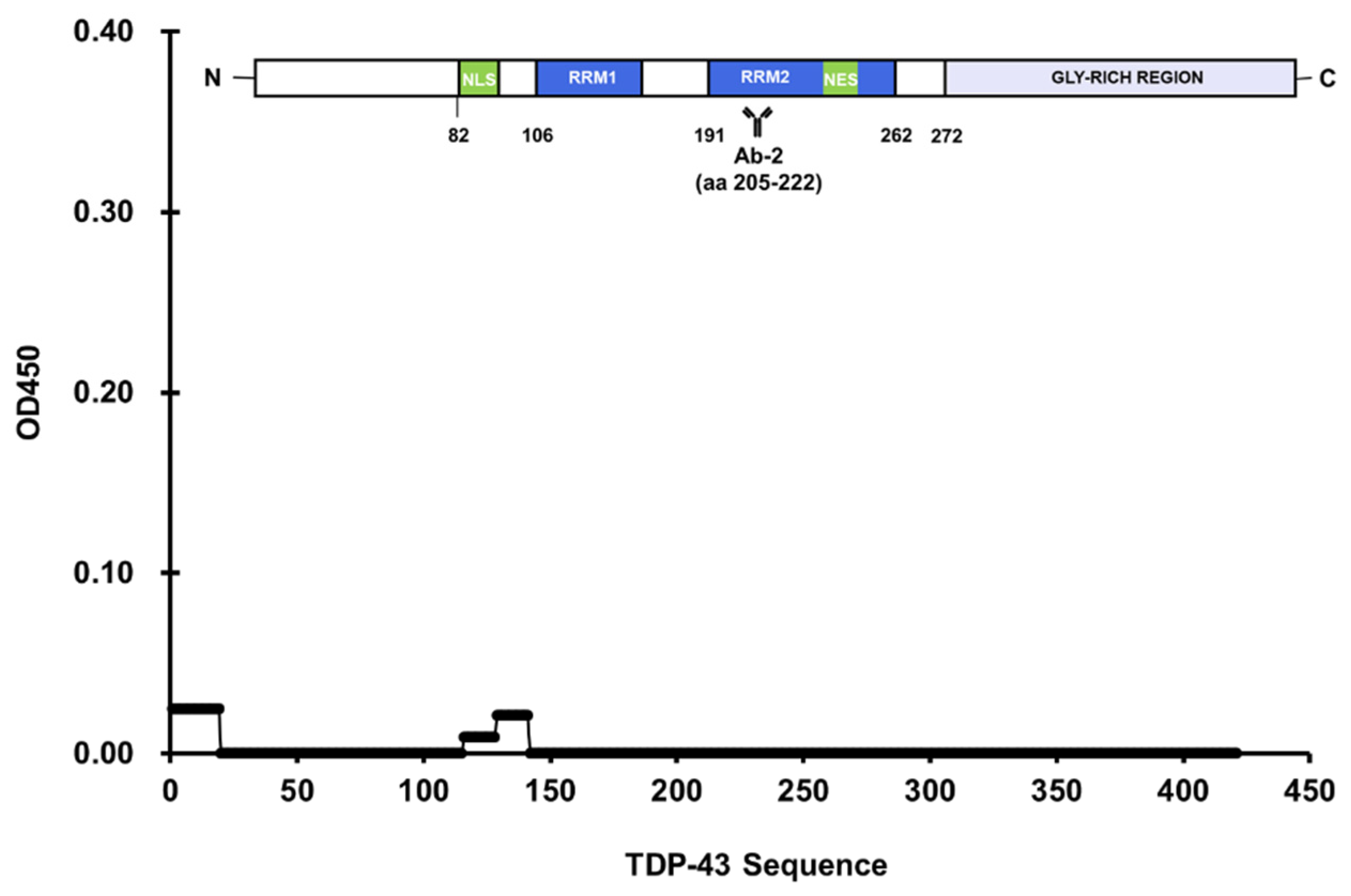
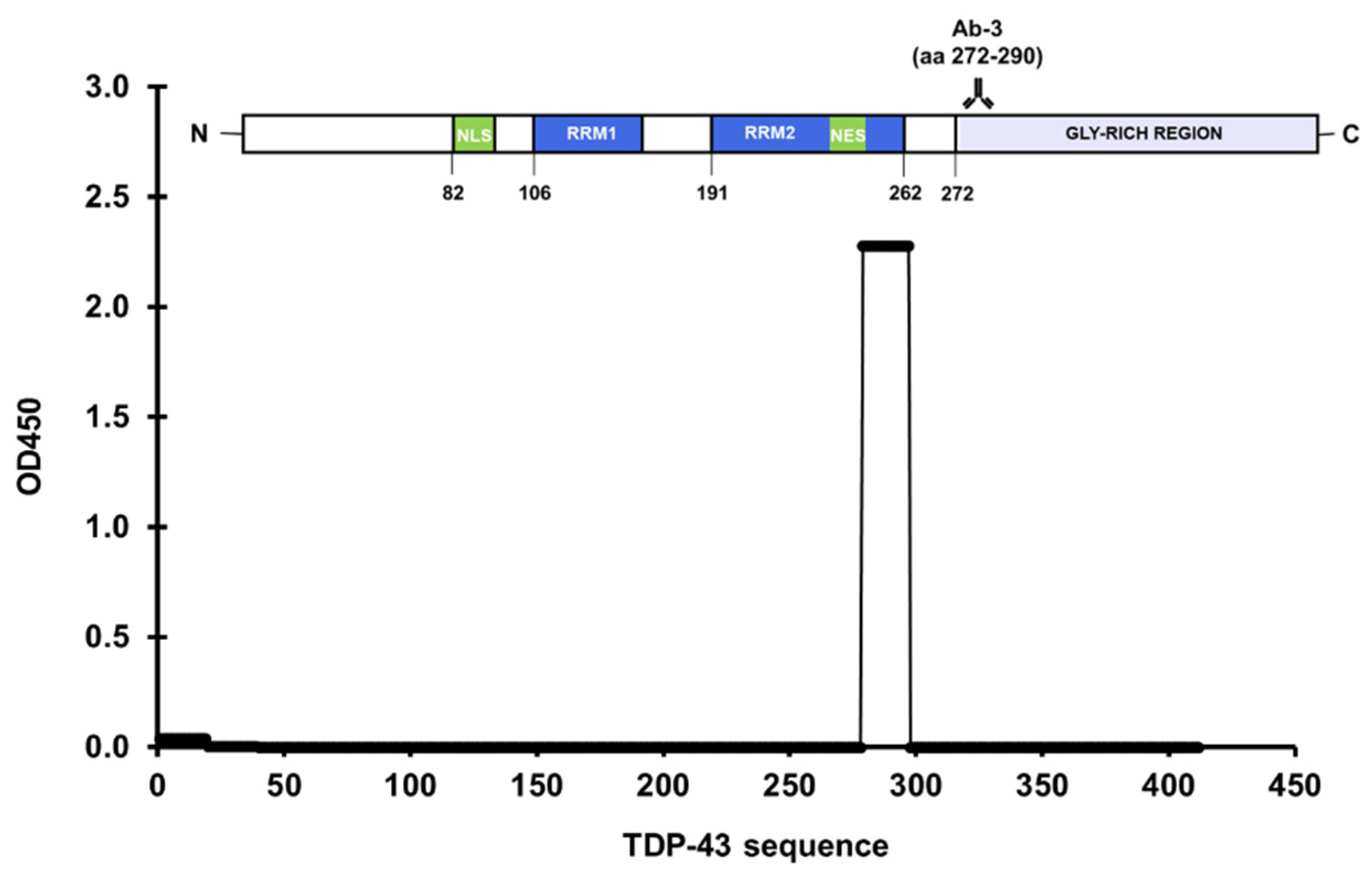
3.1. Epitope Mapping of Anti-TDP-43 Antibodies
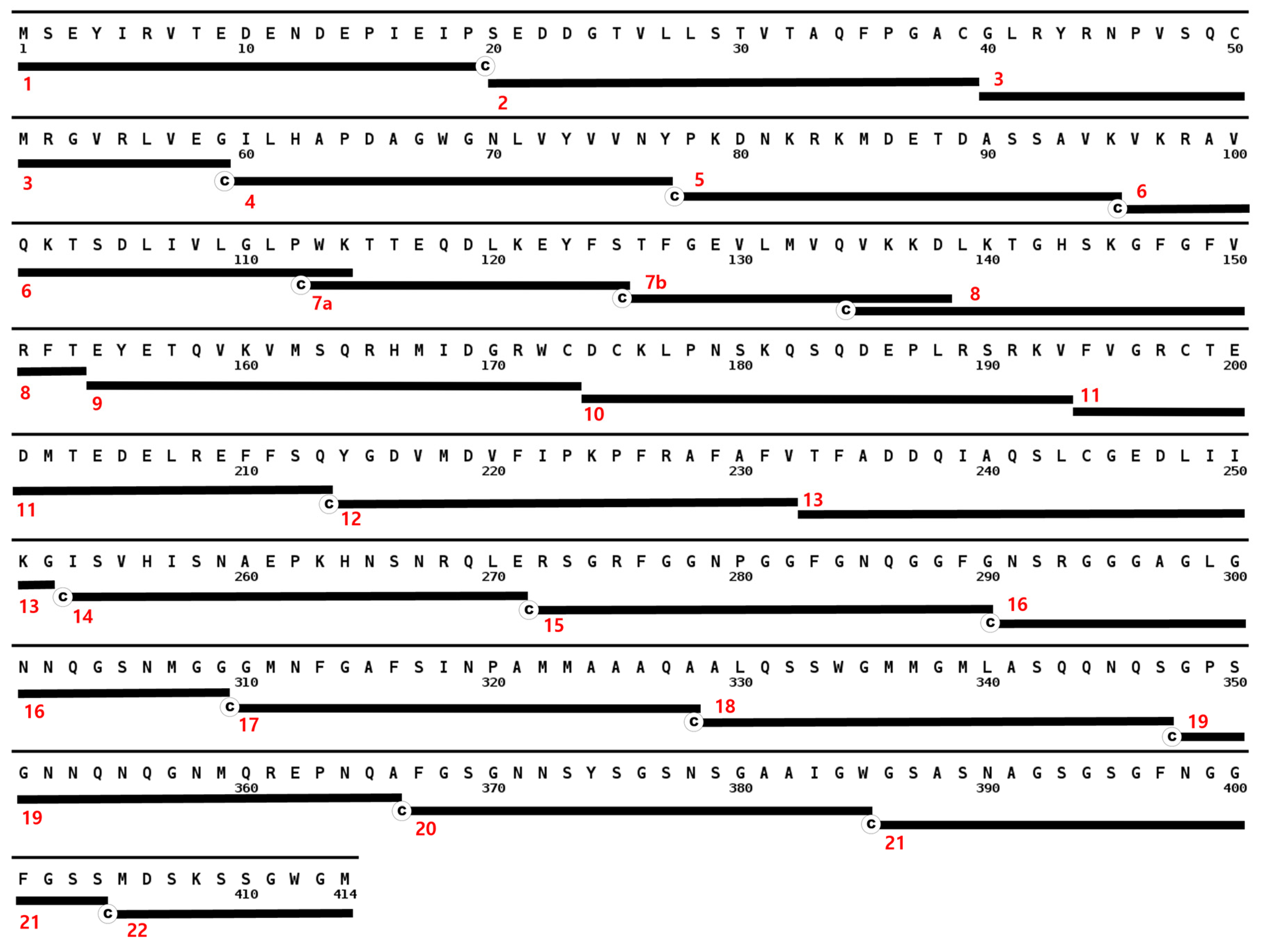
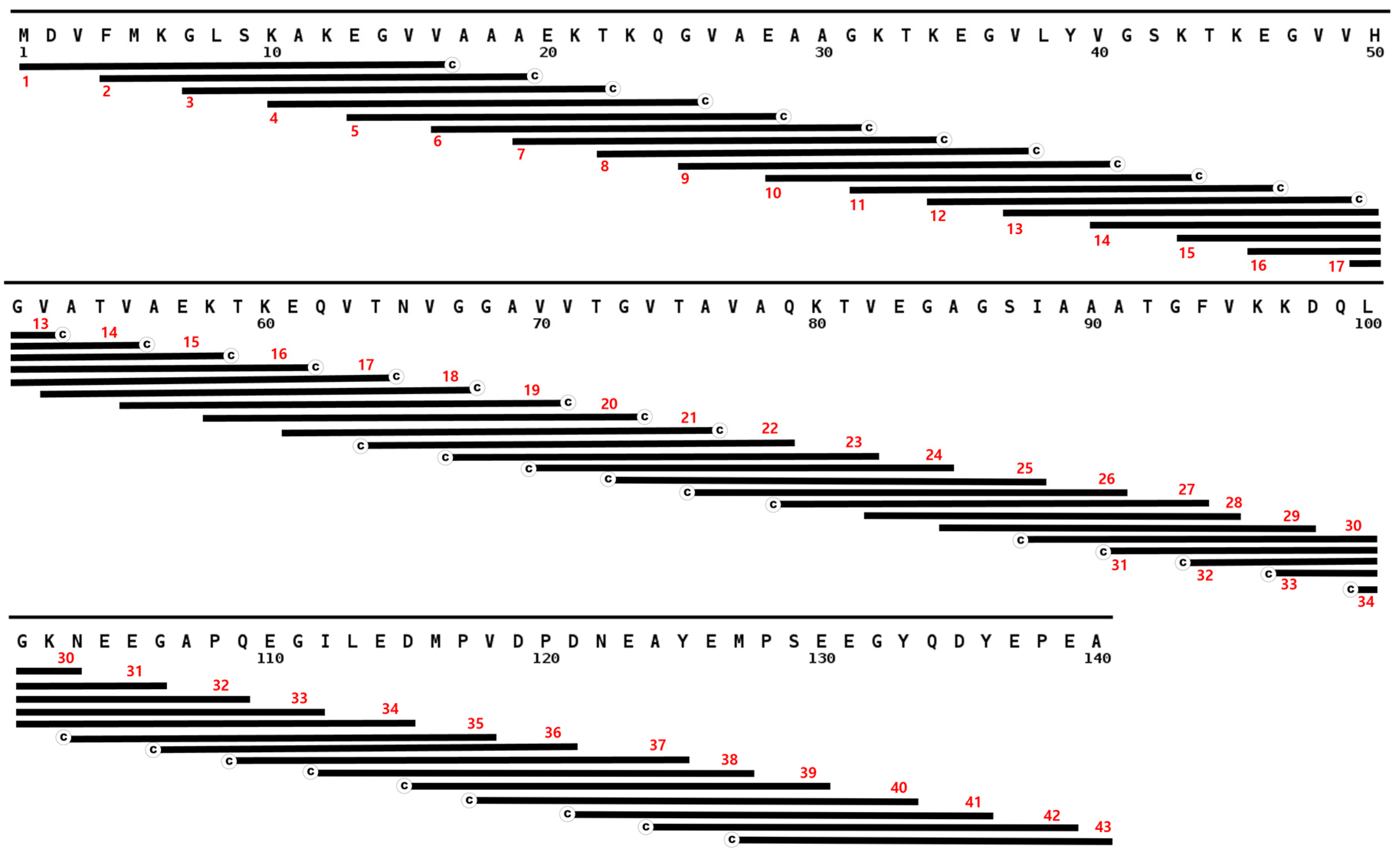
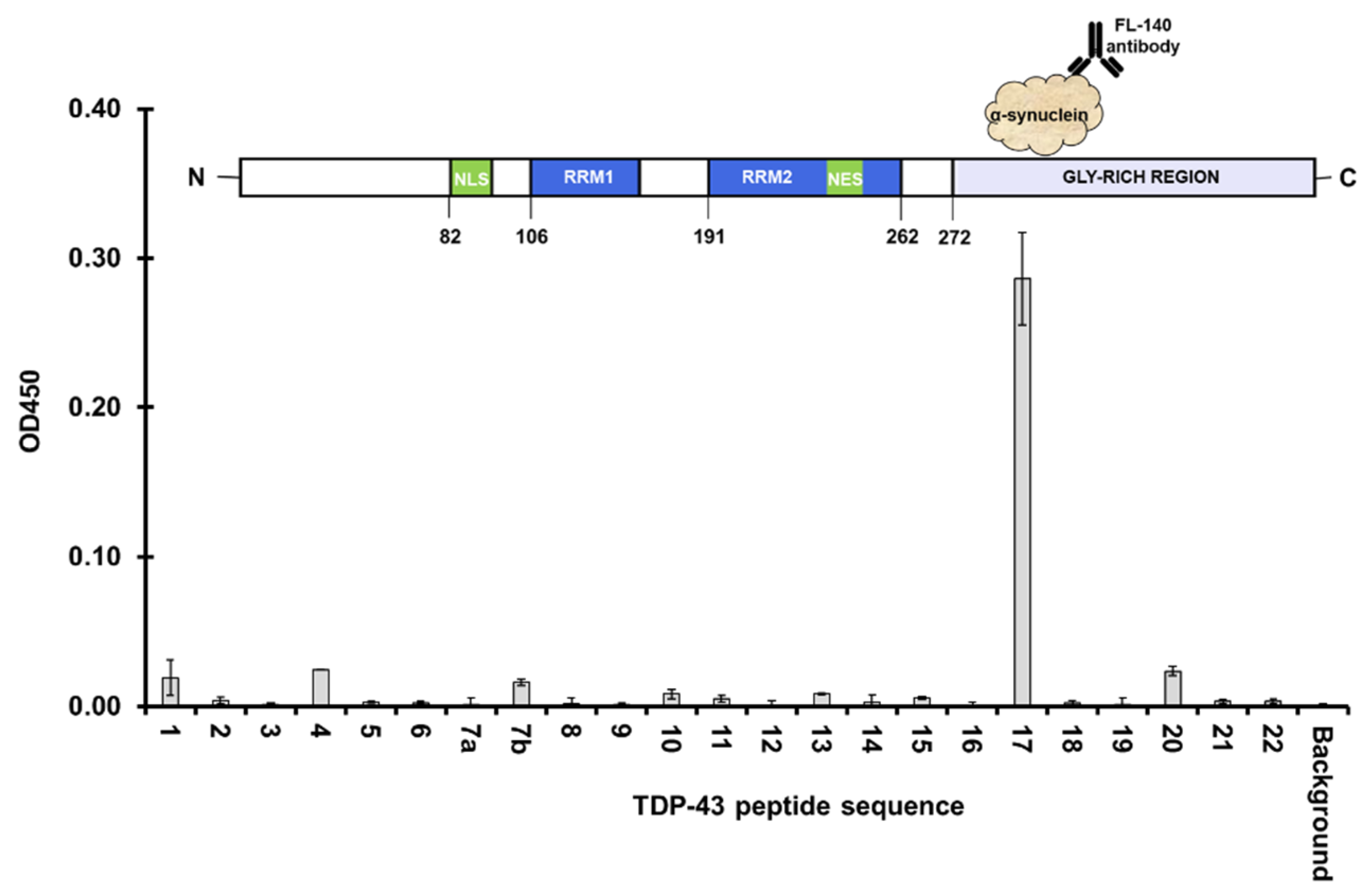
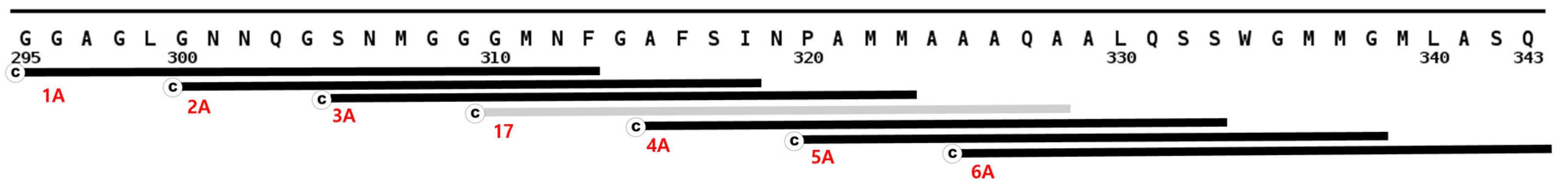
3.2. Full Length Alpha Synuclein Interacts with a Region of the LCD in TDP-43
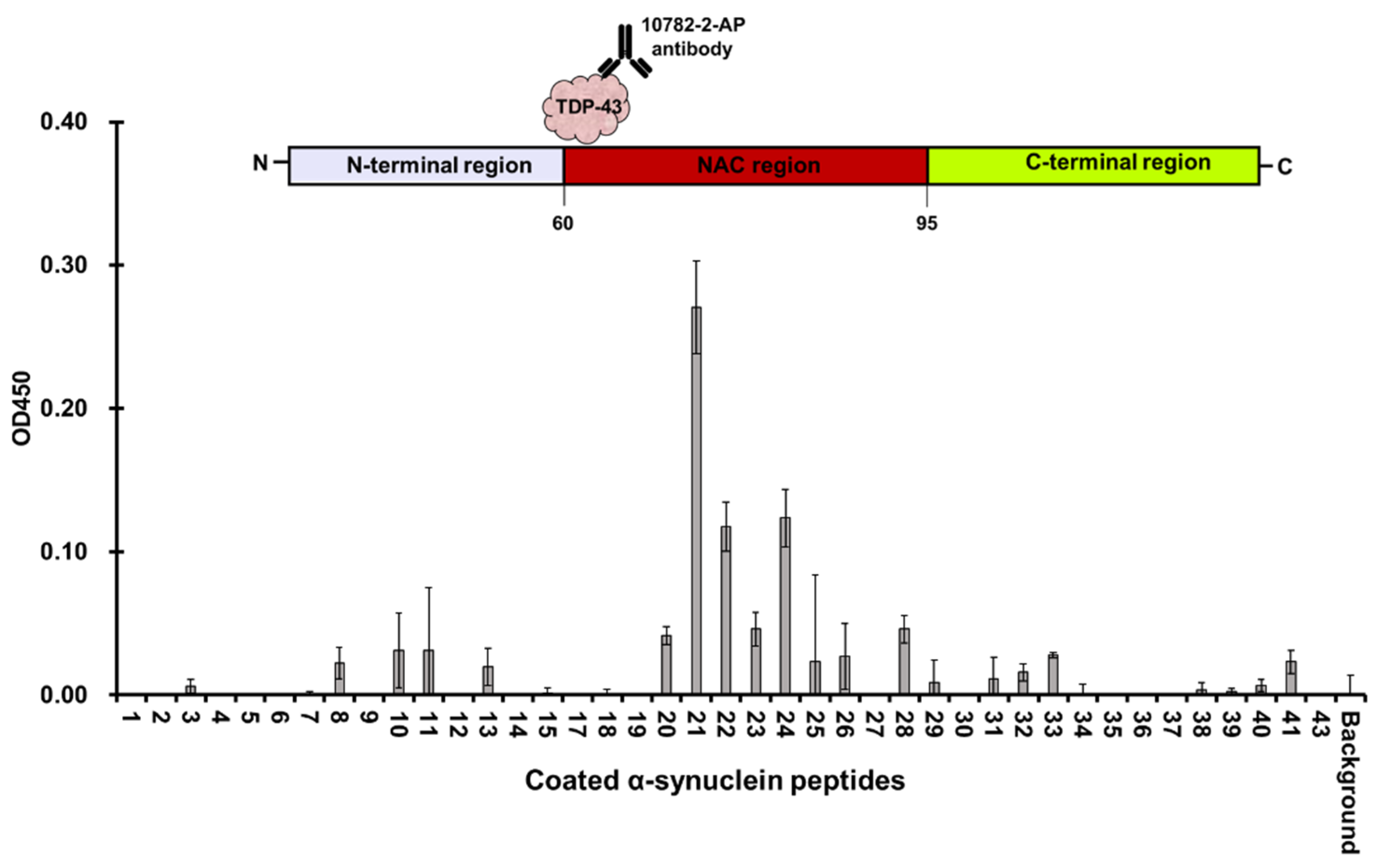
3.3. Full Length TDP-43 Interacts with the NAC Region of α-Synuclein
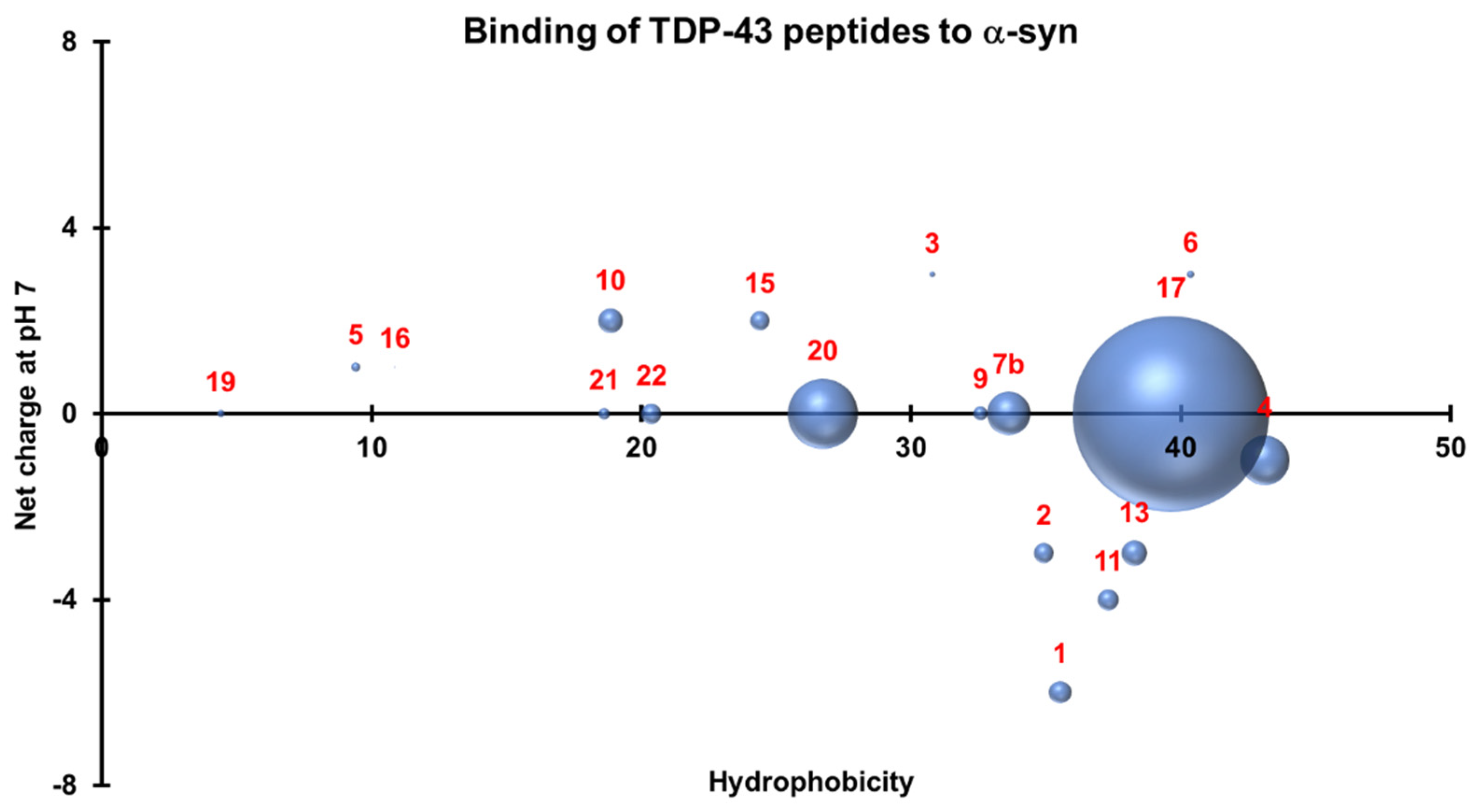
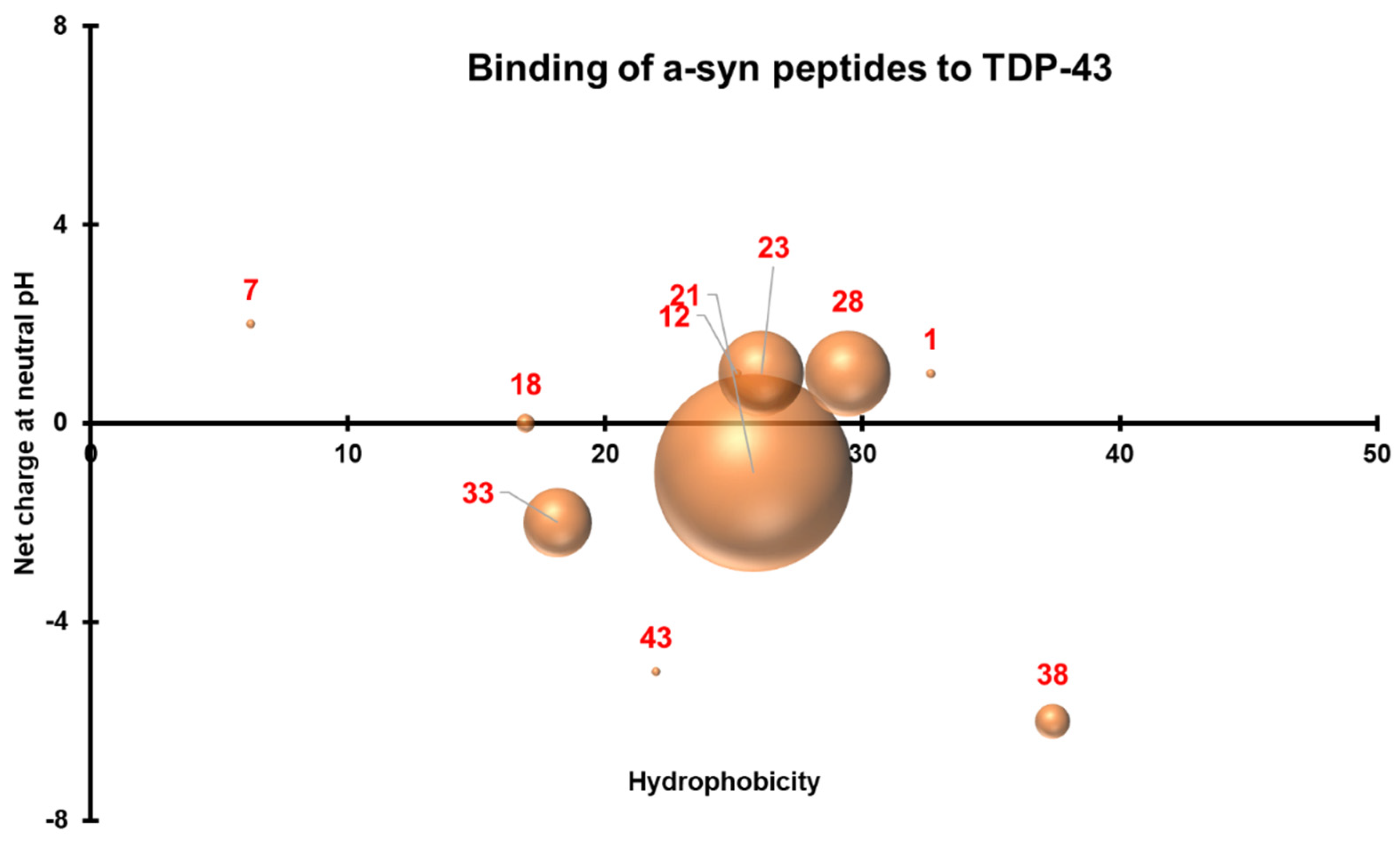
3.4. Three-Dimensional Scatterplots of the Peptides Reveal Similar Properties That Influence Their Interaction
3.5. Conventional ELISA Did Not Show Any Interaction between the Full-Length Proteins
3.6. In Silico Prediction of Protein Binding Affinity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ou, S.H.; Wu, F.; Harrich, D.; García-Martínez, L.F.; Gaynor, R.B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995, 69, 3584–3596. [Google Scholar] [CrossRef] [PubMed]
