Global Transcriptional Profiling of Granulosa Cells from Polycystic Ovary Syndrome Patients: Comparative Analyses of Patients with or without History of Ovarian Hyperstimulation Syndrome Reveals Distinct Biomarkers and Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. RNA Isolation
2.3. Genome-Wide Gene Expression Profiling
2.4. Functional, Pathway and Network Analyses
2.5. Quantitative RT-PCR (qRT-PCR)
3. Results
3.1. Clinical Characteristics of the Patients
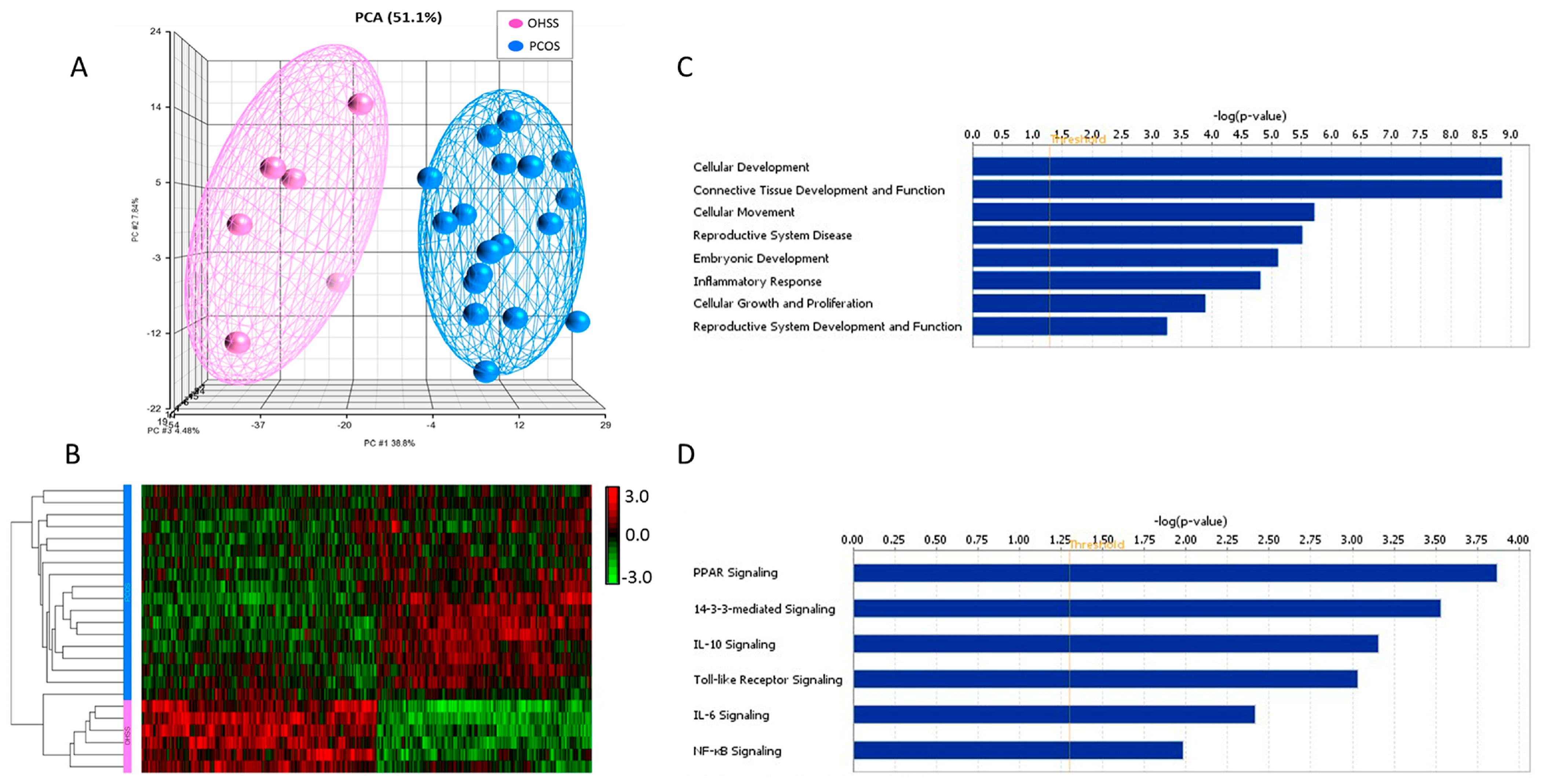
3.2. Identification of Differentially Expressed Genes
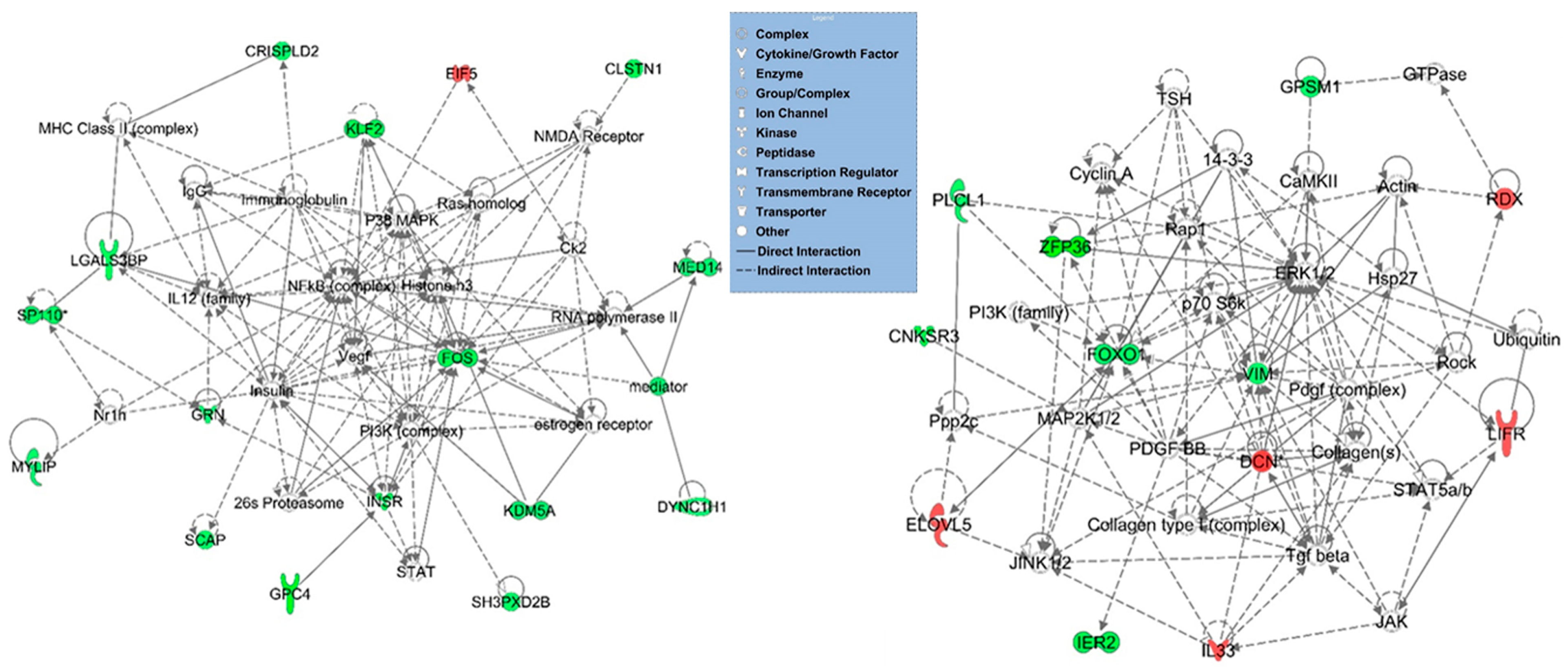
3.3. Functional, Pathway and Gene Network Analysis of Dysregulated Genes
3.4. Confirmation of Gene Expression Using qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Sait, S.F.; Sharma, A.; Kumar, M. Ovarian hyperstimulation syndrome. J. Hum. Reprod. Sci. 2011, 4, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Practice Committee of American Society for Reproductive Medicine. Ovarian hyperstimulation syndrome. Fertil. Steril. 2008, 90, S188–S193. [Google Scholar] [CrossRef] [PubMed]
- Namavar Jahromi, B.M.; Parsanezhad, M.M.; Shomali, Z.M.; Bakhshai, P.M.; Alborzi, M.M.; Moin Vaziri, N.M.D.P.; Anvar, Z.P. Ovarian Hyperstimulation Syndrome: A Narrative Review of Its Pathophysiology, Risk Factors, Prevention, Classification, and Management. Iran. J. Med. Sci. 2018, 43, 248–260. [Google Scholar] [PubMed]
- Sun, B.; Ma, Y.; Li, L.; Hu, L.; Wang, F.; Zhang, Y.; Dai, S.; Sun, Y. Factors Associated with Ovarian Hyperstimulation Syndrome (OHSS) Severity in Women With Polycystic Ovary Syndrome Undergoing IVF/ICSI. Front. Endocrinol. 2020, 11, 615957. [Google Scholar] [CrossRef]
- Soave, I.; Marci, R. Ovarian stimulation in patients in risk of OHSS. Minerva Ginecol. 2014, 66, 165–178. [Google Scholar]
- Lan, C.W.; Chen, M.J.; Tai, K.Y.; Yu, D.C.; Yang, Y.C.; Jan, P.S.; Yang, Y.S.; Chen, H.F.; Ho, H.N. Functional microarray analysis of differentially expressed genes in granulosa cells from women with polycystic ovary syndrome related to MAPK/ERK signaling. Sci. Rep. 2015, 5, 14994. [Google Scholar] [CrossRef]
- Brinca, A.T.; Ramalhinho, A.C.; Sousa, A.; Oliani, A.H.; Breitenfeld, L.; Passarinha, L.A.; Gallardo, E. Follicular Fluid: A Powerful Tool for the Understanding and Diagnosis of Polycystic Ovary Syndrome. Biomedicines 2022, 10, 1254. [Google Scholar] [CrossRef]
- Al-Harazi, O.; Kaya, I.H.; Al-Eid, M.; Alfantoukh, L.; Al Zahrani, A.S.; Al Sebayel, M.; Kaya, N.; Colak, D. Identification of Gene Signature as Diagnostic and Prognostic Blood Biomarker for Early Hepatocellular Carcinoma Using Integrated Cross-Species Transcriptomic and Network Analyses. Front. Genet. 2021, 12, 710049. [Google Scholar] [CrossRef]
