New Perspectives on Myeloid-Derived Suppressor Cells and Their Emerging Role in Haematology
Abstract
1. Introduction
2. When to Call a Cell “MDSC”?
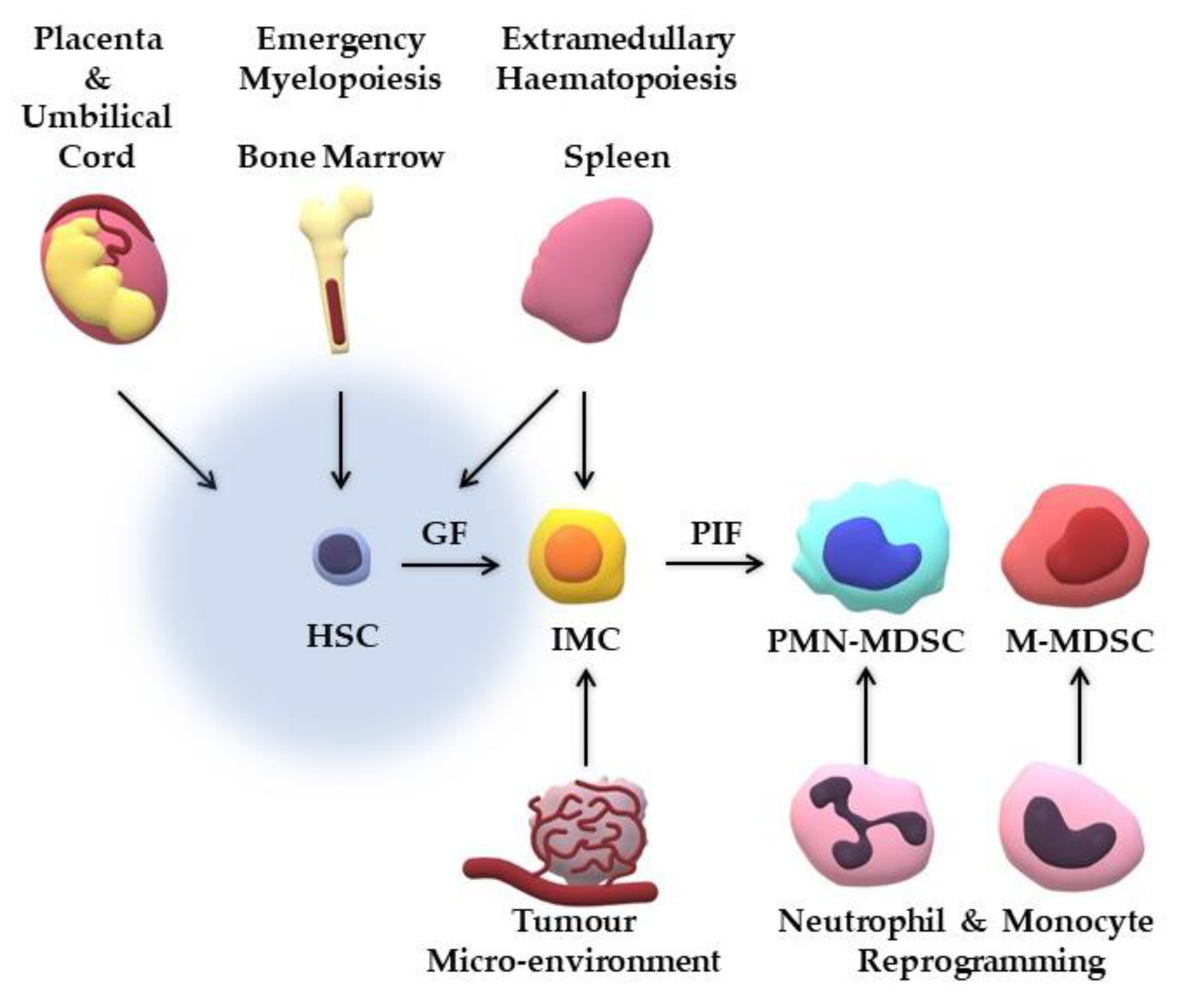
3. Where do MDSCs Originate from?
| Study | Model | Theory | Generated Cells |
|---|---|---|---|
| Park et al. [32] | Human UCB cells cultured with rh-GM-CSF/SCF | MDSCs derived from UCB progenitor cells | Compatible with M-MDSCs |
| Pena et al. [57] | Endotoxin sepsis model; LPS-treated PBMCs | Re-programming of mature monocytes | Compatible with M-MDSCs |
| Singel et al. [58] | Human ovarian cancer cells | Re-programming of mature neutrophils | Compatible with PMN-MDSCs |
| Sinha et al. [59] | Tumour-bearing mice; PGE2-treated BM cells | BM haematopoiesis | Compatible with MDSCs |
| Song et al. [60] | Tumour-bearing mice | Extramedullary haematopoiesis | Compatible with MDSCs |
| Wynn [55] | Tumour-bearing mice | M-MDCs transformation to PMN-MDSCs | Compatible with PMN-MDSCs |
| Yu et al. [61] | Human UCB cells co-cultured with human breast cancer cells | MDSCs derived from UCB progenitor cells | Compatible with eMDSCs |
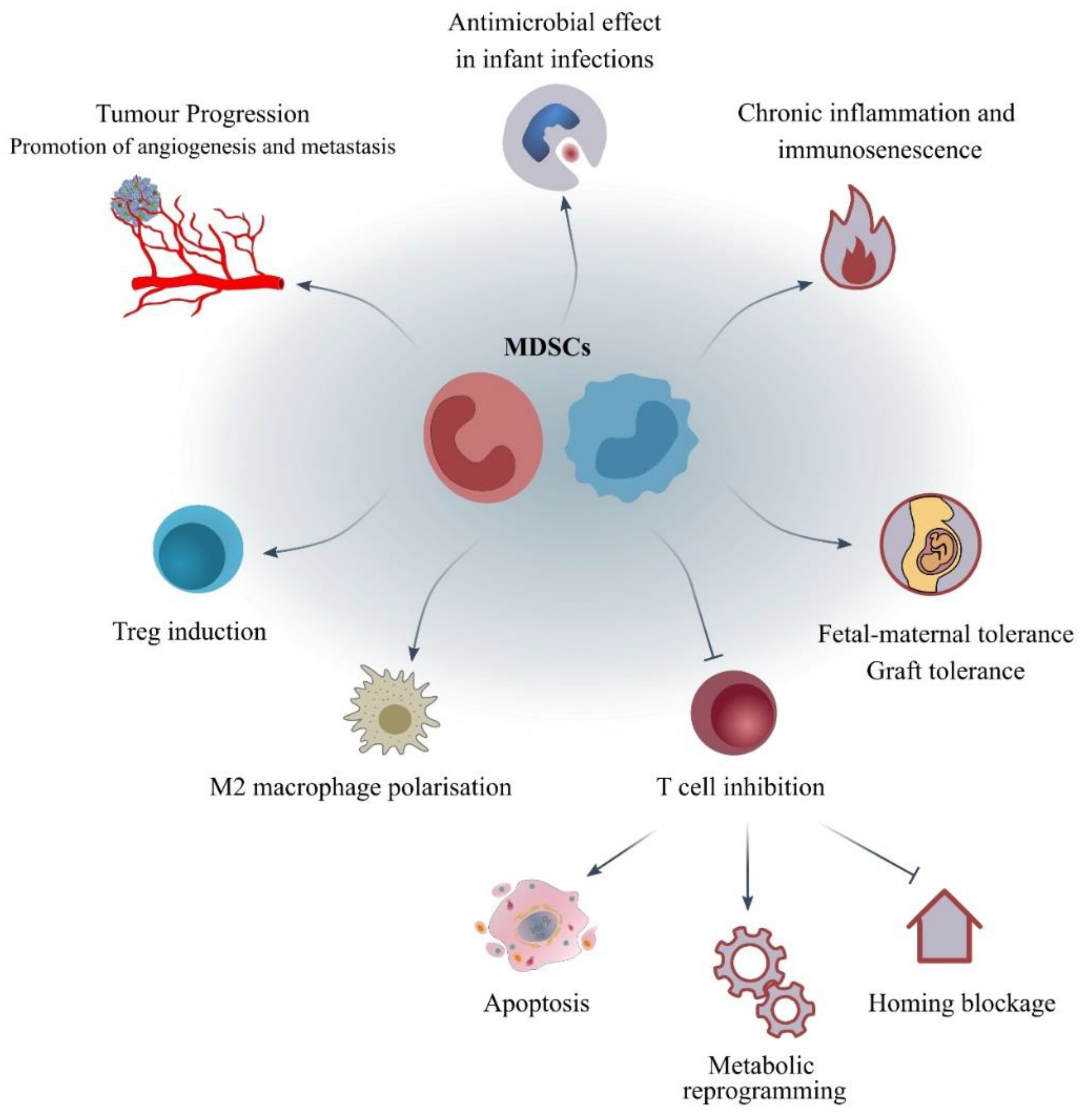
4. What Are the Mechanisms of Action and the Molecular Pathways Correlated to MDSCs?
| Mechanisms of Action of MDSCs | Pathways/Molecules | Cells Affected/Impacts |
|---|---|---|
| Energy metabolic reprogramming | Arg-1 and iNOS: deprivation of Arg, IDO1: deprivation of Trp, Cys importer: deprivation of Cys | Deprivation of metabolites leading to T-cell suppression [14,68] |
| Autophagy | mTORC1, STAT3 | Induction of the immaturity, suppressive character and accumulation of MDSCs [8,79] |
| Extracellular vesicle (EV) cargo | Exosome formation containing: miRNAs, mRNAs, dsDNA | Tumour development and evasion [64,65] |
| Induction of immunosuppressive cells | IFN-γ, IL-10, TGF-β | Induction of T-regs, polarisation of M2-like phenotype of macrophages [66] |
| Oxidative stress environment | iNOS: production of NO; eNOS: production of peroxynitrites; NOX: production of ROS | T-cell apoptosis [66,67] |
| T-cell homing blockage | ADAM17: a metalloprotease that cleaves L-selectin (CD62L) | Impairment of T-cell actions [66] |
| Angiogenesis and metastasis | MMP9, CCL2, S100A8/9, VEGF, BV8 | Formation of the pre-metastatic niche [63] |
| Expression of negative immune checkpoint molecules | PD-L1/PD-1 | T-cell suppression in a contact-dependent manner [73,76] |