- Ayala, Y.M.; Zago, P.; D’Ambrogio, A.; Xu, Y.-F.; Petrucelli, L.; Buratti, E.; Baralle, F.E. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 2008, 121, 3778–3785. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.-H.; Doudeva, L.G.; Wang, Y.-T.; Shen, C.-K.J.; Yuan, H.S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009, 37, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.; Rademakers, R. The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr. Opin. Neurol. 2008, 21, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.; Rademakers, R.; Neumann, M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010, 9, 995–1007. [Google Scholar] [CrossRef]
- Kawakami, I.; Arai, T.; Hasegawa, M. The basis of clinicopathological heterogeneity in TDP-43 proteinopathy. Acta Neuropathol. 2019, 138, 751–770. [Google Scholar] [CrossRef]
- Schwab, C.; Arai, T.; Hasegawa, M.; Yu, S.; McGeer, P.L. Colocalization of Transactivation-Responsive DNA-Binding Protein 43 and Huntingtin in Inclusions of Huntington Disease. J. Neuropathol. Exp. Neurol. 2008, 67, 1159–1165. [Google Scholar] [CrossRef]
- Nakashima-Yasuda, H.; Uryu, K.; Robinson, J.; Xie, S.X.; Hurtig, H.; Duda, J.E.; Arnold, S.E.; Siderowf, A.; Grossman, M.; Leverenz, J.; et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007, 114, 221–229. [Google Scholar] [CrossRef]
- Shih, Y.H.; Tu, L.H.; Chang, T.Y.; Ganesan, K.; Chang, W.W.; Chang, P.S.; Fang, Y.S.; Lin, Y.T.; Jin, L.W.; Chen, Y.R. TDP-43 interacts with amyloid-β, inhibits fibrillization, and worsens pathology in a model of Alzheimer’s disease. Nat. Commun. 2020, 11, 5950. [Google Scholar] [CrossRef]
- Amador-Ortiz, C.; Lin, W.L.; Ahmed, Z.; Personett, D.; Davies, P.; Duara, R.; Graff-Radford, N.R.; Hutton, M.L.; Dickson, D.W. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann. Neurol. 2007, 61, 435–445. [Google Scholar] [CrossRef]
