MicroRNAs miR-451a and Let-7i-5p Profiles in Circulating Exosomes Vary among Individuals with Different Sickle Hemoglobin Genotypes and Malaria
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Laboratory Evaluation of Blood Samples
2.3. Exosomal RNA Extraction
2.4. Real-Time Quantitative PCR (RT-qPCR)
2.5. Statistical Analysis
2.6. Receiver Operating Characteristic (ROC) Curve Analysis
2.7. Bioinformatics
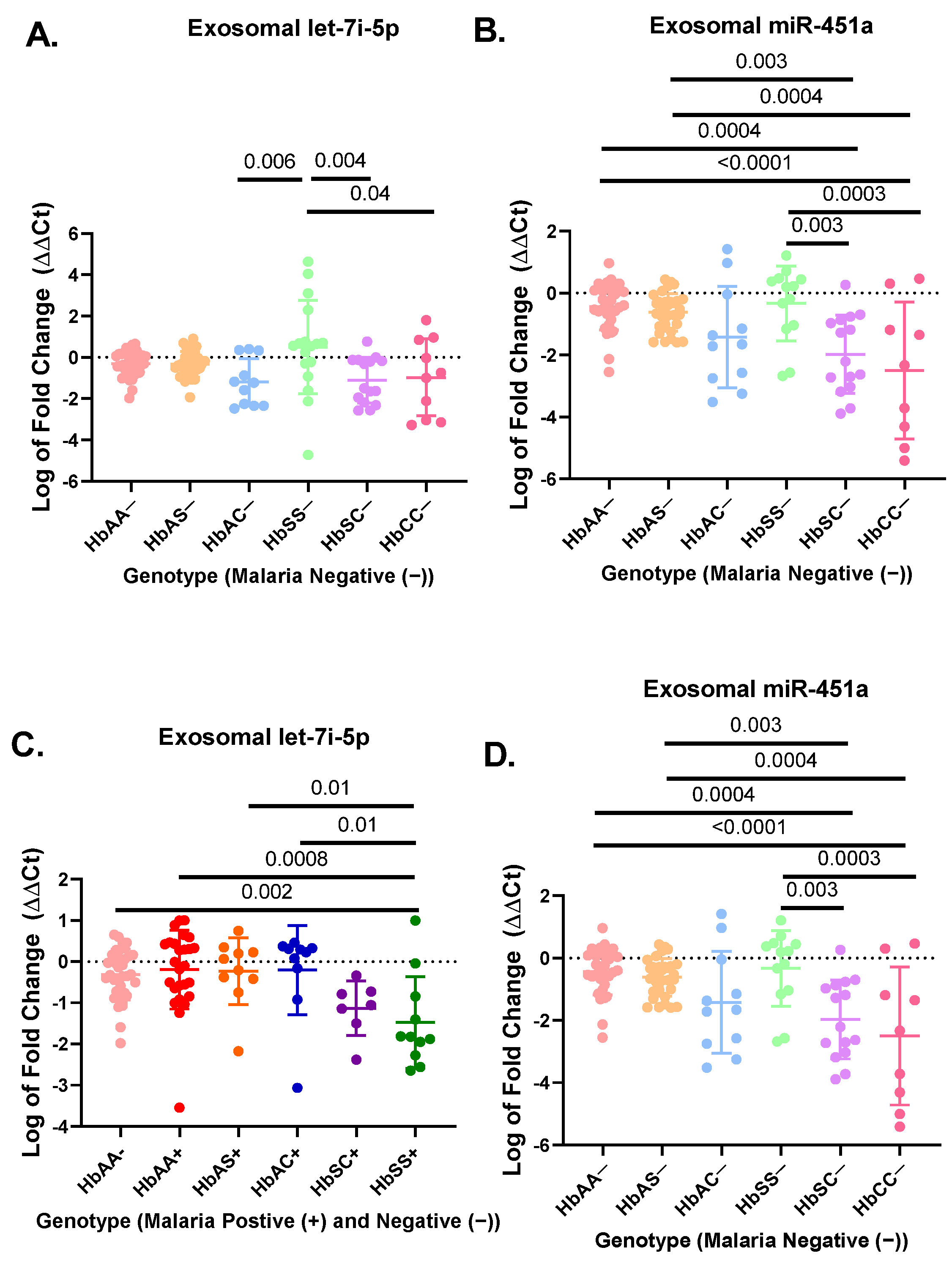
3. Results
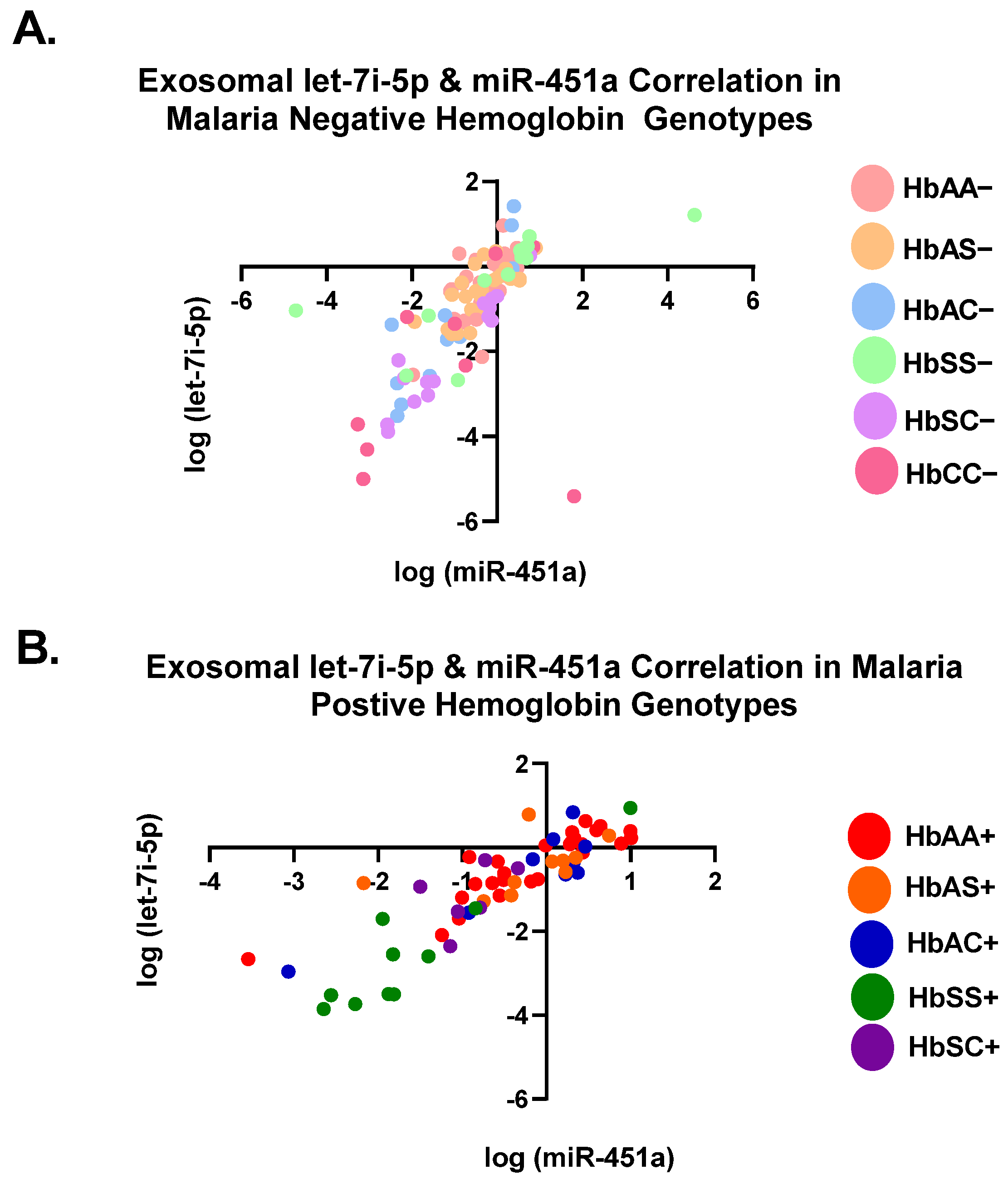
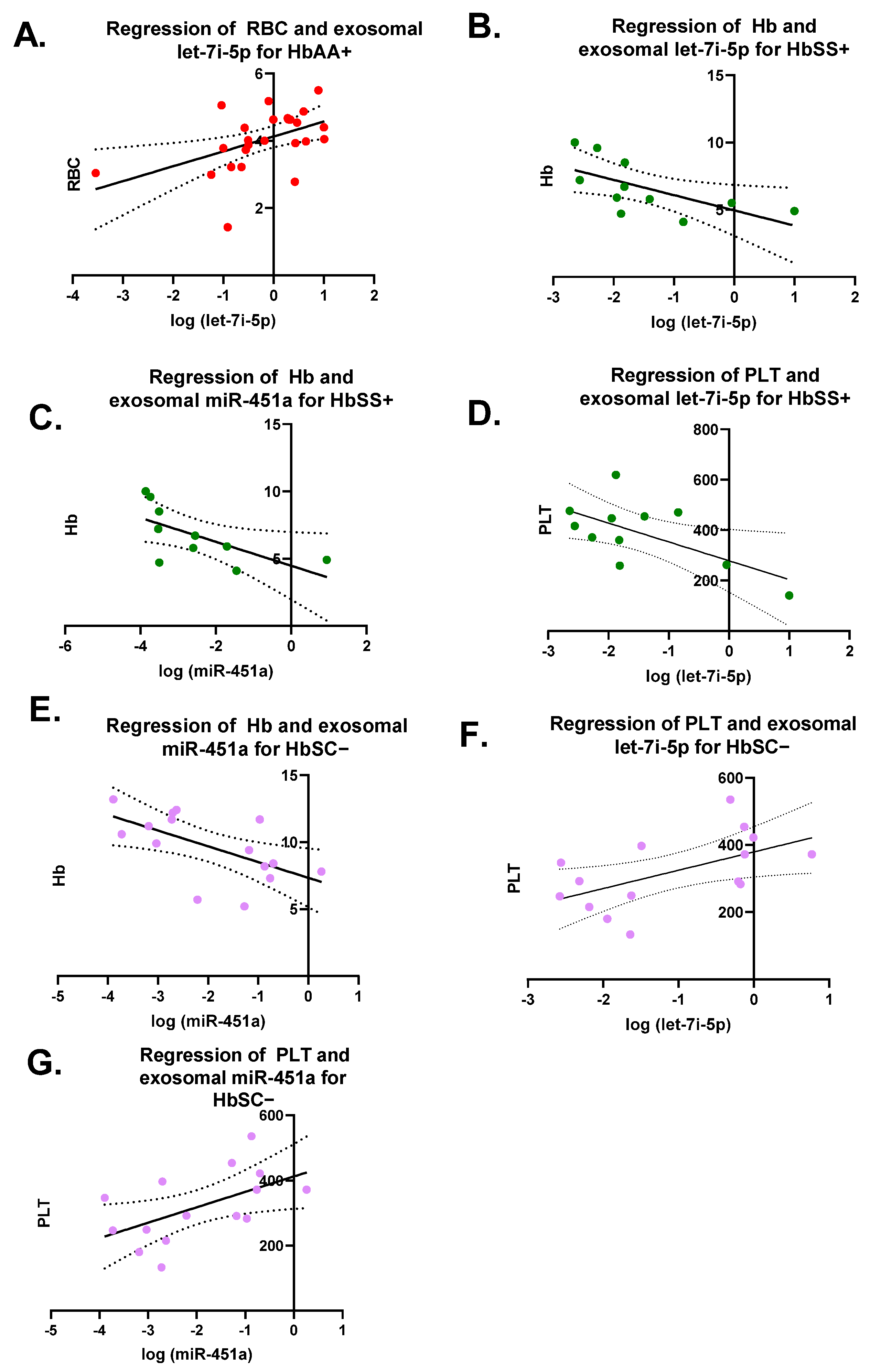
3.1. Let-7i-5p and miR-451a Expression Correlate with Each Other Regardless of Genotype and Malaria Status
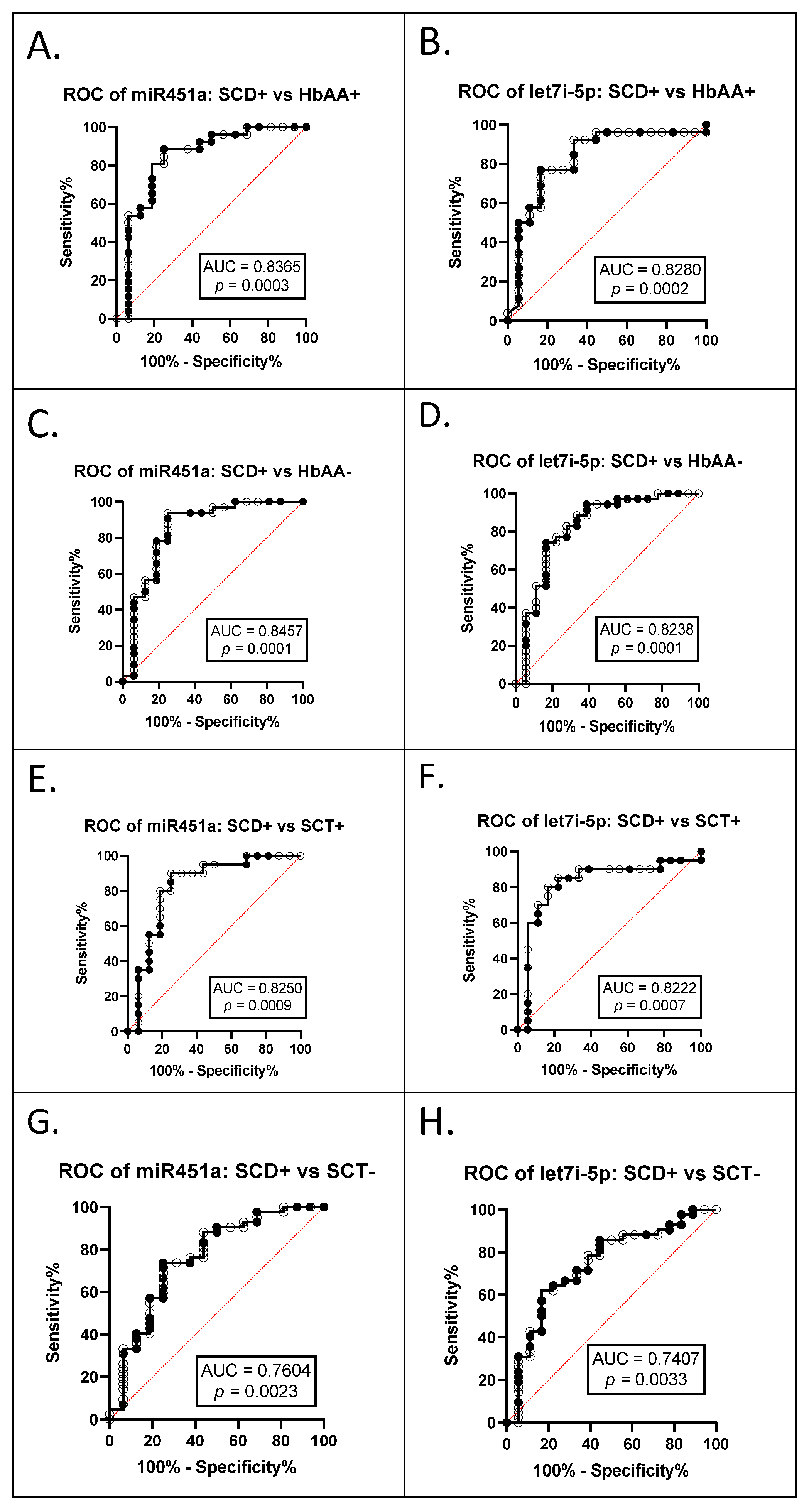