- Al-Harazi, O.; Kaya, I.H.; El Allali, A.; Colak, D. A Network-Based Methodology to Identify Subnetwork Markers for Diagnosis and Prognosis of Colorectal Cancer. Front. Genet. 2021, 12, 721949. [Google Scholar] [CrossRef]
- Coskun, S.; Otu, H.H.; Awartani, K.A.; Al-Alwan, L.A.; Al-Hassan, S.; Al-Mayman, H.; Kaya, N.; Inan, M.S. Gene expression profiling of granulosa cells from PCOS patients following varying doses of human chorionic gonadotropin. J. Assist. Reprod. Genet. 2013, 30, 341–352. [Google Scholar] [CrossRef][Green Version]
- Scarfo, G.; Daniele, S.; Fusi, J.; Gesi, M.; Martini, C.; Franzoni, F.; Cela, V.; Artini, P.G. Metabolic and Molecular Mechanisms of Diet and Physical Exercise in the Management of Polycystic Ovarian Syndrome. Biomedicines 2022, 10, 1305. [Google Scholar] [CrossRef]
- Wu, L.; Sun, Y.; Wan, J.; Luan, T.; Cheng, Q.; Tan, Y. A proteomic analysis identifies candidate early biomarkers to predict ovarian hyperstimulation syndrome in polycystic ovarian syndrome patients. Mol. Med. Rep. 2017, 16, 272–280. [Google Scholar] [CrossRef][Green Version]
- Morgante, G.; Darino, I.; Spano, A.; Luisi, S.; Luddi, A.; Piomboni, P.; Governini, L.; De Leo, V. PCOS Physiopathology and Vitamin D Deficiency: Biological Insights and Perspectives for Treatment. J. Clin. Med. 2022, 11, 4509. [Google Scholar] [CrossRef]
- The Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil. Steril. 2004, 81, 19–25. [Google Scholar] [CrossRef]
- Aldosary, M.; Al-Bakheet, A.; Al-Dhalaan, H.; Almass, R.; Alsagob, M.; Al-Younes, B.; AlQuait, L.; Mustafa, O.M.; Bulbul, M.; Rahbeeni, Z.; et al. Rett Syndrome, a Neurodevelopmental Disorder, Whole-Transcriptome, and Mitochondrial Genome Multiomics Analyses Identify Novel Variations and Disease Pathways. OMICS 2020, 24, 160–171. [Google Scholar] [CrossRef]
- Wu, Z.; Irizarry, R.A. Preprocessing of oligonucleotide array data. Nat. Biotechnol. 2004, 22, 656–658. [Google Scholar] [CrossRef]
- Wu, Z.; Irizarry, R.A. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J. Comput. Biol. 2005, 12, 882–893. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D. Real-time quantitative PCR. Methods 2001, 25, 383–385. [Google Scholar] [CrossRef]
- Lee, N.H.; Saeed, A.I. Microarrays: An overview. Methods Mol. Biol. 2007, 353, 265–300. [Google Scholar] [CrossRef] [PubMed]
- Al-Harazi, O.; El Allali, A.; Colak, D. Biomolecular Databases and Subnetwork Identification Approaches of Interest to Big Data Community: An Expert Review. OMICS 2019, 23, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Colak, D.; Al-Harazi, O.; Mustafa, O.M.; Meng, F.; Assiri, A.M.; Dhar, D.K.; Broering, D.C. RNA-Seq transcriptome profiling in three liver regeneration models in rats: Comparative analysis of partial hepatectomy, ALLPS, and PVL. Sci. Rep. 2020, 10, 5213. [Google Scholar] [CrossRef] [PubMed]