- Youmans, K.L.; Wolozin, B. TDP-43: A new player on the AD field? Exp. Neurol. 2012, 237, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Tomé, S.O.; Vandenberghe, R.; Ospitalieri, S.; Van Schoor, E.; Tousseyn, T.; Otto, M.; Von Arnim, C.A.F.; Thal, D.R. Distinct molecular patterns of TDP-43 pathology in Alzheimer’s disease: Relationship with clinical phenotypes. Acta Neuropathol. Commun. 2020, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Stefanis, L. α-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009399. [Google Scholar] [CrossRef] [PubMed]
- Emamzadeh, F.N. Alpha-synuclein structure, functions, and interactions. J. Res. Med. Sci. 2016, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Iwai, A.; Yoshimoto, M.; Masliah, E.; Saitoh, T. Non-A.beta. Component of Alzheimer’s Disease Amyloid (NAC) is Amyloidogenic. Biochemistry 1995, 34, 10139–10145. [Google Scholar] [CrossRef]
- Török, N.; Majláth, Z.; Szalárdy, L.; Vécsei, L. Investigational α-synuclein aggregation inhibitors: Hope for Parkinson’s disease. Expert Opin. Investig. Drugs 2016, 25, 1281–1294. [Google Scholar] [CrossRef]
- Rajagopalan, S.; Andersen, J.K. Alpha synuclein aggregation: Is it the toxic gain of function responsible for neurodegeneration in Parkinson’s disease? Mech. Ageing Dev. 2001, 122, 1499–1510. [Google Scholar] [CrossRef]
- Kang, M.; Kim, S.Y.; An, S.S.; Ju, Y.R. Erratum: Characterizing affinity epitopes between prion protein and β-amyloid using an epitope mapping immunoassay. Exp. Mol. Med. 2014, 46, e96. [Google Scholar] [CrossRef]
- Biesiadecki, B.J.; Jin, J.-P. A High-Throughput Solid-Phase Microplate Protein-Binding Assay to Investigate Interactions between Myofilament Proteins. J. Biomed. Biotechnol. 2011, 2011, 421701. [Google Scholar] [CrossRef]
- Goossens, J.; Vanmechelen, E.; Trojanowski, J.Q.; Lee, V.M.; Van Broeckhoven, C.; van der Zee, J.; Engelborghs, S. TDP-43 as a possible biomarker for frontotemporal lobar degeneration: A systematic review of existing antibodies. Acta Neuropathol. Commun. 2015, 3, 15. [Google Scholar] [CrossRef][Green Version]
