Global Trends in Research on Cell-Free Nucleic Acids in Obstetrics and Gynecology during 2017–2021
Abstract
1. Introduction
2. Methods
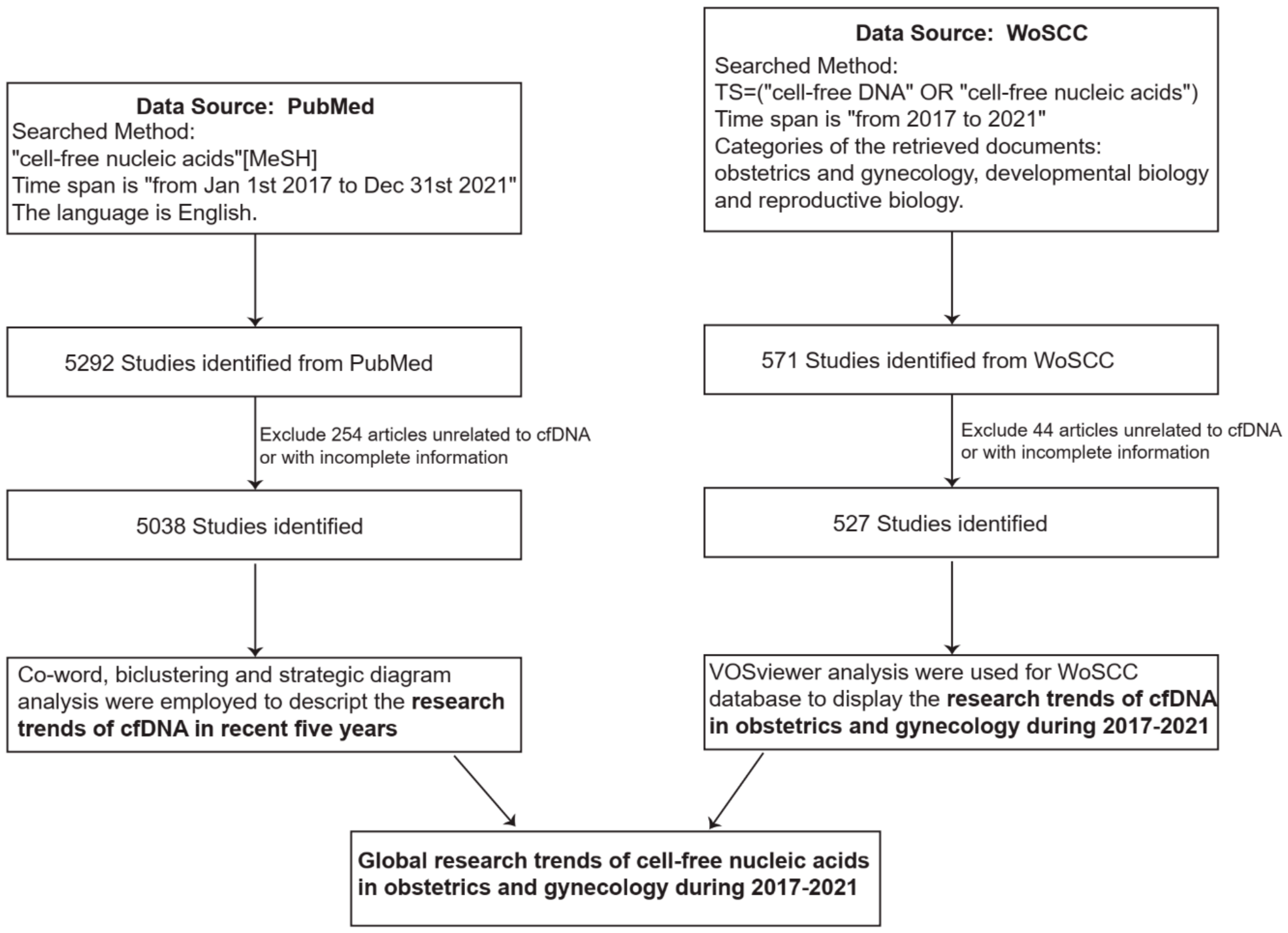
2.1. Data Source and Research Process
- The PubMed database was searched as the source of this research. The Medical Subject Headings (MeSH) phrase “cell-free nucleic acids” was used, and the time span was “from 1 January 2017 to 31 December 2021”. The language was English. This part of the study was performed to describe the trends in research on cfDNA in the selected five years. Two independent investigators screened these articles. The literature was excluded if it was not related to cfDNA study or if the basic information was incomplete. Finally, 5038 articles were chosen (Figure 1).