- Nyegaard, M.; Overgaard, M.T.; Su, Y.Q.; Hamilton, A.E.; Kwintkiewicz, J.; Hsieh, M.; Nayak, N.R.; Conti, M.; Conover, C.A.; Giudice, L.C. Lack of functional pregnancy-associated plasma protein-A (PAPPA) compromises mouse ovarian steroidogenesis and female fertility. Biol. Reprod. 2010, 82, 1129–1138. [Google Scholar] [CrossRef]
- Ramos, S.B.; Stumpo, D.J.; Kennington, E.A.; Phillips, R.S.; Bock, C.B.; Ribeiro-Neto, F.; Blackshear, P.J. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 2004, 131, 4883–4893. [Google Scholar] [CrossRef]
- Rizk, B.; Aboulghar, M.; Smitz, J.; Ron-El, R. The role of vascular endothelial growth factor and interleukins in the pathogenesis of severe ovarian hyperstimulation syndrome. Hum. Reprod. Update 1997, 3, 255–266. [Google Scholar] [CrossRef]
- Tuteja, G.; Kaestner, K.H. Forkhead transcription factors II. Cell 2007, 131, 192. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Z.; Cao, J.; Chen, Y.; Dong, Y. A novel and compact review on the role of oxidative stress in female reproduction. Reprod. Biol. Endocrinol. 2018, 16, 80. [Google Scholar] [CrossRef]
- Shao, X.; Wei, X. FOXP1 enhances fibrosis via activating Wnt/beta-catenin signaling pathway in endometriosis. Am. J. Transl. Res. 2018, 10, 3610–3618. [Google Scholar]
- van der Vos, K.E.; Coffer, P.J. FOXO-binding partners: It takes two to tango. Oncogene 2008, 27, 2289–2299. [Google Scholar] [CrossRef]
- van der Vos, K.E.; Coffer, P.J. The extending network of FOXO transcriptional target genes. Antioxid. Redox Signal. 2011, 14, 579–592. [Google Scholar] [CrossRef]
- Fan, H.Y.; O’Connor, A.; Shitanaka, M.; Shimada, M.; Liu, Z.; Richards, J.S. Beta-catenin (CTNNB1) promotes preovulatory follicular development but represses LH-mediated ovulation and luteinization. Mol. Endocrinol. 2010, 24, 1529–1542. [Google Scholar] [CrossRef]
- Kohan-Ghadr, H.R.; Smith, L.C.; Arnold, D.R.; Murphy, B.D.; Lefebvre, R.C. Aberrant expression of E-cadherin and beta-catenin proteins in placenta of bovine embryos derived from somatic cell nuclear transfer. Reprod. Fertil. Dev. 2012, 24, 588–598. [Google Scholar] [CrossRef]
- Regan, S.L.P.; Knight, P.G.; Yovich, J.L.; Leung, Y.; Arfuso, F.; Dharmarajan, A. Granulosa Cell Apoptosis in the Ovarian Follicle-A Changing View. Front. Endocrinol. 2018, 9, 61. [Google Scholar] [CrossRef]
- Richards, J.S.; Ren, Y.A.; Candelaria, N.; Adams, J.E.; Rajkovic, A. Ovarian Follicular Theca Cell Recruitment, Differentiation, and Impact on Fertility: 2017 Update. Endocr. Rev. 2018, 39, 1–20. [Google Scholar] [CrossRef]