- Tsuji, H.; Nonaka, T.; Yamashita, M.; Masuda-Suzukake, M.; Kametani, F.; Akiyama, H.; Mann, D.M.; Tamaoka, A.; Hasegawa, M. Epitope mapping of antibodies against TDP-43 and detection of protease-resistant fragments of pathological TDP-43 in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Biochem. Biophys. Res. Commun. 2011, 417, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-X.; Tanji, K.; Mori, F.; Wakabayashi, K. Epitope mapping of 2E2-D3, a monoclonal antibody directed against human TDP-43. Neurosci. Lett. 2008, 434, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Koga, S.; Lin, W.-L.; Walton, R.L.; Ross, O.A.; Dickson, D.W. TDP-43 pathology in multiple system atrophy: Colocalization of TDP-43 and α-synuclein in glial cytoplasmic inclusions. Neuropathol. Appl. Neurobiol. 2018, 44, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Mackenzie, I.R.A.; Hasegawa, M.; Nonoka, T.; Niizato, K.; Tsuchiya, K.; Iritani, S.; Onaya, M.; Akiyama, H. Phosphorylated TDP-43 in Alzheimer’s disease and dementia with Lewy bodies. Acta Neuropathol. 2009, 117, 125–136. [Google Scholar] [CrossRef]