3.2. miR-451a and Let-7i-5p Are Significant Biomarkers for Sickle Cell Anemia and Malaria Status
3.3. Predictions of Let-7i-5p and miR-451a Gene Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Driss, A.; Asare, K.O.; Hibbert, J.M.; Gee, B.E.; Adamkiewicz, T.V.; Stiles, J.K. Sickle Cell Disease in the Post Genomic Era: A Monogenic Disease with a Polygenic Phenotype. Genom. Insights 2009, 2009, 23–48. [Google Scholar] [CrossRef]
- Quinn, C.T.; Rogers, Z.R.; Buchanan, G.R. Survival of children with sickle cell disease. Blood 2004, 103, 4023–4027. [Google Scholar] [CrossRef] [PubMed]
- LaMonte, G.; Philip, N.; Reardon, J.; Lacsina, J.R.; Majoros, W.; Chapman, L.; Thornburg, C.D.; Telen, M.J.; Ohler, U.; Nicchitta, C.V.; et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012, 12, 187–199. [Google Scholar] [CrossRef] [PubMed]
- John, N. A review of clinical profile in sickle cell traits. Oman Med. J. 2010, 25, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Mackinnon, M.J.; Mwangi, T.W.; Snow, R.W.; Marsh, K.; Williams, T.N. Heritability of malaria in Africa. PLoS Med. 2005, 2, e340. [Google Scholar] [CrossRef]
- Driss, A.; Hibbert, J.M.; Wilson, N.O.; Iqbal, S.A.; Adamkiewicz, T.V.; Stiles, J.K. Genetic polymorphisms linked to susceptibility to malaria. Malar. J. 2011, 10, 271. [Google Scholar] [CrossRef]
- WHO. Malaria. Available online: https://www.who.int/news-room/fact-sheets/detail/malaria (accessed on 23 May 2021).