- The Web of Science Core Collection (WoSCC) database was searched to identify the trends in research on cfDNA in obstetrics and gynecology. The phrases “cell-free DNA” or “cell-free nucleic acids” were used for the search, and the time span was “from 2017 to 2021”. We refined the categories of the retrieved documents and limited the documents to those related to obstetrics and gynecology, developmental biology, and reproductive biology. The retrieved results were saved as “plain text” and “full record and cited references”. We collected the basic information about each article, such as country, author, institution, journal, references, and keywords. Two investigators independently screened the results of the primary search, and literature not related to cfDNA or missing basic information was excluded. Finally, 527 articles were chosen (Figure 1).
2.2. Analysis Tools and Methods
3. Results
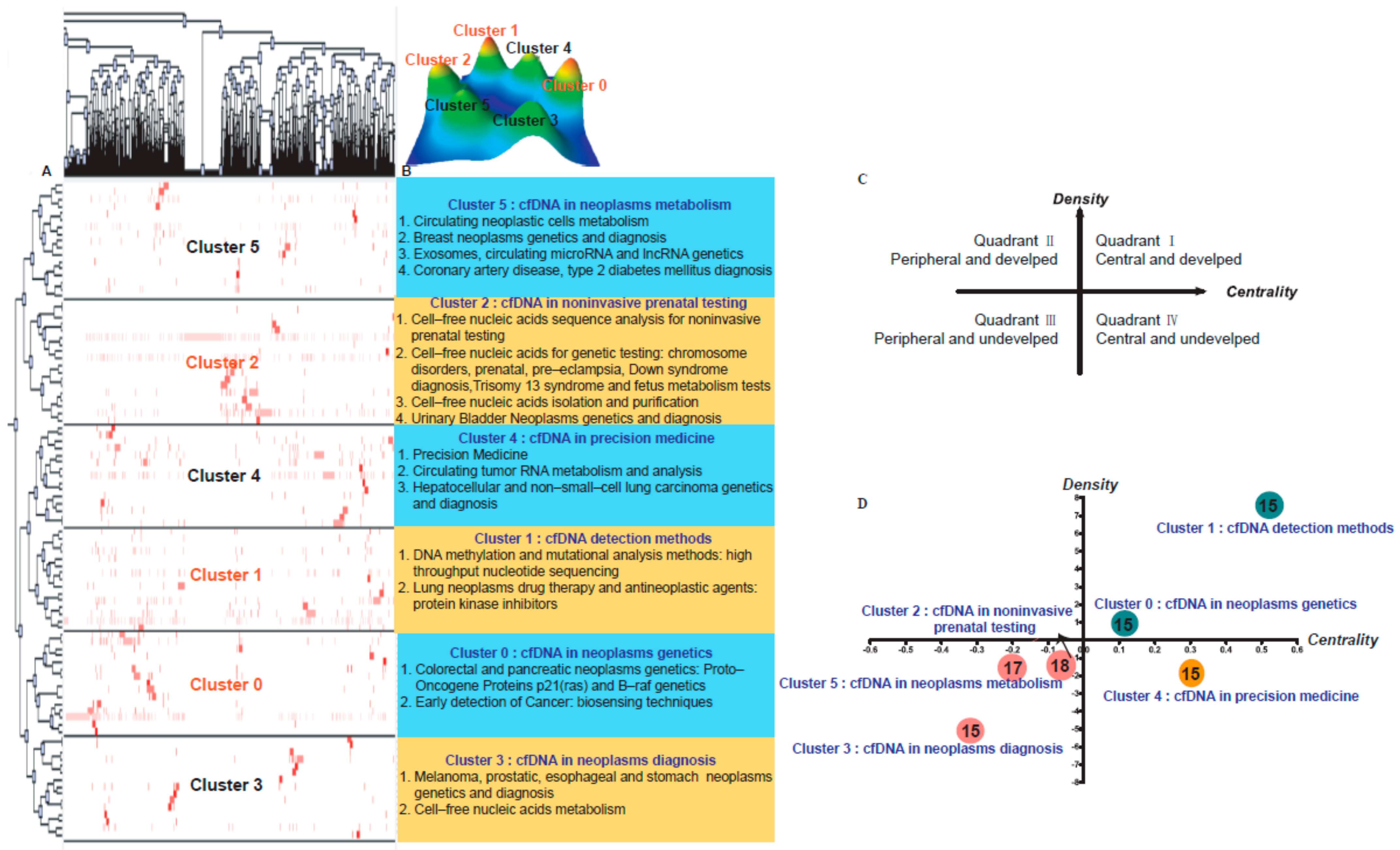
3.1. Research Hotspots Identified on the Basis of MeSH Term Theme Clusters
3.2. cfDNA Theme Trends
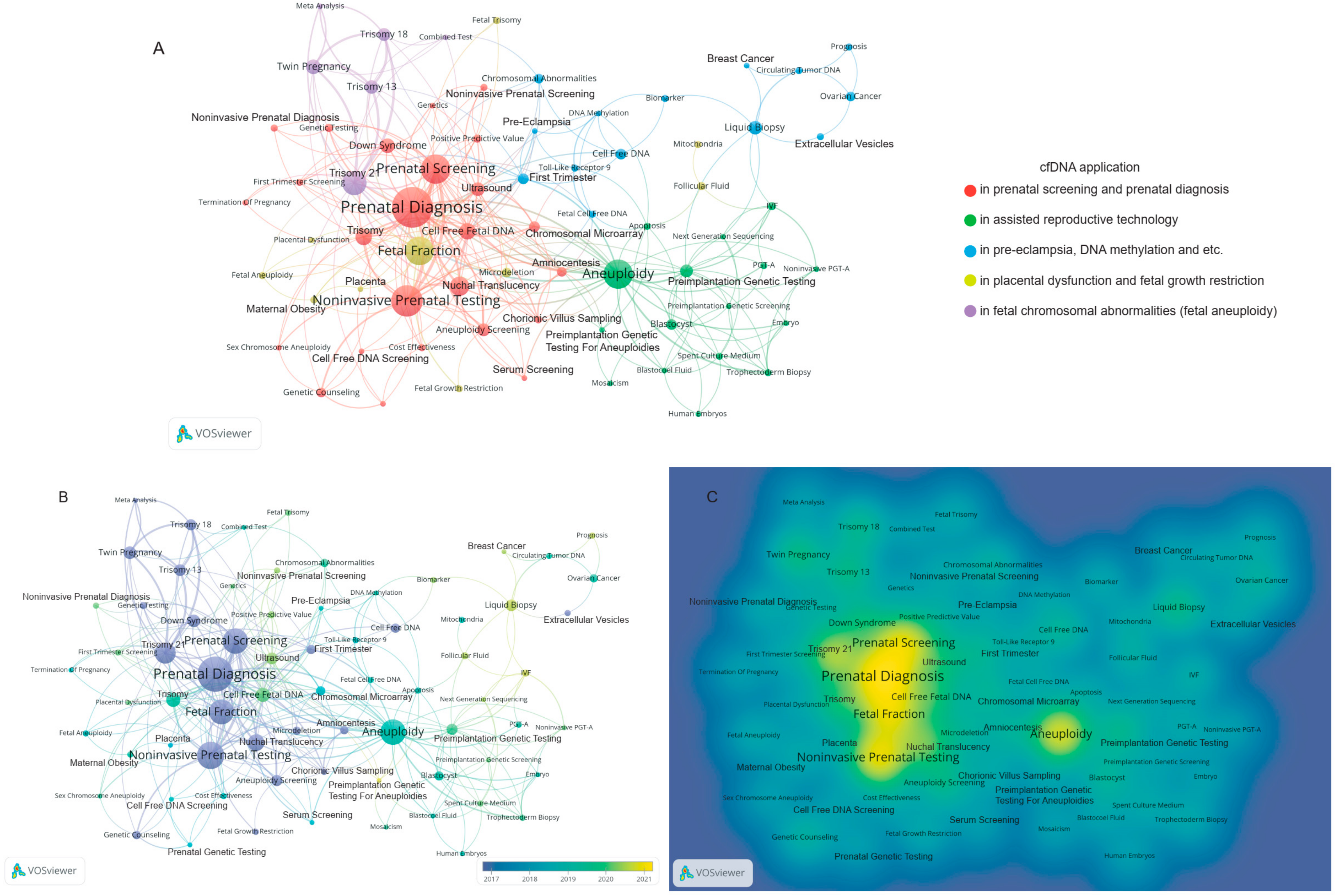
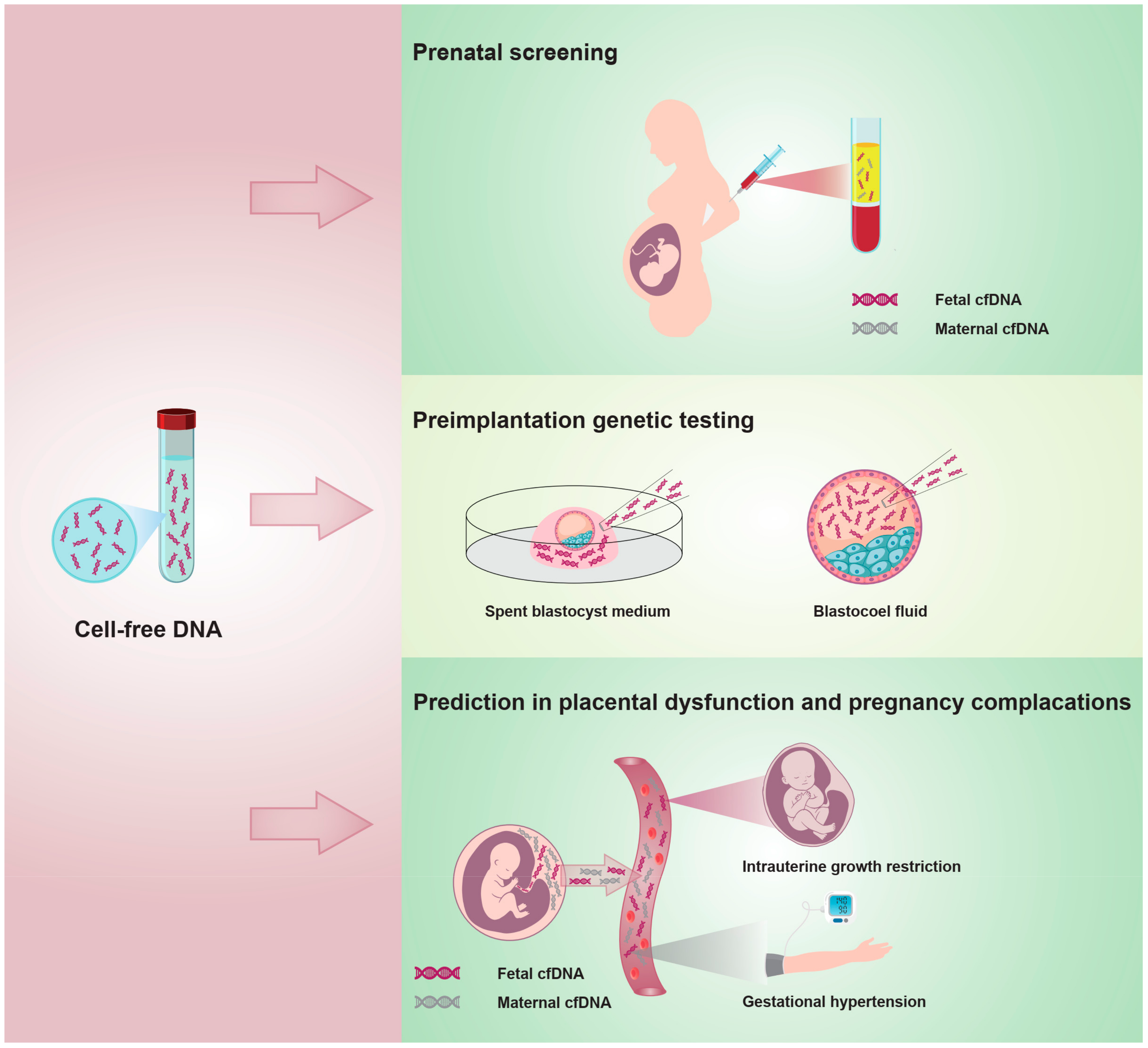
3.3. cfDNA Application in Obstetrics and Gynecology
3.4. Most Influential Authors, Institutions, Countries, and Journals in cfDNA Research on Obstetrics and Gynecology
3.5. Most Influential Keywords in cfDNA Research in Obstetrics and Gynecology
4. Discussion
4.1. Crucial Themes in the Strategy Diagram
4.2. Peripheral and Undeveloped Themes in the Strategy Diagram
4.3. Central and Immature Themes in the Strategy Diagram
4.4. Global cfDNA Trends in Obstetrics and Gynecology
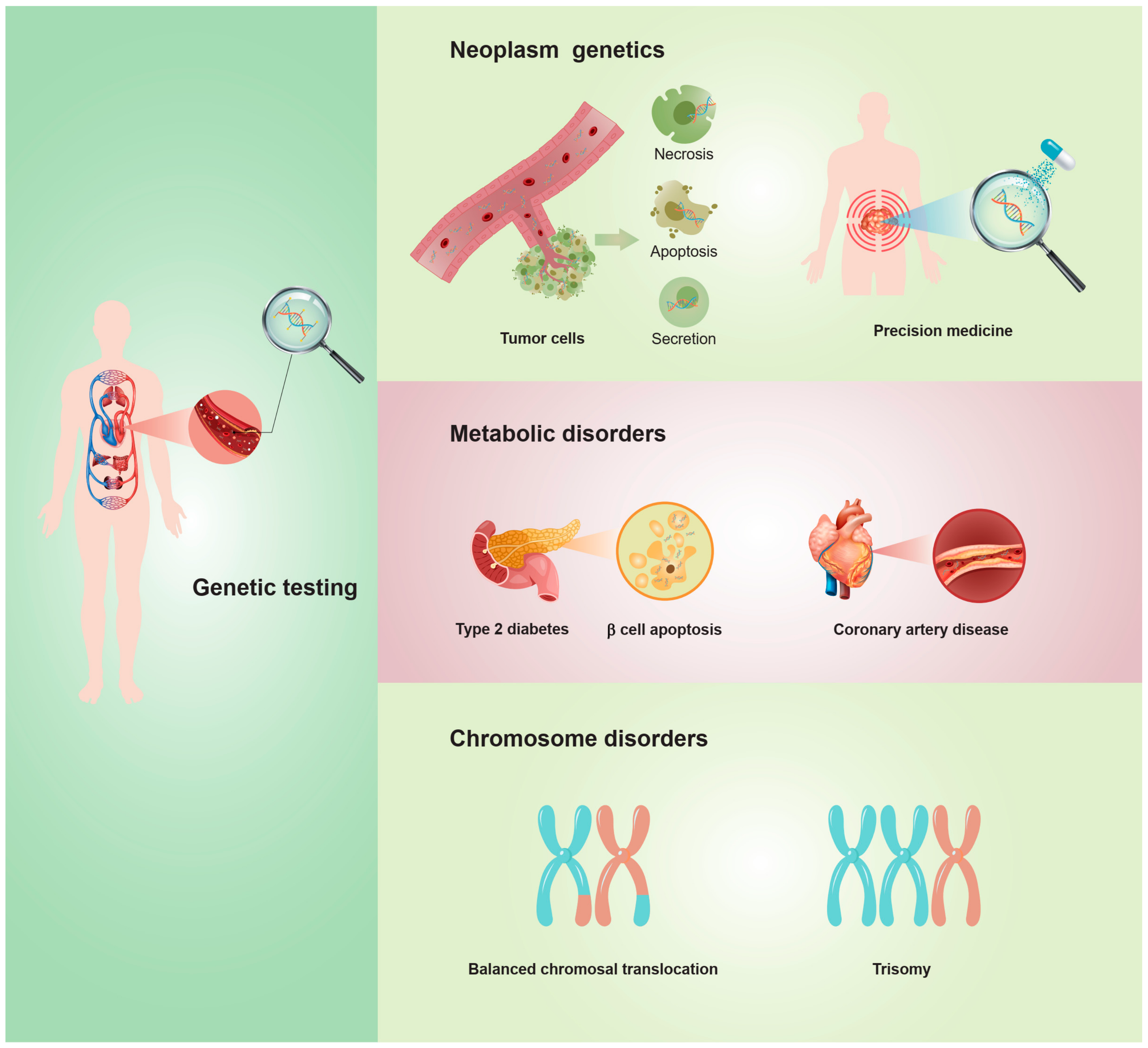
4.5. Knowledge Base, Hotspots, and Emerging Frontiers Related to cfDNA in Obstetrics and Gynecology
5. Limitation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pos, O.; Biro, O.; Szemes, T.; Nagy, B. Circulating cell-free nucleic acids: Characteristics and applications. Eur. J. Hum. Genet. 2018, 26, 937–945. [Google Scholar] [CrossRef]
- Pos, O.; Budis, J.; Szemes, T. Recent trends in prenatal genetic screening and testing. F1000Research 2019, 8, 764. [Google Scholar] [CrossRef] [PubMed]