| Expression of ectoenzymes | Adenosine | T-cell suppression [66] |
5. How Is the Phenotypic and Functional Analysis of Human MDSCs Performed?
5.1. Phenotypic Analysis
5.2. Functional Analysis
6. Which Are the Key Points of Studying MDSCs in Haematology?
6.1. Increased MDSCs: Haematologic Malignancies and Myelodysplasia
6.2. Decreased MDSCs: Immune-Mediated Cytopenias
6.3. MDSCs versus Mesenchymal Stem Cells (MSCs)
6.4. Umbilical Cord Blood Subsets
7. What Are the Potential Clinical Applications of MDSCs in Haematology?
7.1. MDSCs as Biomarkers
7.2. MDSCs as Therapeutic Targets
7.3. Graft-versus-Host Disease
8. Synopsis and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gabrilovich, D.I. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, T.; Shao, S.; Shi, B.; Zhao, Y. Phenotype, Development, and Biological Function of Myeloid-Derived Suppressor Cells. Oncoimmunology 2016, 5, e1004983. [Google Scholar] [CrossRef] [PubMed]
- Pastuła, A.; Marcinkiewicz, J. Myeloid-Derived Suppressor Cells: A Double-Edged Sword? Int. J. Exp. Pathol. 2011, 92, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Budhwar, S.; Verma, P.; Verma, R.; Rai, S.; Singh, K. The Yin and Yang of Myeloid Derived Suppressor Cells. Front. Immunol. 2018, 9, 2776. [Google Scholar] [CrossRef] [PubMed]
- Birbrair, A. (Ed.) Tumor Microenvironment; Springer International Publishing: Cham, Switzerland, 2020; Volume 1224, ISBN 978-3-030-35722-1. [Google Scholar]
- Winfield, R.D.; Delano, M.J.; Pande, K.; Scumpia, P.O.; LaFace, D.; Moldawer, L.L. Myeloid-Derived Suppressor Cells in Cancer Cachexia Syndrome: A New Explanation for an Old Problem. J. Parenter. Enter. Nutr. 2008, 32, 651–655. [Google Scholar] [CrossRef]
- Geis-Asteggiante, L.; Belew, A.T.; Clements, V.K.; Edwards, N.J.; Ostrand-Rosenberg, S.; El-Sayed, N.M.; Fenselau, C. Differential Content of Proteins, MRNAs, and MiRNAs Suggests That MDSC and Their Exosomes May Mediate Distinct Immune Suppressive Functions. J. Proteome Res. 2018, 17, 486–498. [Google Scholar] [CrossRef]
- Alissafi, T.; Hatzioannou, A.; Mintzas, K.; Barouni, R.M.; Banos, A.; Sormendi, S.; Polyzos, A.; Xilouri, M.; Wielockx, B.; Gogas, H.; et al. Autophagy Orchestrates the Regulatory Program of Tumor-Associated Myeloid-Derived Suppressor Cells. J. Clin. Investig. 2018, 128, 3840–3852. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Immunosenescence: The Potential Role of Myeloid-Derived Suppressor Cells (MDSC) in Age-Related Immune Deficiency. Cell. Mol. Life Sci. 2019, 76, 1901–1918. [Google Scholar] [CrossRef]
- Salminen, A.; Kauppinen, A.; Kaarniranta, K. AMPK Activation Inhibits the Functions of Myeloid-Derived Suppressor Cells (MDSC): Impact on Cancer and Aging. J. Mol. Med. 2019, 97, 1049–1064. [Google Scholar] [CrossRef]
- Salminen, A.; Kauppinen, A.; Kaarniranta, K. Myeloid-Derived Suppressor Cells (MDSC): An Important Partner in Cellular/Tissue Senescence. Biogerontology 2018, 19, 325–339. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. The Role of Myeloid-Derived Suppressor Cells (MDSC) in the Inflammaging Process. Ageing Res. Rev. 2018, 48, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Umemura, N.; Sugimoto, M.; Kitoh, Y.; Saio, M.; Sakagami, H. Metabolomic Profiling of Tumor-Infiltrating Macrophages during Tumor Growth. Cancer Immunol. Immunother. 2020, 69, 2357–2369. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Pang, B.; Lin, G.; Zhen, Y.; Yi, H. Energy Metabolism Manipulates the Fate and Function of Tumour Myeloid-Derived Suppressor Cells. Br. J. Cancer 2020, 122, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, N.; Tan, H.; Chueng, F.; Zhang, Z.; Yuen, M.; Feng, Y. Modulation of Gut Microbiota Mediates Berberine-induced Expansion of Immuno-suppressive Cells to against Alcoholic Liver Disease. Clin. Transl. Med. 2020, 10, e112. [Google Scholar] [CrossRef]
- Zheng, Z.M.; Yang, H.L.; Lai, Z.Z.; Wang, C.J.; Yang, S.L.; Li, M.Q.; Shao, J. Myeloid-Derived Suppressor Cells in Obstetrical and Gynecological Diseases. Am. J. Reprod. Immunol. 2020, 84, e13266. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, M.; Mohammadi, M.; Ali-Hassanzadeh, M.; Zare, M.; Gharesi-Fard, B. MDSCs in Pregnancy: Critical Players for a Balanced Immune System at the Feto-Maternal Interface. Cell. Immunol. 2019, 346, 103990. [Google Scholar] [CrossRef]
- Köstlin, N.; Hofstädter, K.; Ostermeir, A.-L.; Spring, B.; Leiber, A.; Haen, S.; Abele, H.; Bauer, P.; Pollheimer, J.; Hartl, D.; et al. Granulocytic Myeloid-Derived Suppressor Cells Accumulate in Human Placenta and Polarize toward a Th2 Phenotype. J. Immunol. 2015, 196, 1132–1145. [Google Scholar] [CrossRef]
- Weber, R.; Umansky, V. Fighting Infant Infections with Myeloid-Derived Suppressor Cells. J. Clin. Investig. 2019, 129, 4080–4082. [Google Scholar] [CrossRef]
- He, Y.-M.; Li, X.; Perego, M.; Nefedova, Y.; Kossenkov, A.V.; Jensen, E.A.; Kagan, V.E.; Liu, Y.-F.; Fu, S.-Y.; Ye, Q.-J.; et al. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. Nat. Med. 2018, 24, 224–231. [Google Scholar] [CrossRef]
- Schrijver, I.T.; Théroude, C.; Roger, T. Myeloid-Derived Suppressor Cells in Sepsis. Front. Immunol. 2019, 10, 327. [Google Scholar] [CrossRef]
- Agrati, C.; Sacchi, A.; Bordoni, V.; Cimini, E.; Notari, S.; Grassi, G.; Casetti, R.; Tartaglia, E.; Lalle, E.; D’Abramo, A.; et al. Expansion of Myeloid-Derived Suppressor Cells in Patients with Severe Coronavirus Disease (COVID-19). Cell Death Differ. 2020, 27, 3196–3207. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I. All Myeloid-Derived Suppressor Cells Are Not Created Equal: How Gender Inequality Influences These Cells and Affects Cancer Therapy. Cancer Discov. 2020, 10, 1100–1102. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Brandau, S.; Chen, S.-H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for Myeloid-Derived Suppressor Cell Nomenclature and Characterization Standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef]
- Bruger, A.M.; Dorhoi, A.; Esendagli, G.; Barczyk-Kahlert, K.; van der Bruggen, P.; Lipoldova, M.; Perecko, T.; Santibanez, J.; Saraiva, M.; van Ginderachter, J.A.; et al. How to Measure the Immunosuppressive Activity of MDSC: Assays, Problems and Potential Solutions. Cancer Immunol. Immunother. 2018, 68, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Condamine, T.; Mastio, J.; Gabrilovich, D.I. Transcriptional Regulation of Myeloid-Derived Suppressor Cells. J. Leukoc. Biol. 2015, 98, 913–922. [Google Scholar] [CrossRef]
- Si, Y.; Merz, S.F.; Jansen, P.; Wang, B.; Bruderek, K.; Altenhoff, P.; Mattheis, S.; Lang, S.; Gunzer, M.; Klode, J.; et al. Multidimensional Imaging Provides Evidence for Down-Regulation of T Cell Effector Function by MDSC in Human Cancer Tissue. Sci. Immunol. 2019, 4, eaaw9159. [Google Scholar] [CrossRef]
- Bizymi, N.; Bjelica, S.; Kittang, A.O.; Mojsilovic, S.; Velegraki, M.; Pontikoglou, C.; Roussel, M.; Ersvær, E.; Santibañez, J.F.; Lipoldová, M.; et al. Myeloid-Derived Suppressor Cells in Hematologic Diseases: Promising Biomarkers and Treatment Targets. HemaSphere 2019, 3, e168. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Bronte, V.; Chen, S.-H.; Colombo, M.P.; Ochoa, A.; Ostrand-Rosenberg, S.; Schreiber, H. The Terminology Issue for Myeloid-Derived Suppressor Cells. Cancer Res. 2007, 67, 425–426. [Google Scholar] [CrossRef]