- Sangokoya, C.; Telen, M.J.; Chi, J.T. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 2010, 116, 4338–4348. [Google Scholar] [CrossRef]
- Chen, S.Y.; Wang, Y.; Telen, M.J.; Chi, J.T. The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS ONE 2008, 3, e2360. [Google Scholar] [CrossRef] [PubMed]
- Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68–74. [Google Scholar] [CrossRef]
- Mohr, A.M.; Mott, J.L. Overview of microRNA biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar] [CrossRef]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Gao, Y.; Li, N.; Shao, F.; Wang, C.; Wang, P.; Yang, Z.; Li, R.; He, J. Exosomes: New players in cancer (Review). Oncol. Rep. 2017, 38, 665–675. [Google Scholar] [CrossRef]
- Li, R.; Chibbar, R.; Xiang, J. Novel EXO-T vaccine using polyclonal CD4(+) T cells armed with HER2-specific exosomes for HER2-positive breast cancer. OncoTargets Ther. 2018, 11, 7089–7093. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef]
- Jalalian, S.H.; Ramezani, M.; Jalalian, S.A.; Abnous, K.; Taghdisi, S.M. Exosomes, new biomarkers in early cancer detection. Anal. Biochem. 2019, 571, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.Y. MicroRNAs in Human Diseases: From Lung, Liver and Kidney Diseases to Infectious Disease, Sickle Cell Disease and Endometrium Disease. Immune Netw. 2011, 11, 309–323. [Google Scholar] [CrossRef]
- Cohen, A.; Combes, V.; Grau, G.E. MicroRNAs and Malaria—A Dynamic Interaction Still Incompletely Understood. J. Neuroinfect. Dis. 2015, 6, 165. [Google Scholar]
- Iqbal, S.A.; Botchway, F.; Badu, K.; Wilson, N.O.; Dei-Adomakoh, Y.; Dickinson-Copeland, C.M.; Chinbuah, H.; Adjei, A.A.; Wilson, M.; Stiles, J.K.; et al. Hematological Differences among Malaria Patients in Rural and Urban Ghana. J. Trop. Pediatr. 2016, 62, 477–486. [Google Scholar] [CrossRef][Green Version]
- Driss, A.; Wilson, N.O.; Mason, K.; Hyacinth, H.I.; Hibbert, J.M.; Serjeant, G.R.; Stiles, J.K. Elevated IL-1α and CXCL10 Serum Levels Occur in Patients with Homozygous Sickle Cell Disease and a History of Acute Splenic Sequestration. Dis. Markers 2012, 32, 295–300. [Google Scholar] [CrossRef]
- Harp, D.; Driss, A.; Mehrabi, S.; Chowdhury, I.; Xu, W.; Liu, D.; Garcia-Barrio, M.; Taylor, R.N.; Gold, B.; Jefferson, S.; et al. Exosomes derived from endometriotic stromal cells have enhanced angiogenic effects in vitro. Cell Tissue Res. 2016, 365, 187–196. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 44, 1741–1748. [Google Scholar] [CrossRef]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vergoulis, T.; Vlachos, I.S.; Alexiou, P.; Georgakilas, G.; Maragkakis, M.; Reczko, M.; Gerangelos, S.; Koziris, N.; Dalamagas, T.; Hatzigeorgiou, A.G. TarBase 6.0: Capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012, 40, D222–D229. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Wang, X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics 2016, 32, 1316–1322. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Wolf, S.; Wu, W.; Jones, C.; Perwitasari, O.; Mahalingam, S.; Tripp, R.A. MicroRNA Regulation of Human Genes Essential for Influenza A (H7N9) Replication. PLoS ONE 2016, 11, e0155104. [Google Scholar] [CrossRef]
- Harp, K.O.; Botchway, F.; Dei-Adomakoh, Y.; Wilson, M.D.; Mubasher, M.; Adjei, A.A.; Thompson, W.E.; Stiles, J.K.; Driss, A. Analysis of clinical presentation, hematological factors, self-reported bed net usage, and malaria burden in sickle cell disease patients. EClinicalMedicine 2021, 39, 101045. [Google Scholar] [CrossRef]
- Gupta, H.; Rubio, M.; Sitoe, A.; Varo, R.; Cisteró, P.; Madrid, L.; Cuamba, I.; Jimenez, A.; Martiáñez-Vendrell, X.; Barrios, D.; et al. Plasma MicroRNA Profiling of Plasmodium falciparum Biomass and Association with Severity of Malaria Disease. Emerg. Infect. Dis. 2021, 27, 430–442. [Google Scholar] [CrossRef]
- Shkurnikov, M.Y.; Knyazev, E.N.; Fomicheva, K.A.; Mikhailenko, D.S.; Nyushko, K.M.; Saribekyan, E.K.; Samatov, T.R.; Alekseev, B.Y. Analysis of Plasma microRNA Associated with Hemolysis. Bull. Exp. Biol. Med. 2016, 160, 748–750. [Google Scholar] [CrossRef] [PubMed]
- Ilesanmi, O.O. Pathological basis of symptoms and crises in sickle cell disorder: Implications for counseling and psychotherapy. Hematol. Rep. 2010, 2, e2. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Peng, R.; Peng, H.; Liu, H.; Wen, L.; Wu, T.; Yi, H.; Li, A.; Zhang, Z. miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell. Endocrinol. 2016, 433, 75–86. [Google Scholar] [CrossRef]