- Nagy, B. Cell-Free Nucleic Acids. Int. J. Mol. Sci. 2019, 20, 5645. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, M.; Guan, P.; Ma, S.; Cui, L. Mapping publication trends and identifying hot spots of research on Internet health information seeking behavior: A quantitative and co-word biclustering analysis. J. Med. Internet Res. 2015, 17, e81. [Google Scholar] [CrossRef] [PubMed]
- Donohue, J.C. Understanding Scientific Literature: A Bibliometric Approach; The Massachusetts Institute of Technology Press: Cambridge, MA, USA, 1974; p. 101. [Google Scholar]
- Shi, B.; Wei, W.; Qin, X.; Zhao, F.; Duan, Y.; Sun, W.; Li, D.; Cao, Y. Mapping theme trends and knowledge structure on adipose-derived stem cells: A bibliometric analysis from 2003 to 2017. Regen. Med. 2019, 14, 33–48. [Google Scholar] [CrossRef]
- Zhao, F.; Shi, B.; Liu, R.; Zhou, W.; Shi, D.; Zhang, J. Theme trends and knowledge structure on choroidal neovascularization: A quantitative and co-word analysis. BMC Ophthalmol. 2018, 18, 86. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids in Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Tong, Y.K.; Lo, Y.M. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin. Chim. Acta 2006, 363, 187–196. [Google Scholar] [CrossRef]
- Stewart, C.M.; Kothari, P.; Mouliere, F.; Mair, R.; Somnay, S.; Benayed, R.; Zehir, A.; Weigelt, B.; Dawson, S.-J.; Arcila, M.E.; et al. The value of cell-free DNA for molecular pathology. J. Pathol. 2018, 244, 616–627. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2021 update. Pharmacol. Res. 2021, 165, 105463. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol. Res. 2022, 175, 106037. [Google Scholar] [CrossRef]
- Qasemi, M.; Mahdian, R.; Amidi, F. Cell-free DNA discoveries in human reproductive medicine: Providing a new tool for biomarker and genetic assays in ART. J. Assist. Reprod. Genet. 2021, 38, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Carbone, L.; Cariati, F.; Sarno, L.; Conforti, A.; Bagnulo, F.; Strina, I.; Pastore, L.; Maruotti, G.M.; Alviggi, C. Non-Invasive Prenatal Testing: Current Perspectives and Future Challenges. Genes 2020, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372, eaaw3616. [Google Scholar] [CrossRef] [PubMed]