- Yang, R.; Roden, R.B.S. Re: The Terminology Issue for Myeloid-Derived Suppressor Cells. Cancer Res. 2007, 67, 426. [Google Scholar] [CrossRef]
- Millrud, C.R.; Bergenfelz, C.; Leandersson, K. On the Origin of Myeloid-Derived Suppressor Cells. Oncotarget 2017, 8, 3649–3665. [Google Scholar] [CrossRef]
- Park, M.-Y.; Lim, B.-G.; Kim, S.; Sohn, H.-J.; Kim, T.-G. GM-CSF Promotes the Expansion and Differentiation of Cord Blood Myeloid-Derived Suppressor Cells, Which Attenuate Xenogeneic Graft-vs.-Host Disease. Front. Immunol. 2019, 10, 183. [Google Scholar] [CrossRef] [PubMed]
- Zoso, A.; Mazza, E.M.C.; Bicciato, S.; Mandruzzato, S.; Bronte, V.; Serafini, P.; Inverardi, L. Human Fibrocytic Myeloid-Derived Suppressor Cells Express IDO and Promote Tolerance via Treg-Cell Expansion. Eur. J. Immunol. 2014, 44, 3307–3319. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-Y.; Ryu, D.-B.; Park, M.-Y.; Lee, S.-E.; Park, G.; Kim, T.-G.; Min, C.-K. Ex Vivo Generated Human Cord Blood Myeloid-Derived Suppressor Cells Attenuate Murine Chronic Graft-versus-Host Diseases. Biol. Blood Marrow Transplant. 2018, 24, 2381–2396. [Google Scholar] [CrossRef]
- Wu, W.-C.; Sun, H.-W.; Chen, H.-T.; Liang, J.; Yu, X.-J.; Wu, C.; Wang, Z.; Zheng, L. Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients. Proc. Natl. Acad. Sci. USA 2014, 111, 4221–4226. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Hua, Q.; Zheng, L. Generation of Myeloid Cells in Cancer: The Spleen Matters. Front. Immunol. 2020, 11, 1126. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-Derived Suppressor Cells as Regulators of the Immune System. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Strauss, L.; Sangaletti, S.; Consonni, F.M.; Szebeni, G.; Morlacchi, S.; Totaro, M.G.; Porta, C.; Anselmo, A.; Tartari, S.; Doni, A.; et al. RORC1 Regulates Tumor-Promoting “Emergency” Granulo-Monocytopoiesis. Cancer Cell 2015, 28, 253–269. [Google Scholar] [CrossRef]
- Corzo, C.A.; Condamine, T.; Lu, L.; Cotter, M.J.; Youn, J.-I.; Cheng, P.; Cho, H.-I.; Celis, E.; Quiceno, D.G.; Padhya, T.; et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J. Exp. Med. 2010, 207, 2439–2453. [Google Scholar] [CrossRef]
- Zhang, Y.; Qu, D.; Sun, J.; Zhao, L.; Wang, Q.-J.; Shao, Q.; Kong, B.; Qu, X. Human trophoblast cells induced MDSCs from peripheral blood CD14+ myelomonocytic cells via elevated levels of CCL2. Cell. Mol. Immunol. 2015, 13, 615–627. [Google Scholar] [CrossRef]
- Bergenfelz, C.; Larsson, A.M.; von Stedingk, K.; Gruvberger-Saal, S.; Aaltonen, K.; Jansson, S.; Jernström, H.; Janols, H.; Wullt, M.; Bredberg, A.; et al. Systemic Monocytic-MDSCs Are Generated from Monocytes and Correlate with Disease Progression in Breast Cancer Patients. PLoS ONE 2015, 10, e0127028. [Google Scholar] [CrossRef]
- Kumar, V.; Cheng, P.; Condamine, T.; Mony, S.; Languino, L.R.; McCaffrey, J.C.; Hockstein, N.; Guarino, M.; Masters, G.; Penman, E.; et al. CD45 Phosphatase Inhibits STAT3 Transcription Factor Activity in Myeloid Cells and Promotes Tumor-Associated Macrophage Differentiation. Immunity 2016, 44, 303–315. [Google Scholar] [CrossRef] [PubMed]
- de Vlaeminck, Y.; González-Rascón, A.; Goyvaerts, C.; Breckpot, K. Cancer-Associated Myeloid Regulatory Cells. Front. Immunol. 2016, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J.; van Waes, C.; Allen, C.T. Overcoming Barriers to Effective Immunotherapy: MDSCs, TAMs, and Tregs as Mediators of the Immunosuppressive Microenvironment in Head and Neck Cancer. Oral Oncol. 2016, 58, 59–70. [Google Scholar] [CrossRef]
- Okła, K.; Wertel, I.; Polak, G.; Surówka, J.; Wawruszak, A.; Kotarski, J. Tumor-Associated Macrophages and Myeloid-Derived Suppressor Cells as Immunosuppressive Mechanism in Ovarian Cancer Patients: Progress and Challenges. Int. Rev. Immunol. 2016, 35, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Ugel, S.; de Sanctis, F.; Mandruzzato, S.; Bronte, V. Tumor-Induced Myeloid Deviation: When Myeloid-Derived Suppressor Cells Meet Tumor-Associated Macrophages. J. Clin. Investig. 2015, 125, 3365–3376. [Google Scholar] [CrossRef] [PubMed]
- Tcyganov, E.; Mastio, J.; Chen, E.; Gabrilovich, D.I. Plasticity of Myeloid-Derived Suppressor Cells in Cancer. Curr. Opin. Immunol. 2018, 51, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Pillay, J.; Tak, T.; Kamp, V.M.; Koenderman, L. Immune Suppression by Neutrophils and Granulocytic Myeloid-Derived Suppressor Cells: Similarities and Differences. Cell. Mol. Life Sci. 2013, 70, 3813–3827. [Google Scholar] [CrossRef]
- Bergenfelz, C.; Leandersson, K. The Generation and Identity of Human Myeloid-Derived Suppressor Cells. Front. Oncol. 2020, 10, 109. [Google Scholar] [CrossRef]
- Wynn, T.A. Myeloid-Cell Differentiation Redefined in Cancer. Nat. Immunol. 2013, 14, 197–199. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of Tumor-Associated Neutrophil Phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef]
- Pena, O.M.; Pistolic, J.; Raj, D.; Fjell, C.D.; Hancock, R.E.W. Endotoxin Tolerance Represents a Distinctive State of Alternative Polarization (M2) in Human Mononuclear Cells. J. Immunol. 2011, 186, 7243–7254. [Google Scholar] [CrossRef] [PubMed]
- Singel, K.L.; Emmons, T.R.; Khan, A.N.H.; Mayor, P.C.; Shen, S.; Wong, J.T.; Morrell, K.; Eng, K.H.; Mark, J.; Bankert, R.B.; et al. Mature Neutrophils Suppress T Cell Immunity in Ovarian Cancer Microenvironment. JCI Insight 2019, 4, e122311. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Clements, V.K.; Fulton, A.M.; Ostrand-Rosenberg, S. Prostaglandin E2 Promotes Tumor Progression by Inducing Myeloid-Derived Suppressor Cells. Cancer Res. 2007, 67, 4507–4513. [Google Scholar] [CrossRef]
- Song, X.; Krelin, Y.; Dvorkin, T.; Bjorkdahl, O.; Segal, S.; Dinarello, C.A.; Voronov, E.; Apte, R.N. CD11b + /Gr-1 + Immature Myeloid Cells Mediate Suppression of T Cells in Mice Bearing Tumors of IL-1β-Secreting Cells. J. Immunol. 2005, 175, 8200–8208. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Du, W.; Yan, F.; Wang, Y.; Li, H.; Cao, S.; Yu, W.; Shen, C.; Liu, J.; Ren, X. Myeloid-Derived Suppressor Cells Suppress Antitumor Immune Responses through IDO Expression and Correlate with Lymph Node Metastasis in Patients with Breast Cancer. J. Immunol. 2013, 190, 3783–3797. [Google Scholar] [CrossRef]
- Vanhaver, C.; van der Bruggen, P.; Bruger, A.M. MDSC in Mice and Men: Mechanisms of Immunosuppression in Cancer. J. Clin. Med. 2021, 10, 2872. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, Y.; Guo, N.; Wang, S. MDSCs: Key Criminals of Tumor Pre-Metastatic Niche Formation. Front. Immunol. 2019, 10, 172. [Google Scholar] [CrossRef]
- Sonda, N.; Simonato, F.; Peranzoni, E.; Calì, B.; Bortoluzzi, S.; Bisognin, A.; Wang, E.; Marincola, F.M.; Naldini, L.; Gentner, B.; et al. MiR-142-3p Prevents Macrophage Differentiation during Cancer-Induced Myelopoiesis. Immunity 2013, 38, 1236–1249. [Google Scholar] [CrossRef]
- Li, L.; Zhang, J.; Diao, W.; Wang, D.; Wei, Y.; Zhang, C.-Y.; Zen, K. MicroRNA-155 and MicroRNA-21 Promote the Expansion of Functional Myeloid-Derived Suppressor Cells. J. Immunol. 2014, 192, 1034–1043. [Google Scholar] [CrossRef]
- Groth, C.; Hu, X.; Weber, R.; Fleming, V.; Altevogt, P.; Utikal, J.; Umansky, V. Immunosuppression Mediated by Myeloid-Derived Suppressor Cells (MDSCs) during Tumour Progression. Br. J. Cancer 2019, 120, 16–25. [Google Scholar] [CrossRef]