- Eyler, C.E.; Jackson, T.; Elliott, L.E.; De Castro, L.M.; Jonassaint, J.; Ashley-Koch, A.; Telen, M.J. beta(2)-Adrenergic receptor and adenylate cyclase gene polymorphisms affect sickle red cell adhesion. Br. J. Haematol. 2008, 141, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Hines, P.C.; Zen, Q.; Burney, S.N.; Shea, D.A.; Ataga, K.I.; Orringer, E.P.; Telen, M.J.; Parise, L.V. Novel epinephrine and cyclic AMP-mediated activation of BCAM/Lu-dependent sickle (SS) RBC adhesion. Blood 2003, 101, 3281–3287. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, C.; Klitz, W.; Cheng, S.; Apple, R.; Steiner, L.; Robles, L.; Girard, T.; Vichinsky, E.; Styles, L. Gene interactions and stroke risk in children with sickle cell anemia. Blood 2004, 103, 2391–2396. [Google Scholar] [CrossRef] [PubMed]
- Gladek, I.; Ferdin, J.; Horvat, S.; Calin, G.A.; Kunej, T. HIF1A gene polymorphisms and human diseases: Graphical review of 97 association studies. Genes Chromosomes Cancer 2017, 56, 439–452. [Google Scholar] [CrossRef] [PubMed]
- MacKinney, A.; Woska, E.; Spasojevic, I.; Batinic-Haberle, I.; Zennadi, R. Disrupting the vicious cycle created by NOX activation in sickle erythrocytes exposed to hypoxia/reoxygenation prevents adhesion and vasoocclusion. Redox Biol. 2019, 101097. [Google Scholar] [CrossRef]
- Payen, E.; Leboulch, P. Advances in stem cell transplantation and gene therapy in the beta-hemoglobinopathies. Hematol. Am. Soc. Hematol. Educ. Program. 2012, 2012, 276–283. [Google Scholar] [CrossRef]
- Ecsedi, M.; Grosshans, H. LIN-41/TRIM71: Emancipation of a miRNA target. Genes Dev. 2013, 27, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; de Vasconcellos, J.F.; Yuan, J.; Byrnes, C.; Noh, S.J.; Meier, E.R.; Kim, K.S.; Rabel, A.; Kaushal, M.; Muljo, S.A.; et al. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood 2013, 122, 1034–1041. [Google Scholar] [CrossRef] [PubMed]
- Fathallah, H.; Atweh, G.F. Induction of fetal hemoglobin in the treatment of sickle cell disease. Hematol. Am. Soc. Hematol. Educ. Program. 2006, 58–62. [Google Scholar] [CrossRef]
- Kahle, K.T.; Rinehart, J.; Lifton, R.P. Phosphoregulation of the Na-K-2Cl and K-Cl cotransporters by the WNK kinases. Biochim. Biophys. Acta 2010, 1802, 1150–1158. [Google Scholar] [CrossRef]
- Luo, N.; Li, G.; Li, Y.; Fan, X.; Wang, Y.; Ye, X.; Mo, X.; Zhou, J.; Yuan, W.; Tan, M.; et al. SAMD4B, a novel SAM-containing protein, inhibits AP-1-, p53- and p21-mediated transcriptional activity. BMB Rep. 2010, 43, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Alawadhi, A.; Saint-Martin, C.; Bhanji, F.; Srour, M.; Atkinson, J.; Sebire, G. Acute Hemorrhagic Encephalitis Responding to Combined Decompressive Craniectomy, Intravenous Immunoglobulin, and Corticosteroid Therapies: Association with Novel RANBP2 Variant. Front. Neurol. 2018, 9, 130. [Google Scholar] [CrossRef]
- Li, S.M.; Wu, H.L.; Yu, X.; Tang, K.; Wang, S.G.; Ye, Z.Q.; Hu, J. The putative tumour suppressor miR-1-3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J. Exp. Clin. Cancer Res. 2018, 37, 219. [Google Scholar] [CrossRef] [PubMed]
- Xing, D.; Suryo Rahmanto, Y.; Zeppernick, F.; Hannibal, C.G.; Kjaer, S.K.; Vang, R.; Shih, I.M.; Wang, T.L. Mutation of NRAS is a rare genetic event in ovarian low-grade serous carcinoma. Hum. Pathol. 2017, 68, 87–91. [Google Scholar] [CrossRef]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef]
- Ro, M.; Park, J.; Nam, M.; Bang, H.J.; Yang, J.W.; Choi, K.S.; Kim, S.K.; Chung, J.H.; Kwack, K. Associations between single-nucleotide polymorphism in the FNDC3A and autism spectrum disorder in a Korean population. Psychiatry Res. 2013, 209, 246–248. [Google Scholar] [CrossRef]