- Wagner, P.K.; Otomo, A.; Christians, J.K. Regulation of pregnancy-associated plasma protein A2 (PAPPA2) in a human placental trophoblast cell line (BeWo). Reprod. Biol. Endocrinol. 2011, 9, 48. [Google Scholar] [CrossRef]
- Ussar, S.; Bezy, O.; Bluher, M.; Kahn, C.R. Glypican-4 enhances insulin signaling via interaction with the insulin receptor and serves as a novel adipokine. Diabetes 2012, 61, 2289–2298. [Google Scholar] [CrossRef]
- Yoo, H.J.; Hwang, S.Y.; Cho, G.J.; Hong, H.C.; Choi, H.Y.; Hwang, T.G.; Kim, S.M.; Bluher, M.; Youn, B.S.; Baik, S.H.; et al. Association of glypican-4 with body fat distribution, insulin resistance, and nonalcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 2013, 98, 2897–2901. [Google Scholar] [CrossRef]
- Ball, C.B.; Rodriguez, K.F.; Stumpo, D.J.; Ribeiro-Neto, F.; Korach, K.S.; Blackshear, P.J.; Birnbaumer, L.; Ramos, S.B. The RNA-binding protein, ZFP36L2, influences ovulation and oocyte maturation. PLoS ONE 2014, 9, e97324. [Google Scholar] [CrossRef]
- Otsuka, H.; Fukao, A.; Funakami, Y.; Duncan, K.E.; Fujiwara, T. Emerging Evidence of Translational Control by AU-Rich Element-Binding Proteins. Front. Genet. 2019, 10, 332. [Google Scholar] [CrossRef]
- Bouchard, L.; Tchernof, A.; Deshaies, Y.; Marceau, S.; Lescelleur, O.; Biron, S.; Vohl, M.C. ZFP36: A promising candidate gene for obesity-related metabolic complications identified by converging genomics. Obes. Surg. 2007, 17, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Diamanti-Kandarakis, E.; Dunaif, A. Insulin resistance and the polycystic ovary syndrome revisited: An update on mechanisms and implications. Endocr. Rev. 2012, 33, 981–1030. [Google Scholar] [CrossRef] [PubMed]
- Diamanti-Kandarakis, E.; Kandarakis, H.A. Conservative management of gynecologic diseases: Insulin sensitizing agents in polycystic ovary syndrome. Ann. N. Y. Acad. Sci. 2003, 997, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Vega, G.L.; Adams-Huet, B.; Peshock, R.; Willett, D.; Shah, B.; Grundy, S.M. Influence of body fat content and distribution on variation in metabolic risk. J. Clin. Endocrinol. Metab. 2006, 91, 4459–4466. [Google Scholar] [CrossRef]
- Eriksson, U.J. The pathogenesis of congenital malformations in diabetic pregnancy. Diabetes Metab. Rev. 1995, 11, 63–82. [Google Scholar] [CrossRef]
- Kjaer, K.; Hagen, C.; Sando, S.H.; Eshoj, O. Epidemiology of menarche and menstrual disturbances in an unselected group of women with insulin-dependent diabetes mellitus compared to controls. J. Clin. Endocrinol. Metab. 1992, 75, 524–529. [Google Scholar] [CrossRef]
- Lanzarini, F.; Pereira, F.A.; Camargo, J.; Oliveira, A.M.; Belaz, K.R.A.; Melendez-Perez, J.J.; Eberlin, M.N.; Brum, M.C.S.; Mesquita, F.S.; Sudano, M.J. ELOVL5 Participates in Embryonic Lipid Determination of Cellular Membranes and Cytoplasmic Droplets. Int. J. Mol. Sci. 2021, 22, 1311. [Google Scholar] [CrossRef]