- Dhakal, S.; Wyant, C.E.; George, H.E.; Morgan, S.E.; Rangachari, V. Prion-like C-Terminal Domain of TDP-43 and α-Synuclein Interact Synergistically to Generate Neurotoxic Hybrid Fibrils. J. Mol. Biol. 2021, 433, 166953. [Google Scholar] [CrossRef]
- Yang, C.; Tan, W.; Whittle, C.; Qiu, L.; Cao, L.; Akbarian, S.; Xu, Z. The C-Terminal TDP-43 Fragments Have a High Aggregation Propensity and Harm Neurons by a Dominant-Negative Mechanism. PLoS ONE 2010, 5, e15878. [Google Scholar] [CrossRef]
- Igaz, L.M.; Kwong, L.K.; Chen-Plotkin, A.; Winton, M.J.; Unger, T.L.; Xu, Y.; Neumann, M.; Trojanowski, J.Q.; Lee, V.M.-Y. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J. Biol. Chem. 2009, 284, 8516–8524. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Xu, Y.-F.; Cook, C.; Gendron, T.F.; Roettges, P.; Link, C.D.; Lin, W.-L.; Tong, J.; Castanedes-Casey, M.; Ash, P.; et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 7607–7612. [Google Scholar] [CrossRef]
- Abbasi, W.A.; Hassan, F.U.; Yaseen, A.; Minhas, F.U.A.A. ISLAND: In-Silico Prediction of Proteins Binding Affinity Using Sequence Descriptors. arXiv 2017, arXiv:1711.10540. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamerlan, A.M.; An, S.S.A. A Microplate-Based Approach to Map Interactions between TDP-43 and α-Synuclein. J. Clin. Med. 2022, 11, 573. https://doi.org/10.3390/jcm11030573
Jamerlan AM, An SSA. A Microplate-Based Approach to Map Interactions between TDP-43 and α-Synuclein. Journal of Clinical Medicine. 2022; 11(3):573. https://doi.org/10.3390/jcm11030573
Chicago/Turabian StyleJamerlan, Angelo M., and Seong Soo A. An. 2022. "A Microplate-Based Approach to Map Interactions between TDP-43 and α-Synuclein" Journal of Clinical Medicine 11, no. 3: 573. https://doi.org/10.3390/jcm11030573
APA StyleJamerlan, A. M., & An, S. S. A. (2022). A Microplate-Based Approach to Map Interactions between TDP-43 and α-Synuclein. Journal of Clinical Medicine, 11(3), 573. https://doi.org/10.3390/jcm11030573