- Arosemena, M.; Meah, F.A.; Mather, K.J.; Tersey, S.A.; Mirmira, R.G. Cell-Free DNA Fragments as Biomarkers of Islet beta-Cell Death in Obesity and Type 2 Diabetes. Int. J. Mol. Sci. 2021, 22, 2151. [Google Scholar] [CrossRef]
- Drag, M.H.; Kilpelainen, T.O. Cell-free DNA and RNA-measurement and applications in clinical diagnostics with focus on metabolic disorders. Physiol. Genomics 2021, 53, 33–46. [Google Scholar] [CrossRef]
- Fazmin, I.T.; Achercouk, Z.; Edling, C.E.; Said, A.; Jeevaratnam, K. Circulating microRNA as a Biomarker for Coronary Artery Disease. Biomolecules 2020, 10, 1354. [Google Scholar] [CrossRef]
- Rahat, B.; Ali, T.; Sapehia, D.; Mahajan, A.; Kaur, J. Circulating Cell-Free Nucleic Acids as Epigenetic Biomarkers in Precision Medicine. Front. Genet. 2020, 11, 844. [Google Scholar] [CrossRef]
- van der Leest, P.; Ketelaar, E.M.; van Noesel, C.J.M.; van den Broek, D.; van Boerdonk, R.A.A.; Deiman, B.; Rifaela, N.; van der Geize, R.; Huijsmans, C.J.J.; Speel, E.J.M.; et al. Dutch National Round Robin Trial on Plasma-Derived Circulating Cell-Free DNA Extraction Methods Routinely Used in Clinical Pathology for Molecular Tumor Profiling. Clin. Chem. 2022, 68, 963–972. [Google Scholar] [CrossRef]
- American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 545: Noninvasive prenatal testing for fetal aneuploidy. Obstet. Gynecol. 2012, 120, 1532–1534. [Google Scholar] [CrossRef]
- Society for Maternal-Fetal Medicine Publications Committee. Electronic address pso. #36: Prenatal aneuploidy screening using cell-free DNA. Am. J. Obstet. Gynecol. 2015, 212, 711–716. [Google Scholar]
- Gregg, A.R.; Skotko, B.G.; Benkendorf, J.L.; Monaghan, K.G.; Bajaj, K.; Best, R.G.; Klugman, S.; Watson, M.S. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: A position statement of the American College of Medical Genetics and Genomics. Genet. Med. 2016, 18, 1056–1065. [Google Scholar] [CrossRef] [PubMed]