- SenBanerjee, S.; Lin, Z.; Atkins, G.B.; Greif, D.M.; Rao, R.M.; Kumar, A.; Feinberg, M.W.; Chen, Z.; Simon, D.I.; Luscinskas, F.W. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 2004, 199, 1305–1315. [Google Scholar] [CrossRef]
- Guangqi, E.; Cao, Y.; Bhattacharya, S.; Dutta, S.; Wang, E.; Mukhopadhyay, D. Endogenous vascular endothelial growth factor-A (VEGF-A) maintains endothelial cell homeostasis by regulating VEGF receptor-2 transcription. J. Biol. Chem. 2012, 287, 3029–3041. [Google Scholar] [CrossRef]
- Kwon, M.; Pavlov, T.S.; Nozu, K.; Rasmussen, S.A.; Ilatovskaya, D.V.; Lerch-Gaggl, A.; North, L.M.; Kim, H.; Qian, F.; Sweeney, W.E., Jr.; et al. G-protein signaling modulator 1 deficiency accelerates cystic disease in an orthologous mouse model of autosomal dominant polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2012, 109, 21462–21467. [Google Scholar] [CrossRef]
- Watanabe, T.M.; Tokuo, H.; Gonda, K.; Higuchi, H.; Ikebe, M. Myosin-X induces filopodia by multiple elongation mechanism. J. Biol. Chem. 2010, 285, 19605–19614. [Google Scholar] [CrossRef]
- Chen, S.U.; Chou, C.H.; Lee, H.; Ho, C.H.; Lin, C.W.; Yang, Y.S. Lysophosphatidic acid up-regulates expression of interleukin-8 and -6 in granulosa-lutein cells through its receptors and nuclear factor-kappaB dependent pathways: Implications for angiogenesis of corpus luteum and ovarian hyperstimulation syndrome. J. Clin. Endocrinol. Metab. 2008, 93, 935–943. [Google Scholar] [CrossRef]
- Wei, L.H.; Chou, C.H.; Chen, M.W.; Rose-John, S.; Kuo, M.L.; Chen, S.U.; Yang, Y.S. The role of IL-6 trans-signaling in vascular leakage: Implications for ovarian hyperstimulation syndrome in a murine model. J. Clin. Endocrinol. Metab. 2013, 98, E472–E484. [Google Scholar] [CrossRef]
- Borgwardt, L.; Olsen, K.W.; Rossing, M.; Helweg-Larsen, R.B.; Toftager, M.; Pinborg, A.; Bogstad, J.; Lossl, K.; Zedeler, A.; Grondahl, M.L. Rare genetic variants suggest dysregulation of signaling pathways in low- and high-risk patients developing severe ovarian hyperstimulation syndrome. J. Assist. Reprod. Genet. 2020, 37, 2883–2892. [Google Scholar] [CrossRef]
- Elchalal, U.; Schenker, J.G. The pathophysiology of ovarian hyperstimulation syndrome—Views and ideas. Hum. Reprod. 1997, 12, 1129–1137. [Google Scholar] [CrossRef]
- Binder, H.; Dittrich, R.; Einhaus, F.; Krieg, J.; Muller, A.; Strauss, R.; Beckmann, M.W.; Cupisti, S. Update on ovarian hyperstimulation syndrome: Part 1—Incidence and pathogenesis. Int. J. Fertil. Womens Med. 2007, 52, 11–26. [Google Scholar]
- Goldsman, M.P.; Pedram, A.; Dominguez, C.E.; Ciuffardi, I.; Levin, E.; Asch, R.H. Increased capillary permeability induced by human follicular fluid: A hypothesis for an ovarian origin of the hyperstimulation syndrome. Fertil. Steril. 1995, 63, 268–272. [Google Scholar] [CrossRef]