- Raber, P.L.; Thevenot, P.; Sierra, R.; Wyczechowska, D.; Halle, D.; Ramirez, M.E.; Ochoa, A.C.; Fletcher, M.; Velasco, C.; Wilk, A.; et al. Subpopulations of Myeloid-Derived Suppressor Cells Impair T Cell Responses through Independent Nitric Oxide-Related Pathways. Int. J. Cancer 2014, 134, 2853–2864. [Google Scholar] [CrossRef] [PubMed]
- Al-Khami, A.A.; Rodriguez, P.C.; Ochoa, A.C. Energy Metabolic Pathways Control the Fate and Function of Myeloid Immune Cells. J. Leukoc. Biol. 2017, 102, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Cramer, T.; Yamanishi, Y.; Clausen, B.E.; Förster, I.; Pawlinski, R.; Mackman, N.; Haase, V.H.; Jaenisch, R.; Corr, M.; Nizet, V.; et al. HIF-1α Is Essential for Myeloid Cell-Mediated Inflammation. Cell 2003, 112, 645–657. [Google Scholar] [CrossRef]
- Palmer, C.S.; Cherry, C.L.; Sada-Ovalle, I.; Singh, A.; Crowe, S.M. Glucose Metabolism in T Cells and Monocytes: New Perspectives in HIV Pathogenesis. eBioMedicine 2016, 6, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Ratter, J.M.; van Heck, J.I.P.; Rooijackers, H.M.M.; Jansen, H.J.; van Poppel, P.C.M.; Tack, C.J.; Stienstra, R. Insulin Acutely Activates Metabolism of Primary Human Monocytes and Promotes a Proinflammatory Phenotype. J. Leukoc. Biol. 2021, 110, 885–891. [Google Scholar] [CrossRef]
- de Sanctis, F.; Solito, S.; Ugel, S.; Molon, B.; Bronte, V.; Marigo, I. MDSCs in Cancer: Conceiving New Prognostic and Therapeutic Targets. Biochim. Biophys. Acta Rev. Cancer 2016, 1865, 35–48. [Google Scholar] [CrossRef]
- Vetsika, E.K.; Koukos, A.; Kotsakis, A. Myeloid-Derived Suppressor Cells: Major Figures That Shape the Immunosuppressive and Angiogenic Network in Cancer. Cells 2019, 8, 1647. [Google Scholar] [CrossRef]
- Lang, S.; Bruderek, K.; Kaspar, C.; Höing, B.; Kanaan, O.; Dominas, N.; Hussain, T.; Droege, F.; Eyth, C.; Hadaschik, B.; et al. Clinical Relevance and Suppressive Capacity of Human Myeloid-Derived Suppressor Cell Subsets. Clin. Cancer Res. 2018, 24, 4834–4844. [Google Scholar] [CrossRef]
- Cassetta, L.; Bruderek, K.; Skrzeczynska-Moncznik, J.; Osiecka, O.; Hu, X.; Rundgren, I.M.; Lin, A.; Santegoets, K.; Horzum, U.; Godinho-Santos, A.; et al. Differential Expansion of Circulating Human MDSC Subsets in Patients with Cancer, Infection and Inflammation. J. Immunother. Cancer 2020, 8, e001223. [Google Scholar] [CrossRef]
- Ballbach, M.; Dannert, A.; Singh, A.; Siegmund, D.M.; Handgretinger, R.; Piali, L.; Rieber, N.; Hartl, D. Expression of Checkpoint Molecules on Myeloid-Derived Suppressor Cells. Immunol. Lett. 2017, 192, 1–6. [Google Scholar] [CrossRef]
- Amaravadi, R.; Kimmelman, A.C.; White, E. Recent Insights into the Function of Autophagy in Cancer. Genes Dev. 2016, 30, 1913–1930. [Google Scholar] [CrossRef] [PubMed]
- Onorati, A.V.; Dyczynski, M.; Ojha, R.; Amaravadi, R.K. Targeting autophagy in cancer. Cancer 2018, 124, 3307–3318. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Si, C.; Zhang, Q.; Yan, F.; Li, C.; Zhang, H.; Ma, Q.; Dai, J.; Li, Z.; Shi, H.; et al. Autophagy Regulates Accumulation and Functional Activity of Granulocytic Myeloid-Derived Suppressor Cells via STAT3 Signaling in Endotoxin Shock. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2796–2807. [Google Scholar] [CrossRef]
- Leiber, A.; Schwarz, J.; Köstlin, N.; Spring, B.; Fehrenbach, B.; Katava, N.; Poets, C.F.; Gille, C. Neonatal Myeloid Derived Suppressor Cells Show Reduced Apoptosis and Immunosuppressive Activity upon Infection with Escherichia Coli. Eur. J. Immunol. 2017, 47, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Mandruzzato, S.; Brandau, S.; Britten, C.M.; Bronte, V.; Damuzzo, V.; Gouttefangeas, C.; Maurer, D.; Ottensmeier, C.; van der Burg, S.H.; Welters, M.J.P.; et al. Toward Harmonized Phenotyping of Human Myeloid-Derived Suppressor Cells by Flow Cytometry: Results from an Interim Study. Cancer Immunol. Immunother. 2016, 65, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Baekkevold, E.S.; Brandau, S.; Bujko, A.; Cassatella, M.A.; Dorhoi, A.; Krieg, C.; Lin, A.; Loré, K.; Marini, O.; et al. Deciphering Myeloid-Derived Suppressor Cells: Isolation and Markers in Humans, Mice and Non-Human Primates. Cancer Immunol. Immunother. 2019, 68, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Chesney, J.A.; Mitchell, R.A.; Yaddanapudi, K. Myeloid-derived Suppressor Cells—a New Therapeutic Target to Overcome Resistance to Cancer Immunotherapy. J. Leukoc. Biol. 2017, 102, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Kotsakis, A.; Harasymczuk, M.; Schilling, B.; Georgoulias, V.; Argiris, A.; Whiteside, T.L. Myeloid-Derived Suppressor Cell Measurements in Fresh and Cryopreserved Blood Samples. J. Immunol. Methods 2012, 381, 14–22. [Google Scholar] [CrossRef]
- Mishalian, I.; Granot, Z.; Fridlender, Z.G. The Diversity of Circulating Neutrophils in Cancer. Immunobiology 2017, 222, 82–88. [Google Scholar] [CrossRef]
- Condamine, T.; Dominguez, G.A.; Youn, J.-I.; Kossenkov, A.V.; Mony, S.; Alicea-Torres, K.; Tcyganov, E.; Hashimoto, A.; Nefedova, Y.; Lin, C.; et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 2016, 1, aaf8943. [Google Scholar] [CrossRef]
- Umansky, V.; Adema, G.J.; Baran, J.; Brandau, S.; Van Ginderachter, J.A.; Hu, X.; Jablonska, J.; Mojsilovic, S.; Papadaki, H.A.; de Coaña, Y.P.; et al. Interactions among myeloid regulatory cells in cancer. Cancer Immunol. Immunother. 2018, 68, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Apodaca, M.C.; Wright, A.E.; Riggins, A.M.; Harris, W.P.; Yeung, R.S.; Yu, L.; Morishima, C. Characterization of a Whole Blood Assay for Quantifying Myeloid-Derived Suppressor Cells. J. Immunother. Cancer 2019, 7, 230. [Google Scholar] [CrossRef] [PubMed]
- Mazza, E.M.C.; Zoso, A.; Mandruzzato, S.; Bronte, V.; Serafini, P.; Inverardi, L.; Bicciato, S. Gene Expression Profiling of Human Fibrocytic Myeloid-Derived Suppressor Cells (f-MDSCs). Genom. Data 2014, 2, 389–392. [Google Scholar] [CrossRef]
- Lewis, D.B. Maturing of the fetal and neonatal immune system. In Clinical Immunology, Principles and Practice (Expert Consult-Online and Print), 4: Clinical Immunology; Elisevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Naeim, F.; Rao, P.N.; Song, S.X.; Phan, R.T. Principles of Immunophenotyping. In Atlas of Hematopathology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 29–56. [Google Scholar]
- Hernández-Caselles, T.; Martínez-Esparza, M.; Pérez-Oliva, A.B.; Quintanilla-Cecconi, A.M.; García-Alonso, A.; Alvarez-López, D.M.R.; García-Peñarrubia, P. A Study of CD33 (SIGLEC-3) Antigen Expression and Function on Activated Human T and NK Cells: Two Isoforms of CD33 Are Generated by Alternative Splicing. J. Leukoc. Biol. 2006, 79, 46–58. [Google Scholar] [CrossRef]
- Yoon, J.; Terada, A.; Kita, H. CD66b Regulates Adhesion and Activation of Human Eosinophils. J. Immunol. 2007, 179, 8454–8462. [Google Scholar] [CrossRef] [PubMed]
- Horzum, U.; Yoyen-Ermis, D.; Taskiran, E.Z.; Yilmaz, K.B.; Hamaloglu, E.; Karakoc, D.; Esendagli, G. CD66b+ Monocytes Represent a Proinflammatory Myeloid Subpopulation in Cancer. Cancer Immunol. Immunother. 2021, 70, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Law, A.M.K.; Valdes-Mora, F.; Gallego-Ortega, D. Myeloid-Derived Suppressor Cells as a Therapeutic Target for Cancer. Cells 2020, 9, 561. [Google Scholar] [CrossRef]
- Woodford-Thomas, T.; Thomas, M.L. The Leukocyte Common Antigen, CD45 and Other Protein Tyrosine Phosphatases in Hematopoietic Cells. Semin. Cell Biol. 1993, 4, 409–418. [Google Scholar] [CrossRef]