- Wapner, R.J.; Martin, C.L.; Levy, B.; Ballif, B.C.; Eng, C.M.; Zachary, J.M.; Savage, M.; Platt, L.D.; Saltzman, D.; Grobman, W.A.; et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 2012, 367, 2175–2184. [Google Scholar] [CrossRef]
- Li, X.; Hao, Y.; Chen, D.; Ji, D.; Zhu, W.; Zhu, X.; Wei, Z.; Cao, Y.; Zhang, Z.; Zhou, P. Non-invasive preimplantation genetic testing for putative mosaic blastocysts: A pilot study. Hum. Reprod. 2021, 36, 2020–2034. [Google Scholar] [CrossRef]
- Kuznyetsov, V.; Madjunkova, S.; Antes, R.; Abramov, R.; Motamedi, G.; Ibarrientos, Z.; Librach, C. Evaluation of a novel non-invasive preimplantation genetic screening approach. PLoS ONE 2018, 13, e0197262. [Google Scholar] [CrossRef]
- Jiao, J.; Shi, B.; Sagnelli, M.; Yang, D.; Yao, Y.; Li, W.; Shao, L.; Lu, S.; Li, D.; Wang, X. Minimally invasive preimplantation genetic testing using blastocyst culture medium. Hum. Reprod. 2019, 34, 1369–1379. [Google Scholar] [CrossRef]
- Lo, Y.M.; Leung, T.N.; Tein, M.S.; Sargent, I.L.; Zhang, J.; Lau, T.K.; Haines, C.J.; Redman, C.W. Quantitative abnormalities of fetal DNA in maternal serum in preeclampsia. Clin. Chem. 1999, 45, 184–188. [Google Scholar] [CrossRef]
- Leung, T.N.; Zhang, J.; Lau, T.K.; Chan, L.Y.; Lo, Y.M. Increased maternal plasma fetal DNA concentrations in women who eventually develop preeclampsia. Clin. Chem. 2001, 47, 137–139. [Google Scholar] [CrossRef]
- Zhong, X.Y.; Holzgreve, W.; Hahn, S. The levels of circulatory cell free fetal DNA in maternal plasma are elevated prior to the onset of preeclampsia. Pregnancy Hypertens. 2002, 21, 77–83. [Google Scholar] [CrossRef]
- Levine, R.J.; Qian, C.; Leshane, E.S.; Yu, K.F.; England, L.J.; Schisterman, E.F.; Wataganara, T.; Romero, R.; Bianchi, D.W. Two-stage elevation of cell-free fetal DNA in maternal sera before onset of preeclampsia. Am. J. Obstet. Gynecol. 2004, 190, 707–713. [Google Scholar] [CrossRef]
- Sifakis, S.; Zaravinos, A.; Maiz, N.; Spandidos, D.A.; Nicolaides, K.H. First-trimester maternal plasma cell-free fetal DNA and preeclampsia. Am. J. Obstet. Gynecol. 2009, 201, 472.e1–472.e7. [Google Scholar] [CrossRef]