- Carriere, V.; Roussel, L.; Ortega, N.; Lacorre, D.A.; Americh, L.; Aguilar, L.; Bouche, G.; Girard, J.P. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc. Natl. Acad. Sci. USA 2007, 104, 282–287. [Google Scholar] [CrossRef]
- Karakose, M.; Demircan, K.; Tutal, E.; Demirci, T.; Arslan, M.S.; Sahin, M.; Celik, H.T.; Kazanci, F.; Karakaya, J.; Cakal, E.; et al. Clinical significance of ADAMTS1, ADAMTS5, ADAMTS9 aggrecanases and IL-17A, IL-23, IL-33 cytokines in polycystic ovary syndrome. J. Endocrinol. Investig. 2016, 39, 1269–1275. [Google Scholar] [CrossRef]
- Prabhu, Y.D.; Valsala Gopalakrishnan, A. gamma-Linolenic acid ameliorates DHEA induced pro-inflammatory response in polycystic ovary syndrome via PPAR-gamma signaling in rats. Reprod. Biol. 2020, 20, 348–356. [Google Scholar] [CrossRef]
- Furqan, M.; Mukhi, N.; Lee, B.; Liu, D. Dysregulation of JAK-STAT pathway in hematological malignancies and JAK inhibitors for clinical application. Biomark. Res. 2013, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Khatib, H.; Huang, W.; Mikheil, D.; Schutzkus, V.; Monson, R.L. Effects of signal transducer and activator of transcription (STAT) genes STAT1 and STAT3 genotypic combinations on fertilization and embryonic survival rates in Holstein cattle. J. Dairy Sci. 2009, 92, 6186–6191. [Google Scholar] [CrossRef] [PubMed]
- Tsurumi, A.; Zhao, C.; Li, W.X. Canonical and non-canonical JAK/STAT transcriptional targets may be involved in distinct and overlapping cellular processes. BMC Genom. 2017, 18, 718. [Google Scholar] [CrossRef] [PubMed]
- Baumer, D.; Trauner, J.; Hollfelder, D.; Cerny, A.; Schoppmeier, M. JAK-STAT signalling is required throughout telotrophic oogenesis and short-germ embryogenesis of the beetle Tribolium. Dev. Biol. 2011, 350, 169–182. [Google Scholar] [CrossRef][Green Version]
- Ferrero, H.; Diaz-Gimeno, P.; Sebastian-Leon, P.; Faus, A.; Gomez, R.; Pellicer, A. Dysregulated genes and their functional pathways in luteinized granulosa cells from PCOS patients after cabergoline treatment. Reproduction 2018, 155, 373–381. [Google Scholar] [CrossRef]
| PCOS (n = 18) | OHSS (n = 6) | p-Value * | |
|---|---|---|---|
| Mean (SD) | Mean (SD) | ||
| Age (years) | 29.0 (3.7) | 30.2 (6.3) | 0.71 |
| BMI (kg/m2) | 24.8 (1.2) | 24.2(1.4) | 0.43 |
| FSH (IU/L) | 6.9 (2.2) | 6.3 (1.1) | 0.47 |
| LH (IU/L) | 13.0 (9.9) | 7.5 (4.9) | 0.14 |
| Testosterone | 2.5 (1.3) | 2.2 (0.9) | 0.76 |
| Androstenedione | 12.3 (6.8) | 15.5 (4.8) | 0.13 |
| Gene | Gene Title | p-Value | FC |
|---|---|---|---|
| PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 0.000798 | 1.86 |