- Haupt, S.; Hernandez, O.M.; Vijayakumaran, R.; Keam, S.; Haupt, Y. The long and the short of it: The MDM4 tail so far. J. Mol. Cell Biol. 2019, 11, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Akinlade, K.S.; Kumuyi, A.S.; Rahamon, S.K.; Olaniyi, J.A. Insulin Sensitivity, Inflammation, and Basal Metabolic Rate in Adults with Sickle Cell Anemia. Int. J. Appl. Basic Med. Res. 2018, 8, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Smiley, D.; Dagogo-Jack, S.; Umpierrez, G. Therapy insight: Metabolic and endocrine disorders in sickle cell disease. Nat. Clin. Practice. Endocrinol. Metab. 2008, 4, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; dos Santos, C.O.; Zhao, G.; Jiang, J.; Amigo, J.D.; Khandros, E.; Dore, L.C.; Yao, Y.; D’Souza, J.; Zhang, Z.; et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010, 24, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Tokumaru, S.; Suzuki, M.; Yamada, H.; Nagino, M.; Takahashi, T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 2008, 29, 2073–2077. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.J.; Miller, S.H.; Lee, Y.T.; Goh, S.H.; Marincola, F.M.; Stroncek, D.F.; Reed, C.; Wang, E.; Miller, J.L. Let-7 microRNAs are developmentally regulated in circulating human erythroid cells. J. Transl. Med. 2009, 7, 98. [Google Scholar] [CrossRef]
- Hou, W.; Tian, Q.; Steuerwald, N.M.; Schrum, L.W.; Bonkovsky, H.L. The let-7 microRNA enhances heme oxygenase-1 by suppressing Bach1 and attenuates oxidant injury in human hepatocytes. Biochim. Biophys. Acta 2012, 1819, 1113–1122. [Google Scholar] [CrossRef]
- Dossing, K.B.; Binderup, T.; Kaczkowski, B.; Jacobsen, A.; Rossing, M.; Winther, O.; Federspiel, B.; Knigge, U.; Kjaer, A.; Friis-Hansen, L. Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1. Genes 2014, 6, 1–21. [Google Scholar] [CrossRef]
- Pamplona, A.; Hanscheid, T.; Epiphanio, S.; Mota, M.M.; Vigario, A.M. Cerebral malaria and the hemolysis/methemoglobin/heme hypothesis: Shedding new light on an old disease. Int. J. Biochem. Cell. Biol. 2009, 41, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Pamplona, A.; Ferreira, A.; Balla, J.; Jeney, V.; Balla, G.; Epiphanio, S.; Chora, A.; Rodrigues, C.D.; Gregoire, I.P.; Cunha-Rodrigues, M.; et al. Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria. Nat. Med. 2007, 13, 703–710. [Google Scholar] [CrossRef]
- Hunt, N.H.; Stocker, R. Heme moves to center stage in cerebral malaria. Nat. Med. 2007, 13, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Epiphanio, S.; Campos, M.G.; Pamplona, A.; Carapau, D.; Pena, A.C.; Ataide, R.; Monteiro, C.A.; Felix, N.; Costa-Silva, A.; Marinho, C.R.; et al. VEGF promotes malaria-associated acute lung injury in mice. PLoS Pathog. 2010, 6, e1000916. [Google Scholar] [CrossRef]
- Geuken, E.; Buis, C.I.; Visser, D.S.; Blokzijl, H.; Moshage, H.; Nemes, B.; Leuvenink, H.G.; de Jong, K.P.; Peeters, P.M.; Slooff, M.J.; et al. Expression of heme oxygenase-1 in human livers before transplantation correlates with graft injury and function after transplantation. Am. J. Transplant. 2005, 5, 1875–1885. [Google Scholar] [CrossRef]
- Datta, D.; Banerjee, P.; Gasser, M.; Waaga-Gasser, A.M.; Pal, S. CXCR3-B can mediate growth-inhibitory signals in human renal cancer cells by down-regulating the expression of heme oxygenase-1. J. Biol. Chem. 2010, 285, 36842–36848. [Google Scholar] [CrossRef]
- Pae, H.O.; Oh, G.S.; Choi, B.M.; Kim, Y.M.; Chung, H.T. A molecular cascade showing nitric oxide-heme oxygenase-1-vascular endothelial growth factor-interleukin-8 sequence in human endothelial cells. Endocrinology 2005, 146, 2229–2238. [Google Scholar] [CrossRef]
- Bussolati, B.; Mason, J.C. Dual role of VEGF-induced heme-oxygenase-1 in angiogenesis. Antioxid. Redox Signal. 2006, 8, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; Dormond, O.; Basu, A.; Briscoe, D.M.; Pal, S. Heme oxygenase-1 modulates the expression of the anti-angiogenic chemokine CXCL-10 in renal tubular epithelial cells. Am. J. Physiol. Renal. Physiol. 2007, 293, F1222–F1230. [Google Scholar] [CrossRef][Green Version]
- Williams, T.N. Human red blood cell polymorphisms and malaria. Curr. Opin. Microbiol. 2006, 9, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Reiter, C.D.; Wang, X.; Tanus-Santos, J.E.; Hogg, N.; Cannon, R.O., 3rd; Schechter, A.N.; Gladwin, M.T. Cell-free hemoglobin limits nitric oxide bioavailability in sickle-cell disease. Nat. Med. 2002, 8, 1383–1389. [Google Scholar] [CrossRef]
- Muller-Eberhard, U.; Javid, J.; Liem, H.H.; Hanstein, A.; Hanna, M. Plasma concentrations of hemopexin, haptoglobin and heme in patients with various hemolytic diseases. Blood 1968, 32, 4. [Google Scholar] [CrossRef]
- Hebbel, R.P. Perspectives series: Cell adhesion in vascular biology. Adhesive interactions of sickle erythrocytes with endothelium. J. Clin. Investig. 1997, 99, 2561–2564. [Google Scholar] [CrossRef] [PubMed]