- Scharfe-Nugent, A.; Corr, S.; Carpenter, S.B.; Keogh, L.; Doyle, B.; Martin, C.; Fitzgerald, K.; Daly, S.; O’Leary, J.J.; O’Neill, L. TLR9 provokes inflammation in response to fetal DNA: Mechanism for fetal loss in preterm birth and preeclampsia. J. Immunol. 2012, 188, 5706–5712. [Google Scholar] [CrossRef]
- Gerson, K.D.; Truong, S.; Haviland, M.J.; O’Brien, B.M.; Hacker, M.R.; Spiel, M.H. Low fetal fraction of cell-free DNA predicts placental dysfunction and hypertensive disease in pregnancy. Pregnancy Hypertens. 2019, 16, 148–153. [Google Scholar] [CrossRef]
- Del Vecchio, G.; Li, Q.; Li, W.; Thamotharan, S.; Tosevska, A.; Morselli, M.; Sung, K.; Janzen, C.; Zhou, X.; Pellegrini, M.; et al. Cell-free DNA Methylation and Transcriptomic Signature Prediction of Pregnancies with Adverse Outcomes. Epigenetics 2021, 16, 642–661. [Google Scholar] [CrossRef]
- Sifakis, S.; Koukou, Z.; Spandidos, D.A. Cell-free fetal DNA and pregnancy-related complications (review). Mol. Med. Rep. 2015, 11, 2367–2372. [Google Scholar] [CrossRef]
- Hopkins, M.K.; Koelper, N.; Bender, W.; Durnwald, C.; Sammel, M.; Dugoff, L. Association between cell-free DNA fetal fraction and gestational diabetes. Prenat. Diagn. 2020, 40, 724–727. [Google Scholar] [CrossRef]
- Cushen, S.C.; Sprouse, M.L.; Blessing, A.; Sun, J.; Jarvis, S.S.; Okada, Y.; Fu, Q.; Romero, S.A.; Phillips, N.R.; Goulopoulou, S. Cell-free mitochondrial DNA increases in maternal circulation during healthy pregnancy: A prospective, longitudinal study. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2020, 318, R445–R452. [Google Scholar] [CrossRef]
- Gil, M.M.; Accurti, V.; Santacruz, B.; Plana, M.N.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol. 2017, 50, 302–314. [Google Scholar] [CrossRef]
- Gil, M.M.; Quezada, M.S.; Revello, R.; Akolekar, R.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol. 2015, 45, 249–266. [Google Scholar] [CrossRef]
- Morris, S.; Karlsen, S.; Chung, N.; Hill, M.; Chitty, L.S. Model-based analysis of costs and outcomes of non-invasive prenatal testing for Down’s syndrome using cell free fetal DNA in the UK National Health Service. PLoS ONE 2014, 9, e93559. [Google Scholar] [CrossRef]