| DCN | Decorin | 0.0044 | 1.76 |
| PTPLA | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member | 0.005688 | 1.71 |
| FKBP1A | FK506 binding protein 1A, 12 kDa | 0.002428 | 1.7 |
| SHOX2 | short stature homeobox 2 | 0.008934 | 1.64 |
| ELOVL5 | ELOVL family member 5, elongation of long-chain fatty acids (FEN1/Elo2, SUR4/Elo) | 0.000683 | 1.55 |
| IL33 | interleukin 33 | 0.003649 | 1.55 |
| EIF5 | eukaryotic translation initiation factor 5 | 0.007926 | 1.52 |
| TMX1 | thioredoxin-related transmembrane protein 1 | 0.000272 | 1.52 |
| LIFR | leukemia inhibitory factor receptor alpha | 0.00038 | 1.51 |
| RDX | Radixin | 0.005259 | 1.51 |
| OBSL1 | obscurin-like 1 | 0.009828 | −1.5 |
| RNPEP | arginylaminopeptidase (aminopeptidase B) | 0.000196 | −1.5 |
| BAT3 | HLA-B associated transcript 3 | 0.001025 | −1.51 |
| DYNC1H1 | dynein, cytoplasmic 1, heavy chain 1 | 0.004793 | −1.51 |
| FOXP1 | forkhead box P1 | 0.001132 | −1.51 |
| MAP4K4 | mitogen-activated protein kinase kinasekinasekinase 4 | 0.000309 | −1.51 |
| GRN | Granulin | 0.008224 | −1.52 |
| MED14 | mediator complex subunit 14 | 0.001012 | −1.52 |
| HS3ST1 | heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | 0.007768 | −1.53 |
| SEC14L1 | SEC14-like 1 (S. cerevisiae) | 0.005745 | −1.53 |
| SNRNP200 | small nuclear ribonucleoprotein 200k Da (U5) | 0.001182 | −1.53 |
| BBS1 | Bardet–Biedl syndrome 1 | 0.000834 | −1.54 |
| MAN2B2 | mannosidase, alpha, class 2B, member 2 | 0.00033 | −1.54 |
| MYLIP | myosin regulatory light chain interacting protein | 0.006807 | −1.55 |
| KIAA0513 | KIAA0513 | 0.004853 | −1.56 |
| KDM5A | lysine (K)-specific demethylase 5A | 0.000431 | −1.57 |
| PCSK5 | proproteinconvertasesubtilisin/kexintype 5 | 0.002482 | −1.58 |
| GAS2L1 | growth arrest-specific 2 like 1 | 0.005994 | −1.59 |
| MGC21881 | hypothetical locus MGC21881 | 0.009274 | −1.59 |
| PLCL1 | phospholipase C-like 1 | 0.009504 | −1.59 |
| ZSWIM5 | zinc finger, SWIM-type containing 5 | 0.005745 | −1.59 |
| VIM | Vimentin | 0.001645 | −1.6 |
| CRISPLD2 | cysteine-rich secretory protein LCCL domain containing 2 | 0.00095 | −1.61 |
| GPSM1 | G-protein signaling modulator 1 | 0.002004 | −1.61 |
| CHST3 | carbohydrate (chondroitin 6) sulfotransferase 3 | 0.001192 | −1.63 |
| SCAP | SREBF chaperone | 0.008736 | −1.63 |
| LAMP1 | lysosomal-associated membrane protein 1 | 0.008512 | −1.64 |
| SH3PXD2B | SH3 and PX domains 2B | 0.004433 | −1.64 |
| SP110 | SP110 nuclear body protein | 0.00000735 | −1.66 |
| PHEX | phosphate regulating endopeptidase homolog, X-linked | 0.004347 | −1.67 |
| FOXO1 | forkhead box O1 | 0.001054 | −1.68 |
| TPP1 | tripeptidyl peptidase I | 0.009215 | −1.69 |
| LGALS3BP | lectin, galactoside-binding, soluble, 3 binding protein | 0.007244 | −1.71 |
| ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 | 0.004646 | −1.76 |
| CADM1 | cell adhesion molecule 1 | 0.003501 | −1.77 |
| CLSTN1 | calsyntenin 1 | 0.002802 | −1.77 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 0.00617 | −1.81 |
| MYO10 | myosin X | 0.004334 | −1.86 |
| KLF2 | Kruppel-like factor 2 (lung) | 0.003583 | −1.87 |
| INSR | insulin receptor | 0.008567 | −1.88 |
| IER2 | immediate early response 2 | 0.006056 | −1.91 |
| CNKSR3 | CNKSR family member 3 | 0.004281 | −1.99 |
| GPC4 | glypican 4 | 0.003565 | −2.08 |
| MYADM | myeloid-associated differentiation marker | 0.002895 | −2.14 |
| PXDN | peroxidasin homolog (Drosophila) | 0.00015 | −2.15 |
| DAPK1 | death-associated protein kinase 1 | 0.001645 | −2.17 |
| ZFP36 | zinc finger protein 36, C3H type, homolog (mouse) | 0.000679 | −2.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daghestani, M.H.; Alqahtani, H.A.; AlBakheet, A.; Al Deery, M.; Awartani, K.A.; Daghestani, M.H.; Kaya, N.; Warsy, A.; Coskun, S.; Colak, D. Global Transcriptional Profiling of Granulosa Cells from Polycystic Ovary Syndrome Patients: Comparative Analyses of Patients with or without History of Ovarian Hyperstimulation Syndrome Reveals Distinct Biomarkers and Pathways. J. Clin. Med. 2022, 11, 6941. https://doi.org/10.3390/jcm11236941
Daghestani MH, Alqahtani HA, AlBakheet A, Al Deery M, Awartani KA, Daghestani MH, Kaya N, Warsy A, Coskun S, Colak D. Global Transcriptional Profiling of Granulosa Cells from Polycystic Ovary Syndrome Patients: Comparative Analyses of Patients with or without History of Ovarian Hyperstimulation Syndrome Reveals Distinct Biomarkers and Pathways. Journal of Clinical Medicine. 2022; 11(23):6941. https://doi.org/10.3390/jcm11236941
Chicago/Turabian StyleDaghestani, Maha H., Huda A. Alqahtani, AlBandary AlBakheet, Mashael Al Deery, Khalid A. Awartani, Mazin H. Daghestani, Namik Kaya, Arjumand Warsy, Serdar Coskun, and Dilek Colak. 2022. "Global Transcriptional Profiling of Granulosa Cells from Polycystic Ovary Syndrome Patients: Comparative Analyses of Patients with or without History of Ovarian Hyperstimulation Syndrome Reveals Distinct Biomarkers and Pathways" Journal of Clinical Medicine 11, no. 23: 6941. https://doi.org/10.3390/jcm11236941
APA StyleDaghestani, M. H., Alqahtani, H. A., AlBakheet, A., Al Deery, M., Awartani, K. A., Daghestani, M. H., Kaya, N., Warsy, A., Coskun, S., & Colak, D. (2022). Global Transcriptional Profiling of Granulosa Cells from Polycystic Ovary Syndrome Patients: Comparative Analyses of Patients with or without History of Ovarian Hyperstimulation Syndrome Reveals Distinct Biomarkers and Pathways. Journal of Clinical Medicine, 11(23), 6941. https://doi.org/10.3390/jcm11236941