- Amin, B.R.; Bauersachs, R.M.; Meiselman, H.J.; Mohandas, N.; Hebbel, R.P.; Bowen, P.E.; Schlegel, R.A.; Williamson, P.; Westerman, M.P. Monozygotic twins with sickle cell anemia and discordant clinical courses: Clinical and laboratory studies. Hemoglobin 1991, 15, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Marguti, I.; Bechmann, I.; Jeney, V.; Chora, A.; Palha, N.R.; Rebelo, S.; Henri, A.; Beuzard, Y.; Soares, M.P. Sickle hemoglobin confers tolerance to Plasmodium infection. Cell 2011, 145, 398–409. [Google Scholar] [CrossRef]
- Hebbel, R.P.; Morgan, W.T.; Eaton, J.W.; Hedlund, B.E. Accelerated autoxidation and heme loss due to instability of sickle hemoglobin. Proc. Natl. Acad. Sci. USA 1988, 85, 237–241. [Google Scholar] [CrossRef]
- Seixas, E.; Gozzelino, R.; Chora, A.; Ferreira, A.; Silva, G.; Larsen, R.; Rebelo, S.; Penido, C.; Smith, N.R.; Coutinho, A.; et al. Heme oxygenase-1 affords protection against noncerebral forms of severe malaria. Proc. Natl. Acad. Sci. USA 2009, 106, 15837–15842. [Google Scholar] [CrossRef]
- Gozzelino, R.; Jeney, V.; Soares, M.P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 323–354. [Google Scholar] [CrossRef]
- Balla, J.; Jacob, H.S.; Balla, G.; Nath, K.; Vercellotti, G.M. Endothelial cell heme oxygenase and ferritin induction by heme proteins: A possible mechanism limiting shock damage. Trans. Assoc. Am. Physicians 1992, 105, 1–6. [Google Scholar]
- Harp, K.O.; Botchway, F.; Dei-Adomakoh, Y.; Wilson, M.D.; Hood, J.L.; Adjei, A.A.; Stiles, J.K.; Driss, A. Hemoglobin Genotypes Modulate Inflammatory Response to Plasmodium Infection. Front. Immunol. 2020, 11, 593546. [Google Scholar] [CrossRef]
- Komba, A.N.; Makani, J.; Sadarangani, M.; Ajala-Agbo, T.; Berkley, J.A.; Newton, C.R.; Marsh, K.; Williams, T.N. Malaria as a cause of morbidity and mortality in children with homozygous sickle cell disease on the coast of Kenya. Clin. Infect. Dis. 2009, 49, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Albiti, A.H.; Nsiah, K. Comparative haematological parameters of HbAA and HbAS genotype children infected with Plasmodium falciparum malaria in Yemen. Hematology 2014, 19, 169–174. [Google Scholar] [CrossRef] [PubMed]
| Characteristics (Mean ± SD) | HbAA− n = 35 | HbAS− n = 31 | HbAC− n = 11 | HbSS− n = 17 | HbSC− n = 15 | HbCC− n = 10 | HbAA+ n = 26 | HbAS+ n = 10 | HbAC+ n = 10 | HbSS+ n = 11 | HbSC+ n = 7 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 37.9 ± 15 | 38.2 ± 13.1 | 31.5 ± 11.3 | 26.3 ± 9.4 | 32.9 ± 11 | 34 ± 9.2 | 33.1 ± 20.7 | 22 ± 22.4 | 22.5 ± 12.8 | 22.5 ± 10.4 | 38.4 ± 18.9 |
| Sex (male) | 17 | 7 | 4 | 8 | 6 | 5 | 13 | 3 | 3 | 6 | 3 |
| WBC (×103/mm3) | 6.4 ± 3.5 | 5.6 ± 1.7 | 6 ± 1.7 | 12.11 ± 4.5 | 10 ± 4.2 | 8 ± 1.8 | 6.2 ± 3.5 | 5.6 ± 2.4 | 6.2 ± 2 | 14.1 ± 4.1 | 10.2 ± 9.6 |
| RBC (×106/µL) | 4.4 ± 0.6 | 4.2 ± 0.6 | 4.7 ± 1.2 | 3.3 ± 0.9 | 3.7 ± 1.1 | 4.5 ± 0.5 | 4 ± 0.2 | 4.3 ± 0.7 | 4.3 ± 0.6 | 2.5 ± 1 | 3.5 ± 1.5 |
| Hemoglobin (g/dL) | 12.1 ± 2.6 | 11.9 ± 1.5 | 12.3 ± 1.5 | 8.1 ± 1.1 | 9.6 ± 2.5 | 11.8 ± 1.6 | 11 ± 2.7 | 11.3 ± 2.1 | 11.5 ± 1.3 | 6.6 ± 2 | 8.6 ± 3.7 |
| Platelets (×103/µL) | 266.3 ± 95.5 | 238.8 ± 69.3 | 280.1 ± 121.3 | 435.8 ± 120.4 | 319.3 ± 108.8 | 209.5 ± 63 | 149 ± 89.7 | 190.6 ± 115.4 | 167.2 ± 39.7 | 388.5 ± 131 | 279.9 ± 139.4 |
| Genotype | R2 | p-Value |
|---|---|---|
| HbAA−n = 35 | 0.44 | <0.0001 |
| HbAS−n = 31 | 0.53 | <0.0001 |
| HbAC−n = 11 | 0.78 | 0.0003 |
| HbSS−n = 17 | 0.49 | 0.007 |
| HbSC−n = 15 | 0.9 | <0.0001 |
| HbCC−n = 10 | 0.1 | 0.4 |
| HbAA+n = 26 | 0.8 | <0.0001 |
| HbAS+n = 10 | 0.24 | 0.15 |
| HbAC+n = 10 | 0.8 | 0.0005 |
| HbSS+n = 11 | 0.86 | 0.0001 |
| HbSC+n = 7 | 0.21 | 0.36 |
| All Malaria Negativen = 119 | 0.45 | <0.0001 |
| All Malaria Positiven = 64 | 0.76 | <0.0001 |
| All Groupsn = 183 | 0.54 | <0.0001 |
| miR-451a | Let-7i-5p | |||
|---|---|---|---|---|
| Groups | AUC | p-Value | AUC | p-Value |
| SCA− vs. SCA+ | 0.6586 | 0.0832 | 0.6927 | 0.0249 |
| SCA− vs. HbAA− | 0.6395 | 0.0640 | 0.5397 | 0.6059 |
| SCA− vs. HbAA+ | 0.6490 | 0.0603 | 0.5152 | 0.8310 |
| SCA− vs. SCT+ | 0.6268 | 0.1376 | 0.5281 | 0.7349 |
| SCA− vs. SCT− | 0.5531 | 0.4537 | 0.5595 | 0.3827 |
| SCA− vs. HbCC− | 0.6825 | 0.1034 | 0.5781 | 0.3470 |
| SCA+ vs. HbAA− | 0.8457 | 0.0001 | 0.8238 | 0.0002 |