| Rank | First Author | Articles | Co-Cited Author | Citations of Co-Cited Author |
|---|---|---|---|---|
| 1 | Dugoff, Lorraine | 15 | Levy, Brynn | 119 |
| 2 | Norton, Mary E | 14 | Norton, Mary E | 74 |
| 3 | Caughey, Aaron B | 9 | Hui, Lisa | 68 |
| 4 | Hui, Lisa | 8 | Dugoff, Lorraine | 67 |
| 5 | Menezes, Melody | 8 | Costa, Fabricio Da Silva | 67 |
| 6 | Chitty, Lyn S | 7 | Menezes, Melody | 57 |
| 7 | Costa, Fabricio Da Silva | 7 | Bianchi, Diana W | 53 |
| 8 | Avram, Carmen M | 6 | Mclennan, Andrew | 51 |
| 9 | Bromley, Bryann | 6 | Scott, Fergus | 51 |
| 10 | Benn, Peter | 5 | Nisbet, Debbie | 50 |
| Rank | Institutions | Countries | Number of Articles Published | Citations |
|---|---|---|---|---|
| 1 | Univ Melbourne | Australia | 24 | 181 |
| 2 | Guangzhou Med Univ | China | 17 | 32 |
| 3 | Kings Coll Hosp London | England | 16 | 215 |
| 4 | Monash Ultrasound Women | Australia | 16 | 99 |
| 5 | Monash Univ | Australia | 16 | 116 |
| 6 | Massachusetts Gen Hosp | USA | 15 | 50 |
| 7 | Murdoch Childrens Res Inst | Australia | 15 | 121 |
| 8 | Mercy Hosp Women | Australia | 14 | 108 |
| 9 | Univ Libre Bruxelles | Belgium | 14 | 153 |
| 10 | Univ Sydney | Australia | 14 | 148 |
| Rank | Countries | Articles | Citations |
|---|---|---|---|
| 1 | USA | 189 | 1012 |
| 2 | China | 82 | 255 |
| 3 | England | 42 | 378 |
| 4 | Australia | 41 | 292 |
| 5 | Spain | 36 | 332 |
| 6 | Belgium | 29 | 240 |
| 7 | Italy | 29 | 226 |
| 8 | France | 24 | 62 |
| 9 | Germany | 19 | 64 |
| 10 | Netherlands | 16 | 134 |
| Rank | Journal | Countries | Publications of Articles |
|---|---|---|---|
| 1 | Prenatal Diagnosis | England | 90 |
| 2 | Ultrasound in Obstetrics & Gynecology | England | 51 |
| 3 | American Journal of Obstetrics and Gynecology | USA | 48 |
| 4 | Journal of Maternal-Fetal & Neonatal Medicine | England | 28 |
| 5 | Fertility and Sterility | USA | 27 |
| 6 | Frontiers in Cell and Developmental Biology | Switzerland | 17 |
| 7 | Reproductive Sciences | USA | 17 |
| 8 | Fetal Diagnosis and Therapy | Switzerland | 15 |