- Tesfaigzi, Y.; Daheshia, M. CD14. In Encyclopedia of Respiratory Medicine; Elsevier: Amsterdam, The Netherlands, 2006; pp. 343–347. [Google Scholar]
- Zhang, J.; Xu, X.; Shi, M.; Chen, Y.; Yu, D.; Zhao, C.; Gu, Y.; Yang, B.; Guo, S.; Ding, G.; et al. CD13hi Neutrophil-like Myeloid-Derived Suppressor Cells Exert Immune Suppression through Arginase 1 Expression in Pancreatic Ductal Adenocarcinoma. Oncoimmunology 2017, 6, e1258504. [Google Scholar] [CrossRef]
- Dondossola, E.; Rangel, R.; Guzman-Rojas, L.; Barbu, E.M.; Hosoya, H.; John, L.S.; Molldrem, J.J.; Corti, A.; Sidman, R.L.; Arap, W.; et al. CD13-Positive Bone Marrow-Derived Myeloid Cells Promote Angiogenesis, Tumor Growth, and Metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 20717–20722. [Google Scholar] [CrossRef]
- Shen, J.; Chen, X.; Wang, Z.; Zhang, G.; Chen, W. Downregulation of CD40 Expression Contributes to the Accumulation of Myeloid-Derived Suppressor Cells in Gastric Tumors. Oncol. Lett. 2014, 8, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-I.; Nagaraj, S.; Collazo, M.; Gabrilovich, D.I. Subsets of Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. J. Immunol. 2008, 181, 5791–5802. [Google Scholar] [CrossRef] [PubMed]
- Kiss, M.; van Gassen, S.; Movahedi, K.; Saeys, Y.; Laoui, D. Myeloid Cell Heterogeneity in Cancer: Not a Single Cell Alike. Cell. Immunol. 2018, 330, 188–201. [Google Scholar] [CrossRef]
- van Roy, F.; Berx, G. The Cell-Cell Adhesion Molecule E-Cadherin. Cell. Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef] [PubMed]
- Colin, D.J.; Cottet-Dumoulin, D.; Faivre, A.; Germain, S.; Triponez, F.; Serre-Beinier, V. Experimental Model of Human Malignant Mesothelioma in Athymic Mice. Int. J. Mol. Sci. 2018, 19, 1881. [Google Scholar] [CrossRef] [PubMed]
- Tarrant, J. Emerging Translatable Safety Biomarkers. In Comprehensive Medicinal Chemistry III; Elsevier: Amsterdam, The Netherlands, 2017; pp. 255–284. [Google Scholar]
- Ray, P.D.; Huang, B.-W.; Tsuji, Y. Reactive Oxygen Species (ROS) Homeostasis and Redox Regulation in Cellular Signaling. Cell. Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef]
- Sinha, P.; Okoro, C.; Foell, D.; Freeze, H.H.; Ostrand-Rosenberg, S.; Srikrishna, G. Proinflammatory S100 Proteins Regulate the Accumulation of Myeloid-Derived Suppressor Cells. J. Immunol. 2008, 181, 4666–4675. [Google Scholar] [CrossRef]
- Awasthi, N.; Liongue, C.; Ward, A.C. STAT Proteins: A Kaleidoscope of Canonical and Non-Canonical Functions in Immunity and Cancer. J. Hematol. Oncol. 2021, 14, 198. [Google Scholar] [CrossRef]
- Waight, J.D.; Netherby, C.; Hensen, M.L.; Miller, A.; Hu, Q.; Liu, S.; Bogner, P.N.; Farren, M.R.; Lee, K.P.; Liu, K.; et al. Myeloid-Derived Suppressor Cell Development Is Regulated by a STAT/IRF-8 Axis. J. Clin. Investig. 2013, 123, 4464–4478. [Google Scholar] [CrossRef]
- Marigo, I.; Bosio, E.; Solito, S.; Mesa, C.; Fernandez, A.; Dolcetti, L.; Ugel, S.; Sonda, N.; Bicciato, S.; Falisi, E.; et al. Tumor-Induced Tolerance and Immune Suppression Depend on the C/EBPβ Transcription Factor. Immunity 2010, 32, 790–802. [Google Scholar] [CrossRef]
- Agresta, L.; Hoebe, K.H.N.; Janssen, E.M. The Emerging Role of CD244 Signaling in Immune Cells of the Tumor Microenvironment. Front. Immunol. 2018, 9, 2809. [Google Scholar] [CrossRef] [PubMed]
- Onofre, G.; Kolácková, M.; Jankovicová, K.; Krejsek, J. Scavenger Receptor CD163 and Its Biological Functions. Acta Med. 2009, 52, 57–61. [Google Scholar] [CrossRef]
- Ryzhov, S.; Novitskiy, S.V.; Goldstein, A.E.; Biktasova, A.; Blackburn, M.R.; Biaggioni, I.; Dikov, M.M.; Feoktistov, I. Adenosinergic Regulation of the Expansion and Immunosuppressive Activity of CD11b+Gr1+Cells. J. Immunol. 2011, 187, 6120–6129. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; De Tenbossche, C.G.P.; Canè, S.; Colau, D.; Van Baren, N.; Lurquin, C.; Schmitt-Verhulst, A.-M.; Liljeström, P.; Uyttenhove, C.; Van den Eynde, B.J. Resistance to cancer immunotherapy mediated by apoptosis of tumor-infiltrating lymphocytes. Nat. Commun. 2017, 8, 1404. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Wick, W.; van den Eynde, B.J. Tryptophan Catabolism in Cancer: Beyond IDO and Tryptophan Depletion. Cancer Res. 2012, 72, 5435–5440. [Google Scholar] [CrossRef] [PubMed]
- Lašťovička, J.; Rataj, M.; Bartůňková, J. Assessment of Lymphocyte Proliferation for Diagnostic Purpose: Comparison of CFSE Staining, Ki-67 Expression and 3H-Thymidine Incorporation. Hum. Immunol. 2016, 77, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Solito, S.; Pinton, L.; de Sanctis, F.; Ugel, S.; Bronte, V.; Mandruzzato, S.; Marigo, I. Methods to Measure MDSC Immune Suppressive Activity In Vitro and In Vivo. Curr. Protoc. Immunol. 2019, 124, e61. [Google Scholar] [CrossRef]
- Bruger, A.M.; Vanhaver, C.; Bruderek, K.; Amodio, G.; Tavukçuoğlu, E.; Esendagli, G.; Gregori, S.; Brandau, S.; van der Bruggen, P. Protocol to Assess the Suppression of T-Cell Proliferation by Human MDSC. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 632, pp. 155–192. ISBN 9780128186756. [Google Scholar]
- Wang, J.C.; Kundra, A.; Andrei, M.; Baptiste, S.; Chen, C.; Wong, C.; Sindhu, H. Myeloid-Derived Suppressor Cells in Patients with Myeloproliferative Neoplasm. Leuk Res. 2016, 43, 39–43. [Google Scholar] [CrossRef]
- Giallongo, C.; Romano, A.; Parrinello, N.L.; La Cava, P.; Brundo, M.V.; Bramanti, V.; Stagno, F.; Vigneri, P.; Chiarenza, A.; Palumbo, G.A.; et al. Mesenchymal Stem Cells (MSC) Regulate Activation of Granulocyte-Like Myeloid Derived Suppressor Cells (G-MDSC) in Chronic Myeloid Leukemia Patients. PLoS ONE 2016, 11, e0158392. [Google Scholar] [CrossRef]
- Gunes, E.G.; Rosen, S.T.; Querfeld, C. The Role of Myeloid-Derived Suppressor Cells in Hematologic Malignancies. Curr. Opin. Oncol. 2020, 32, 518–526. [Google Scholar] [CrossRef]
- Hyun, S.Y.; Na, E.J.; Jang, J.E.; Chung, H.; Kim, S.J.; Kim, J.S.; Kong, J.H.; Shim, K.Y.; Lee, J.I.; Min, Y.H.; et al. Immunosuppressive role of CD11b+ CD33+ HLA-DR − myeloid-derived suppressor cells-like blast subpopulation in acute myeloid leukemia. Cancer Med. 2020, 9, 7007–7017. [Google Scholar] [CrossRef] [PubMed]
- Hanna, B.S.; Öztürk, S.; Seiffert, M. Beyond Bystanders: Myeloid Cells in Chronic Lymphocytic Leukemia. Mol. Immunol. 2019, 110, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Zarobkiewicz, M.; Kowalska, W.; Chocholska, S.; Tomczak, W.; Szymańska, A.; Morawska, I.; Wojciechowska, A.; Bojarska-Junak, A. High M-MDSC Percentage as a Negative Prognostic Factor in Chronic Lymphocytic Leukaemia. Cancers 2020, 12, 2614. [Google Scholar] [CrossRef]
- Kowalska, W.; Bojarska-Junak, A. Monocytic MDSC as a Source of Immunosuppressive Cytokines in Chronic Lymphocytic Leukemia (CLL) Microenvironment. Folia Histochem. Cytobiol. 2020, 58, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Jitschin, R.; Braun, M.; Dettmer-Wilde, K.; Bricks, J.; Berger, J.; Eckart, M.J.; Krause, S.W.; Oefner, P.J.; le Blanc, K.; Mackensen, A.; et al. CLL-cells induce IDOhi CD14+ HLA-DRlo myeloid-derived suppressor cells that inhibit T-cell responses and promote TRegs. Blood 2014, 124, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Zahran, A.M.; Moeen, S.M.; Thabet, A.F.; Rayan, A.; Abdel-Rahim, M.H.; Mohamed, W.M.Y.; Hetta, H.F. Monocytic Myeloid-Derived Suppressor Cells in Chronic Lymphocytic Leukemia Patients: A Single Center Experience. Leuk. Lymphoma 2020, 61, 1645–1652. [Google Scholar] [CrossRef] [PubMed]