| SCA+ vs. HbAA+ | 0.8365 | 0.0003 | 0.8280 | 0.0001 |
| SCA+ vs. SCT+ | 0.8250 | 0.0009 | 0.8222 | 0.0007 |
| SCA+ vs. SCT − | 0.7604 | 0.0023 | 0.7407 | 0.0033 |
| SCA+ vs. HbCC− | 0.5556 | 0.6506 | 0.5500 | 0.5987 |
| HbAA− vs. HbAA+ | 0.5282 | 0.7133 | 0.5747 | 0.3213 |
| HbAA− vs. SCT+ | 0.5414 | 0.6482 | 0.5077 | 0.9294 |
| HbAA− vs. SCT− | 0.6328 | 0.0515 | 0.6241 | 0.0873 |
| HbAA− vs. HbCC− | 0.7639 | 0.0167 | 0.6154 | 0.1837 |
| HbAA+ vs. SCT+ | 0.5529 | 0.5423 | 0.6164 | 0.1539 |
| HbAA+ vs. SCT− | 0.6351 | 0.0627 | 0.5724 | 0.2759 |
| HbAA+ vs. HbCC− | 0.7735 | 0.0157 | 0.5971 | 0.2341 |
| SCT+ vs. SCT− | 0.5982 | 0.2141 | 0.6452 | 0.0662 |
| SCT+ vs. HbCC− | 0.7667 | 0.2370 | 0.6150 | 0.2134 |
| SCT− vs. HbCC− | 0.7143 | 0.0454 | 0.5571 | 0.4698 |
| miRNA Species | Target Gene Name | Abbreviation | Entrez Gene ID | Function | Ref. |
|---|---|---|---|---|---|
| miR-451a | Odd-Skipped Related Transcription Factor 1 | OSR1 | 130497 | Key component in regulation of intracellular concentration of chloride which is required for cell volume regulation. | [47] |
| Sterile Alpha Motif Domain- Containing 4B | SAMD4B | 55095 | Regulator of transcriptional signaling activity. | [48] | |
| Protease Subunit Beta 8 | PSMB8 | 5696 | Inhibits NFҡB when miR-451a levels are raised. Also known as LMP7. | [37] | |
| Let-7i-5p | Adrenoceptor Beta 2 | ADRB2 | 154 | Can increase the adhesion of HbSS RBCS | [38,39,40] |
| Hypoxia Inducible Factor 1 Subunit Alpha Inhibitor | HIF1AN | 55662 | Helps maintain cell viability during oxygen deprivation | [41,42] | |
| RAN Binding Protein 2 | RANBP2 | 5903 | Is a nuclear pore protein that is involved in the cell cycle | [49] | |
| E2F Transcription Factor 5 | E2F5 | 1875 | Has an important role in cell cycle and tumor suppression | [50] | |
| NRAS Proto-Oncogene, GTPase | NRAS | 4893 | Involved in the RAS signaling pathway | [51] | |
| Lysine methyltransferase 2D | KMT2D | 8085 | Involved in multiple functions like differentiation & metabolism | [52] | |
| Fibronectin Type III Domain Containing 3A | FNDC3A | 22862 | Encodes for Fibronectin module type III (FN3) which mediates protein-protein interactions | [53] | |
| High Mobility Group AT-Hook 2 | HMGA2 | 8091 | A report of a viral vector integrating with an intragenic site in a SCD patient and the SCD patient became transfusion independent | [43] | |
| MDM4, P53 Regulator | MDM4 | 4194 | Regulates tumor suppressor p53 | [54] | |
| Tripartite motif Containing 71 | TRIM71 | 131405 | Is an ortholog of Lineage Variant 41 (LIN-41). Lin-41regulates fetal hemoglobin (HbF) via let-7 miRNAs | [44] | |
| Lin-28 Homolog B | LIN28B | 389421 | Regulates HbF via let-7 miRNAs | [45,46] | |
| Insulin Like Growth Factor Binding Protein 1 | IGFBP1 | 3484 | Associated with metabolism | [55,56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oxendine Harp, K.; Bashi, A.; Botchway, F.; Dei-Adomakoh, Y.; Iqbal, S.A.; Wilson, M.D.; Adjei, A.A.; Stiles, J.K.; Driss, A. MicroRNAs miR-451a and Let-7i-5p Profiles in Circulating Exosomes Vary among Individuals with Different Sickle Hemoglobin Genotypes and Malaria. J. Clin. Med. 2022, 11, 500. https://doi.org/10.3390/jcm11030500
Oxendine Harp K, Bashi A, Botchway F, Dei-Adomakoh Y, Iqbal SA, Wilson MD, Adjei AA, Stiles JK, Driss A. MicroRNAs miR-451a and Let-7i-5p Profiles in Circulating Exosomes Vary among Individuals with Different Sickle Hemoglobin Genotypes and Malaria. Journal of Clinical Medicine. 2022; 11(3):500. https://doi.org/10.3390/jcm11030500
Chicago/Turabian StyleOxendine Harp, Keri, Alaijah Bashi, Felix Botchway, Yvonne Dei-Adomakoh, Shareen A. Iqbal, Michael D. Wilson, Andrew A. Adjei, Jonathan K. Stiles, and Adel Driss. 2022. "MicroRNAs miR-451a and Let-7i-5p Profiles in Circulating Exosomes Vary among Individuals with Different Sickle Hemoglobin Genotypes and Malaria" Journal of Clinical Medicine 11, no. 3: 500. https://doi.org/10.3390/jcm11030500
APA StyleOxendine Harp, K., Bashi, A., Botchway, F., Dei-Adomakoh, Y., Iqbal, S. A., Wilson, M. D., Adjei, A. A., Stiles, J. K., & Driss, A. (2022). MicroRNAs miR-451a and Let-7i-5p Profiles in Circulating Exosomes Vary among Individuals with Different Sickle Hemoglobin Genotypes and Malaria. Journal of Clinical Medicine, 11(3), 500. https://doi.org/10.3390/jcm11030500