| 9 | European Journal of Obstetrics & Gynecology and Reproductive Biology | Netherland | 13 |
| 10 | Human Reproduction | England | 12 |
| I (Red) | II (Green) | III (Blue) | IV (Yellow) | V (Purple) |
|---|---|---|---|---|
| Prenatal Diagnosis | Aneuploidy | First Trimester | Fetal Fraction | Trisomy 21 |
| Prenatal Screening | Preimplantation Genetic Testing (PGT) | Liquid Biopsy | Microdeletion | Trisomy 13 |
| Non-invasive Prenatal Testing | Blastocyst | Cell-Free DNA | Fetal Trisomy | Twin Pregnancy |
| Nuchal Translucency | Spent Culture Medium | Chromosomal Abnormalities | Fetal Aneuploidy | Trisomy 18 |
| Cell-Free Fetal DNA | Trophectoderm Biopsy | Pre-Eclampsia | Maternal Obesity | Meta-Analysis |
| Ultrasound | Blastocoel Fluid | Biomarker | Follicular Fluid | Combined Test |
| Amniocentesis | IVF | DNA Methylation | Fetal Growth Restriction | |
| Chorionic Villus Sampling | Preimplantation Genetic Screening | Fetal Cell-Free DNA | Placenta | |
| Chromosomal Microarray | Apoptosis | Circulating Tumor DNA | Mitochondria | |
| Genetic Testing | PGT-A | Toll-Like Receptor 9 | Placental Dysfunction |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, W.; Yang, H.; Cheng, W.; Wang, X.; Li, D.; Shi, B. Global Trends in Research on Cell-Free Nucleic Acids in Obstetrics and Gynecology during 2017–2021. J. Clin. Med. 2022, 11, 5545. https://doi.org/10.3390/jcm11195545
Gao W, Yang H, Cheng W, Wang X, Li D, Shi B. Global Trends in Research on Cell-Free Nucleic Acids in Obstetrics and Gynecology during 2017–2021. Journal of Clinical Medicine. 2022; 11(19):5545. https://doi.org/10.3390/jcm11195545
Chicago/Turabian StyleGao, Wenyan, Hongyue Yang, Wanting Cheng, Xiao Wang, Da Li, and Bei Shi. 2022. "Global Trends in Research on Cell-Free Nucleic Acids in Obstetrics and Gynecology during 2017–2021" Journal of Clinical Medicine 11, no. 19: 5545. https://doi.org/10.3390/jcm11195545
APA StyleGao, W., Yang, H., Cheng, W., Wang, X., Li, D., & Shi, B. (2022). Global Trends in Research on Cell-Free Nucleic Acids in Obstetrics and Gynecology during 2017–2021. Journal of Clinical Medicine, 11(19), 5545. https://doi.org/10.3390/jcm11195545