- Pyzer, A.R.; Stroopinsky, D.; Rajabi, H.; Washington, A.; Tagde, A.; Coll, M.; Fung, J.; Bryant, M.P.; Cole, L.; Palmer, K.; et al. MUC1-Mediated Induction of Myeloid-Derived Suppressor Cells in Patients with Acute Myeloid Leukemia. Blood 2017, 129, 1791–1801. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Y.; Zhang, Z.-F.; Ju, Y.; Li, L.; Zhang, B.-C.; Liu, B. Increase in myeloid-derived suppressor cells (MDSCs) associated with minimal residual disease (MRD) detection in adult acute myeloid leukemia. Int. J. Hematol. 2015, 102, 579–586. [Google Scholar] [CrossRef]
- Kittang, A.O.; Kordasti, S.; Sand, K.E.; Costantini, B.; Kramer, A.M.; Perezabellan, P.; Seidl, T.; Rye, K.P.; Hagen, K.M.; Kulasekararaj, A.; et al. Expansion of Myeloid Derived Suppressor Cells Correlates with Number of T Regulatory Cells and Disease Progression in Myelodysplastic Syndrome. Ncoimmunology 2016, 5, e1062208. [Google Scholar] [CrossRef]
- Silva, S.D.; Rajadhyaksha, S.B.; Singh, M. Immune Dysregulation in MDS: The Role of Cytokines and Immune Cells. In Recent Dev. Myelodysplastic Syndr; IntechOpen: London, UK, 2019; Volume 4, p. 45. [Google Scholar] [CrossRef]
- Chen, X.; Eksioglu, E.A.; Zhou, J.; Zhang, L.; Djeu, J.; Fortenbery, N.; Epling-Burnette, P.; van Bijnen, S.; Dolstra, H.; Cannon, J.; et al. Induction of Myelodysplasia by Myeloid-Derived Suppressor Cells. J. Clin. Investig. 2013, 123, 4595–4611. [Google Scholar] [CrossRef]
- de Veirman, K.; Menu, E.; Maes, K.; de Beule, N.; de Smedt, E.; Maes, A.; Vlummens, P.; Fostier, K.; Kassambara, A.; Moreaux, J.; et al. Myeloid-Derived Suppressor Cells Induce Multiple Myeloma Cell Survival by Activating the AMPK Pathway. Cancer Lett. 2019, 442, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Parrinello, N.L.; la Cava, P.; Tibullo, D.; Giallongo, C.; Camiolo, G.; Puglisi, F.; Parisi, M.; Pirosa, M.C.; Martino, E.; et al. PMN-MDSC and Arginase Are Increased in Myeloma and May Contribute to Resistance to Therapy. Expert Rev. Mol. Diagn. 2018, 18, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, I.R.; Condamine, T.; Lin, C.; Herlihy, S.E.; Garfall, A.; Vogl, D.T.; Gabrilovich, D.I.; Nefedova, Y. Bone Marrow PMN-MDSCs and Neutrophils Are Functionally Similar in Protection of Multiple Myeloma from Chemotherapy. Cancer Lett. 2016, 371, 117–124. [Google Scholar] [CrossRef]
- Binsfeld, M.; Muller, J.; Lamour, V.; de Veirman, K.; de Raeve, H.; Bellahcène, A.; van Valckenborgh, E.; Baron, F.; Beguin, Y.; Caers, J.; et al. Granulocytic Myeloid-Derived Suppressor Cells Promote Angiogenesis in the Context of Multiple Myeloma. Oncotarget 2016, 7, 37931. [Google Scholar] [CrossRef]
- Marini, O.; Spina, C.; Mimiola, E.; Cassaro, A.; Malerba, G.; Todeschini, G.; Perbellini, O.; Scupoli, M.; Carli, G.; Facchinelli, D.; et al. Identification of Granulocytic Myeloid-Derived Suppressor Cells (G-MDSCs) in the Peripheral Blood of Hodgkin and Non-Hodgkin Lymphoma Patients. Oncotarget 2016, 7, 27676–27688. [Google Scholar] [CrossRef]
- Lin, Y.; Gustafson, M.P.; Bulur, P.A.; Gastineau, D.A.; Witzig, T.E.; Dietz, A.B. Immunosuppressive CD14+ HLA-DRlow/- Monocytes in B-Cell Non-Hodgkin Lymphoma. Blood 2011, 117, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Betsch, A.; Rutgeerts, O.; Fevery, S.; Sprangers, B.; Verhoef, G.; Dierickx, D.; Beckers, M. Myeloid-Derived Suppressor Cells in Lymphoma: The Good, the Bad and the Ugly. Blood Rev. 2018, 32, 490–498. [Google Scholar] [CrossRef]
- Romano, A.; Parrinello, N.L.; Vetro, C.; Forte, S.; Chiarenza, A.; Figuera, A.; Motta, G.; Palumbo, G.A.; Ippolito, M.; Consoli, U.; et al. Circulating Myeloid-Derived Suppressor Cells Correlate with Clinical Outcome in Hodgkin Lymphoma Patients Treated up-Front with a Risk-Adapted Strategy. Br. J. Haematol. 2015, 168, 689–700. [Google Scholar] [CrossRef]
- Azzaoui, I.; Uhel, F.; Rossille, D.; Pangault, C.; Dulong, J.; le Priol, J.; Lamy, T.; Houot, R.; le Gouill, S.; Cartron, G.; et al. T-Cell Defect in Diffuse Large B-Cell Lymphomas Involves Expansion of Myeloid-Derived Suppressor Cells. Blood 2016, 128, 1081–1092. [Google Scholar] [CrossRef]
- Bizymi, N.; Damianaki, A.; Velegraki, M.; Zavitsanou, K.; Karasachinidis, A.; Georgopoulou, A.; Mavroudi, I.; Sperelakis, J.; Kontakis, G.; Pontikoglou, C.; et al. Frequency and Functional Analysis of Myeloid-Derived Suppressor Cells (MDSCs) in the Peripheral Blood and Bone Marrow of Patients with Chronic Idiopathic Neutropenia (CIN). Blood 2020, 136, 26–27. [Google Scholar] [CrossRef]
- Bizymi, N.; Velegraki, M.; Damianaki, A.; Koutala, H.; Papadaki, H.A. Altered Monocyte Subsets in Patients with Chronic Idiopathic Neutropenia. J. Clin. Immunol. 2019, 39, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Semple, J.W. Move over Tregs, MDSCs Are Here. Blood 2016, 127, 1526–1528. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, J.; Zhou, Y.; Wen, J.; Sun, X.; Zhang, X. Circulating Myeloid-Derived Suppressor Cells Predict Disease Activity and Treatment Response in Patients with Immune Thrombocytopenia. Braz. J. Med. Biol. Res. 2017, 50, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Wu, B.; Cheng, L.; Li, F.; Zhan, Y.; Liu, C.; Ji, L.; Min, Z.; Ke, Y.; Sun, L.; et al. Distinct Alterations of CD68 + CD163 + M2-like Macrophages and Myeloid-Derived Suppressor Cells in Newly Diagnosed Primary Immune Thrombocytopenia with or without CR after High-Dose Dexamethasone Treatment. J. Transl. Med. 2018, 16, 48. [Google Scholar] [CrossRef]
- Liu, Y.-W.; Qu, W.; Wang, H.-Q.; Xing, L.-M.; Wu, Y.-H.; Liu, Z.-Y.; Zhang, Y.; Liu, H.; Don, X.-F.; Tao, J.-L.; et al. Number and Function of Myeloid-Derived Suppressor Cells in Patients with Adult Primary Immune Thrombocytopenia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2018, 26, 1151–1155. [Google Scholar] [CrossRef]
- Wen, R.; Wang, Y.; Hong, Y.; Yang, Z. Cellular Immune Dysregulation in the Pathogenesis of Immune Thrombocytopenia. Blood Coagul. Fibrinolysis 2020, 31, 113–120. [Google Scholar] [CrossRef]
- Vladimirovna, I.L.; Sosunova, E.; Nikolaev, A.; Nenasheva, T. Mesenchymal Stem Cells and Myeloid Derived Suppressor Cells: Common Traits in Immune Regulation. J. Immunol. Res. 2016, 2016, 7121580. [Google Scholar] [CrossRef]
- Kapor, S.; Santibanez, J.F. Myeloid-derived Suppressor Cells and Mesenchymal Stem/Stromal Cells in Myeloid Malignancies. J. Clin. Med. 2021, 10, 2788. [Google Scholar] [CrossRef]
- Gantt, S.; Gervassi, A.; Jaspan, H.; Horton, H. The Role of Myeloid-Derived Suppressor Cells in Immune Ontogeny. Front. Immunol. 2014, 5, 387. [Google Scholar] [CrossRef]
- Nair, R.R.; Sinha, P.; Khanna, A.; Singh, K. Reduced Myeloid-Derived Suppressor Cells in the Blood and Endometrium Is Associated with Early Miscarriage. Am. J. Reprod. Immunol. 2015, 73, 479–486. [Google Scholar] [CrossRef]
- Rieber, N.; Gille, C.; Köstlin, N.; Schäfer, I.; Spring, B.; Ost, M.; Spieles, H.; Kugel, H.A.; Pfeiffer, M.; Heininger, V.; et al. Neutrophilic Myeloid-Derived Suppressor Cells in Cord Blood Modulate Innate and Adaptive Immune Responses. Clin. Exp. Immunol. 2013, 174, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Köstlin, N.; Kugel, H.; Spring, B.; Leiber, A.; Marmé, A.; Henes, M.; Rieber, N.; Hartl, D.; Poets, C.F.; Gille, C. Granulocytic Myeloid Derived Suppressor Cells Expand in Human Pregnancy and Modulate T-Cell Responses. Eur. J. Immunol. 2014, 44, 2582–2591. [Google Scholar] [CrossRef] [PubMed]
- Gervassi, A.; Lejarcegui, N.; Dross, S.; Jacobson, A.; Itaya, G.; Kidzeru, E.; Gantt, S.; Jaspan, H.; Horton, H. Myeloid Derived Suppressor Cells Are Present at High Frequency in Neonates and Suppress in Vitro T Cell Responses. PLoS ONE 2014, 9, e107816. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.; Scheckenbach, V.; Kugel, H.; Spring, B.; Pagel, J.; Härtel, C.; Pauluschke-Fröhlich, J.; Peter, A.; Poets, C.F.; Gille, C.; et al. Granulocytic Myeloid-Derived Suppressor Cells (GR-MDSC) Accumulate in Cord Blood of Preterm Infants and Remain Elevated during the Neonatal Period. Clin. Exp. Immunol. 2017, 191, 328–337. [Google Scholar] [CrossRef]
- Bizymi, N.; Georgopoulou, A.; Mastrogamvraki, N.; Matheakakis, A.; Gontika, I.; Fragiadaki, I.; Mavroudi, I.; Papadaki, H.A. Myeloid-Derived Suppressor Cells (MDSC) in the Umbilical Cord Blood: Biological Significance and Possible Therapeutic Applications. J. Clin. Med. 2022, 11, 727. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, R.; Li, Q.; Wang, H.; Tao, Q.; Zhai, Z. Elevated M-MDSCs in Circulation Are Indicative of Poor Prognosis in Diffuse Large B-Cell Lymphoma Patients. J. Clin. Med. 2021, 10, 1768. [Google Scholar] [CrossRef]
- Wu, C.; Wu, X.; Liu, X.; Yang, P.; Xu, J.; Chai, Y.; Guo, Q.; Wang, Z.; Zhang, L. Prognostic Significance of Monocytes and Monocytic Myeloid-Derived Suppressor Cells in Diffuse Large B-Cell Lymphoma Treated with R-CHOP. Cell. Physiol. Biochem. 2016, 39, 521–530. [Google Scholar] [CrossRef]
- Papafragkos, I.; Markaki, E.; Kalpadakis, C.; Verginis, P. Decoding the Myeloid-derived Suppressor Cells in Lymphoid Malignancies. J. Clin. Med. 2021, 10, 3462. [Google Scholar] [CrossRef]
- Geskin, L.J.; Akilov, O.E.; Kwon, S.; Schowalter, M.; Watkins, S.; Whiteside, T.L.; Butterfield, L.H.; Falo, L.D. Therapeutic Reduction of Cell-Mediated Immunosuppression in Mycosis Fungoides and Sézary Syndrome. Cancer Immunol. Immunother. 2017, 67, 423–434. [Google Scholar] [CrossRef]
- Marvel, D.; Gabrilovich, D.I. Myeloid-Derived Suppressor Cells in the Tumor Microenvironment: Expect the Unexpected. J. Clin. Investig. 2015, 125, 3356–3364. [Google Scholar] [CrossRef]
- Musolino, C.; Allegra, A.; Pioggia, G.; Gangemi, S. Immature Myeloid-Derived Suppressor Cells: A Bridge between Inflammation and Cancer. Oncol. Rep. 2016, 37, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yao, Y.; Shen, Q.; Li, G.; Hu, L.; Zhang, X. Demethylating Agent Decitabine Disrupts Tumor-Induced Immune Tolerance by Depleting Myeloid-Derived Suppressor Cells. J. Cancer Res. Clin. Oncol. 2017, 143, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Krejcik, J.; Casneuf, T.; Nijhof, I.S.; Verbist, B.; Bald, J.; Plesner, T.; Syed, K.; Liu, K.; van de Donk, N.W.C.J.; Weiss, B.M.; et al. Daratumumab Depletes CD38 + Immune-Regulatory Cells, Promotes T-Cell Expansion, and Skews T-Cell Repertoire in Multiple Myeloma. Blood 2016, 128, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Younos, I.H.; Abe, F.; Talmadge, J.E. Myeloid-Derived Suppressor Cells: Their Role in the Pathophysiology of Hematologic Malignancies and Potential as Therapeutic Targets. Leuk. Lymphoma 2015, 56, 2251–2263. [Google Scholar] [CrossRef]
- De Veirman, K.; Valckenborgh, E.E.; Elahmar, Q.; Geeraerts, X.; De Bruyne, E.; Menu, E.; Riet, I.E.; Evanderkerken, K.; Van Ginderachter, J.A. Myeloid-Derived Suppressor Cells as Therapeutic Target in Hematological Malignancies. Front. Oncol. 2014, 4, 349. [Google Scholar] [CrossRef]
- Stiff, A.; Trikha, P.; Wesolowski, R.; Kendra, K.; Hsu, V.; Uppati, S.; McMichael, E.; Duggan, M.; Campbell, A.; Keller, K.; et al. Myeloid-Derived Suppressor Cells Express Bruton’s Tyrosine Kinase and Can Be Depleted in Tumor-Bearing Hosts by Ibrutinib Treatment. Cancer Res. 2016, 76, 2125–2136. [Google Scholar] [CrossRef]
- Arina, A.; Corrales, L.; Bronte, V. Enhancing T Cell Therapy by Overcoming the Immunosuppressive Tumor Microenvironment. Semin. Immunol. 2016, 28, 54–63. [Google Scholar] [CrossRef]
- Olivares-Hernández, A.; Figuero-Pérez, L.; Terán-Brage, E.; López-Gutiérrez, Á.; Velasco, Á.T.; Sarmiento, R.G.; Cruz-Hernández, J.J.; Miramontes-González, J.P. Resistance to Immune Checkpoint Inhibitors Secondary to Myeloid-Derived Suppressor Cells: A New Therapeutic Targeting of Haematological Malignancies. J. Clin. Med. 2021, 10, 1919. [Google Scholar] [CrossRef]
- Hou, Y.; Feng, Q.; Xu, M.; Li, G.S.; Liu, X.N.; Sheng, Z.; Zhou, H.; Ma, J.; Wei, Y.; Sun, Y.X.; et al. High-Dose Dexamethasone Corrects Impaired Myeloid-Derived Suppressor Cell Function via Ets1 in Immune Thrombocytopenia. Blood 2016, 127, 1587–1597. [Google Scholar] [CrossRef]
- Aslam, R.; Burack, W.R.; Segel, G.B.; Mcvey, M.; Spence, S.A.; Semple, J.W. Intravenous Immunoglobulin Treatment of Spleen Cells from Patients with Immune Thrombocytopenia Significantly Increases the Percentage of Myeloid-Derived Suppressor Cells. Br. J. Haematol. 2017, 181, 262–264. [Google Scholar] [CrossRef]
- Eksioglu, E.A.; Chen, X.; Heider, K.H.; Rueter, B.; McGraw, K.L.; Basiorka, A.A.; Wei, M.; Burnette, A.; Cheng, P.; Lancet, J.; et al. Novel Therapeutic Approach to Improve Hematopoiesis in Low Risk MDS by Targeting MDSCs with the Fc-Engineered CD33 Antibody BI 836858. Leukemia 2017, 31, 2172–2180. [Google Scholar] [CrossRef] [PubMed]
- Blanc, K.; Jitschin, R.; Mougiakakos, D. Myeloid-Derived Suppressor Cells in Allogeneic Hematopoietic Stem Cell Transplantation: A Double-Edged Sword? Oncoimmunology 2013, 2, 7–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yin, J.; Wang, C.; Huang, M.; Mao, X.; Zhou, J.; Zhang, Y. Circulating CD14(+) HLA-DR(-/Low) Myeloid-Derived Suppressor Cells in Leukemia Patients with Allogeneic Hematopoietic Stem Cell Transplantation: Novel Clinical Potential Strategies for the Prevention and Cellular Therapy of Graft-versus-Host Disease. Cancer Med. 2016, 5, 1654–1669. [Google Scholar] [CrossRef] [PubMed]
- Scalea, J.R.; Lee, Y.; Davila, E.; Bromberg, J.S. Myeloid-Derived Suppressor Cells and Their Potential Application in Transplantation. Transplantation 2018, 102, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Koehn, B.H.; Blazar, B.R. Role of Myeloid-Derived Suppressor Cells in Allogeneic Hematopoietic Cell Transplantation. J. Leukoc. Biol. 2017, 102, 335–341. [Google Scholar] [CrossRef]
- Demosthenous, C.; Sakellari, I.; Douka, V.; Papayanni, P.G.; Anagnostopoulos, A.; Gavriilaki, E. The Role of Myeloid-Derived Suppressor Cells (MDSCs) in Graft-versus-Host Disease (GvHD). J. Clin. Med. 2021, 10, 2050. [Google Scholar] [CrossRef]
| Marker/Molecule | Cell | Function | Importance in MDSCs |
|---|---|---|---|
| HLA-DR: Major histocompatibility complex (MHC) II cell surface receptor | Antigen-presenting cells | Antigen presentation to T-cells | Absent or low in MDSCs [24,90] |
| CD33 | Expression decreases in mature cells, expressed in myeloid stem cells (CFU-GEMM, CFU-GM, CFU-G, E-BFU), myeloblasts, monoblasts, monocytes/macrophages, granulocyte precursors, and mast cells | Sialoadhesin | Present [24,91,92] |
| CD66b or CEACAM8 or CGM6 or NCA-95 | Granulocytes, eosinophils (granulocyte lineage cells) | Regulation of adhesion and activation | Present in PMN-MDSCs [24,91,93,94] |
| CD11b | Granulocytes, monocytes/macrophages, NK cells, and subsets of B- and T-cells, pan-myeloid marker | Adhesion | Present [86] |
| CD11c | Monocytes/macrophages, NK cells, and hairy cells | Adhesion | Mostly absent [86,90] |
| CD45 (lymphocyte common antigen) | Leukocytes | Receptor-linked protein tyrosine phosphatase, activation, signal transduction | Present [42,91] |
| CD15 (Lewis x or Lex) | Present in cells past the myeloblast stage in the granulocytic lineage | Neutrophil adhesion to dendritic cells | Present in PMN-MDSCs [24,86] |
| CD14 | Expressed in the myelomonocyte lineage (monocytes, macrophages) | Endotoxin receptor, activation of innate immunity | Present in M-MDSCs [24,92] |
| CD3 | Mature T-cells | Part of the TCR complex | Absent [86] |
| CD19 | B-cells, precursors of B-cells, follicular dendritic cells | Regulation of development, activation, and differentiation | Absent [86] |
| CD20 | B-cells | Development and differentiation of B-cells into plasma cells | Absent [86] |
| CD16 | Subsets of NK and T-cell types, granulocytes, tissue macrophages, CD16+ subsets of monocytes, eosinophils, DCs | Fc gamma receptor; antibody-dependent cell-mediated cytotoxicity | Absent [86] |
| CD56 | Neural cells, NK cells, subset of T-cells | Adhesion molecule | Absent [86] |
| CD13 | Mature neutrophils | Aminopeptidase N; mediates tumour angiogenesis | Present in CD13high PMN-MDSCs [86,93,94] |
| CD40 | Antigen-presenting cells, epithelial cells, hematopoietic progenitor cells, activated T-cells | Member of the tumour necrosis factor (TNF) receptor superfamily; regulates tumour growth and apoptosis dependently on the level of its expression; promotes the immunosuppression of MDSCs and T-regs induction but not their accumulation | Present in MDSCs [86,95] |
| CD80 | Myeloid cells | Protein B7-1; suppression of T-cell immune responses; binds both CD28 (stimulatory) and CTLA-4 (inhibitory) on T-cells | Present in MDSCs [2,96] |
| CD86 | Antigen-presenting cells | Protein B7-2; activation of T-cells; binds both CD28 (stimulatory) and CTLA-4 (inhibitory) on T-cells | Absent or low in MDSCs [96] |
| CD83 | Antigen-presenting cells, mature DCs | Immune responses | Present in M-MDSCs [31,90] |
| CD36 | Widespread presence | Fatty acid translocase; immunosuppressive activity of MDSCs | Present in MDSCs [14] |
| HLA-ABC: Major histocompatibility complex (MHC) I antigens | Widespread presence | Immune response | Present in MDSCs [2] |
| CD54 or ICAM-1 (Intercellular Adhesion Molecule 1) | Endothelial cells, leukocytes | Cell–cell interactions, transmigration | High in M-MDSCs, low in PMN-MDSCs [90] |
| CD195 or CCR5 | Leukocytes | C-C chemokine receptor type 5 | Present [86,90] |
| CD197 or CCR7 | Mature DCs, B-cells, T-cells, cancer cells | C-C chemokine receptor type 7 | Present [86,97] |
| CD62L or L-selectin | Leukocytes | Cell adhesion molecule | Present [2,86] |
| E-Cadherin | Epithelial cells, cancer cells | Cell adhesion molecule | Downregulated by MDSCs [98,99] |
| N-Cadherin | Mesenchymal cells, cancer cells | Cell adhesion molecule | Upregulated by MDSCs [99] |
| CD124 or IL-4Ra | B-cells, T-cells, NK cells, macrophages | Activation of STAT6, Th2 response | Present [2,26] |
| CD34 or Sialomucin | Early haemopoietic cells, endothelial cells | Cell adhesion molecule | Absent [2,86] |
| LOX-1 | Endothelial cells, low expression in normal neutrophils | Receptor oxidised LDL receptor 1; endoplasmic reticulum (ER) stress, lipid metabolism | Present in PMN-MDSCs [81] |
| CD192 or CCR2 | Leukocytes | C-C chemokine receptor type 2 | High in M-MDSCs, low in PMN-MDSCs [90] |
| CXCR4 | T-cells | Recruitment | Present [86,90] |
| Arg1 | Erythrocytes, lymphocytes, macrophages | Deprivation of arginine that is crucial for TCR expression and TCR-mediated signal transduction | Present [100] |
| NO | Widespread presence | Nitration of TCRs and production of chemokines important for T-cell migration or induction of T-cell apoptosis | Present mainly in M-MDSCs [62] |
| ROS | Widespread presence | Oxidative stress induction, apoptosis of T-cells | Present [101] |
| S100A8/9 | Myeloid cells, tumour cells | Interaction with CD33, NF-kB pathway induction, MDSCs migration | Present [102] |
| STATs | Immune cells, tumour cells | Transcription factors important for MDSCs development | Present [103,104] |
| PD-L1 or B7-H1 or CD274 | Myeloid cells, lymphoid cells, epithelial cells, tumour cells | Co-inhibitory receptor leading to T-cell activity suppression | Present in M-MDSCs (mainly) and PMN-MDSCs [71] |
| C/EBPb | BM cells in differentiation of myeloid lineage and emergency granulopoiesis | Transcription factor important for immunosuppressive properties of MDSCs | Present [67,105] |
| TGF-β | Secreted by most cells | Inhibition of immune effector cell functions | Secretion by MDSCs [67] |
| IL-10 | Secreted by immune cells | Inhibition of immune effector cell functions | Secretion by MDSCs [67] |
| VEGF | Secreted in TME | Angiogenesis | Induction of its secretion by MDSCs [67] |
| CD115 | Monocyte and macrophage lineage cells | Receptor of M-CSF | Present [86] |
| CD244 | NK cells, T-cells, monocytes, dendritic cells | Important for immunosuppressive properties of MDSCs | Present in PMN-MDSCs [106] |
| CD163 | Activated T-cells, macrophages | Scavenger receptor | Present in M-MDSCs [41,107] |
| CD39 | Leukocytes | Increased production of adenosine that suppresses effector T-cell function | Present [86,108] |
| CD73 | B-cells, T-cells | Increased production of adenosine that suppresses effector T-cell function | Present [86,108] |
| Fas Ligand or CD95L or CD178 | T-cells, NK cells | Mediator of T-cell apoptosis | Present in PMN-MDSCs [86,109] |
| IDO | Antigen-presenting cells, tumour cells | Degradation of L-tryptophan leading to cell cycle arrest and anergy of T-cells and differentiation of T-cells into T-regs | Present [110] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bizymi, N.; Matthaiou, A.M.; Matheakakis, A.; Voulgari, I.; Aresti, N.; Zavitsanou, K.; Karasachinidis, A.; Mavroudi, I.; Pontikoglou, C.; Papadaki, H.A. New Perspectives on Myeloid-Derived Suppressor Cells and Their Emerging Role in Haematology. J. Clin. Med. 2022, 11, 5326. https://doi.org/10.3390/jcm11185326
Bizymi N, Matthaiou AM, Matheakakis A, Voulgari I, Aresti N, Zavitsanou K, Karasachinidis A, Mavroudi I, Pontikoglou C, Papadaki HA. New Perspectives on Myeloid-Derived Suppressor Cells and Their Emerging Role in Haematology. Journal of Clinical Medicine. 2022; 11(18):5326. https://doi.org/10.3390/jcm11185326
Chicago/Turabian StyleBizymi, Nikoleta, Andreas M. Matthaiou, Angelos Matheakakis, Ioanna Voulgari, Nikoletta Aresti, Konstantina Zavitsanou, Anastasios Karasachinidis, Irene Mavroudi, Charalampos Pontikoglou, and Helen A. Papadaki. 2022. "New Perspectives on Myeloid-Derived Suppressor Cells and Their Emerging Role in Haematology" Journal of Clinical Medicine 11, no. 18: 5326. https://doi.org/10.3390/jcm11185326
APA StyleBizymi, N., Matthaiou, A. M., Matheakakis, A., Voulgari, I., Aresti, N., Zavitsanou, K., Karasachinidis, A., Mavroudi, I., Pontikoglou, C., & Papadaki, H. A. (2022). New Perspectives on Myeloid-Derived Suppressor Cells and Their Emerging Role in Haematology. Journal of Clinical Medicine, 11(18), 5326. https://doi.org/10.3390/jcm11185326








