Abstract
In infectious diseases, extracellular vesicles (EVs) released from a pathogen or pathogen-infected cells can transfer pathogen-derived biomolecules, especially proteins, to target cells and consequently regulate these target cells. For example, malaria is an important tropical infectious disease caused by Plasmodium spp. Previous studies have identified the roles of Plasmodium falciparum-infected red blood cell-derived EVs (Pf-EVs) in the pathogenesis, activation, and modulation of host immune responses. This study investigated the proteomic profiles of Pf-EVs isolated from four P. falciparum strains. We also compared the proteomes of EVs from (i) different EV types (microvesicles and exosomes) and (ii) different parasite growth stages (early- and late-stage). The proteomic analyses revealed that the human proteins carried in the Pf-EVs were specific to the type of Pf-EVs. By contrast, most of the P. falciparum proteins carried in Pf-EVs were common across all types of Pf-EVs. As the proteomics results revealed that Pf-EVs contained invasion-associated proteins, the effect of Pf-EVs on parasite invasion was also investigated. Surprisingly, the attenuation of parasite invasion efficiency was found with the addition of Pf-MVs. Moreover, this effect was markedly increased in culture-adapted isolates compared with laboratory reference strains. Our evidence supports the concept that Pf-EVs play a role in quorum sensing, which leads to parasite growth-density regulation.
1. Introduction
Malaria is an important tropical infectious disease leading to an estimated 240 million malaria cases in 2020 in 85 malaria-endemic countries [1]. The disease is caused by an apicomplexan protozoan, Plasmodium spp., transmitted to humans by the bite of female Anopheles mosquitos. Among five Plasmodium spp. that cause malaria in humans, P. falciparum is the most important because it is associated with severe syndromes and high mortality. The life cycle of P. falciparum in humans includes both the exo- and intraerythrocytic cycles in which the parasite grows and develops in hepatocytes or red blood cells (RBCs), respectively. During intraerythrocytic growth and development, the merozoites released from hepatocytes infect RBCs and asexually develop inside a parasitophorous vacuole from the ring stage to the trophozoite stage and schizont stage (containing many merozoites). At the end of schizogony, around 48 h after the invasion (for P. falciparum), infected RBCs rupture, and merozoites egress and, in turn, invade new RBCs, with the cycle repeating.
To infect the RBCs, the merozoite invasion progresses through a multi-step process consisting of initial attachment, apical reorientation, merozoite internalization, and parasitophorous vacuole membrane formation. Many ligand-receptor interactions between P. falciparum proteins take place on the merozoite surface and RBC receptors. In addition, signaling transduction-associated proteins sequentially participate in this process, facilitating parasite invasion. The list of essential merozoite proteins for the invasion process is extensive, including merozoite surface proteins (MSPs), erythrocyte-binding-like (EBL) and reticulocyte-binding-like (RBL) families, erythrocyte-binding antigen (EBA) and Rh protein family, P. falciparum reticulocyte-binding homolog 5 (PfRh5)/PfRh5-interacting protein (PfRipr)/cysteine-rich protective antigen (CyRPA) complex, rhoptry neck (RON) proteins, apical membrane antigen-1 (AMA-1), glycosylphosphatidylinositol-anchored micronemal antigen (GAMA), S-antigen, glutamate-rich protein (GLURP), serine-repeat antigen protein (SERA), claudin-like apicomplexan microneme protein (CLAMP), subtilisin-like serine protease (SUB), calcium-dependent protein kinases (CDPKs), glideosome-associated proteins (GAPs), myosin A, profilin, and cofilin [2,3,4,5,6,7,8,9]. Alterations in the function of these proteins by knockout gene expression or blocking (with inhibitors or antibodies) affect merozoite invasion efficiency and may inhibit parasite invasion [10,11].
During the intraerythrocytic growth cycle of Plasmodium spp., extracellular vesicles (EVs), which are nano-sized membrane-bound vesicles, are released from both uninfected and infected red blood cells (iRBCs) [12]. The EVs can be classified as an ectosome (microvesicles (MV) or microparticle (MP)) or exosome (Exo), based on their biogenesis and physical characteristics. These EVs function as mediators of intercellular communication via their cargos of biomolecules (proteins, lipids, metabolites, and genetic materials) from originated cells to recipient cells. EVs have also been reported to have essential roles in signal transduction, transcriptional regulation, and inflammatory reactions during the immune response in both pro- and anti-inflammatory phenomena [13,14,15,16]. The biological significance of EVs has been increasingly demonstrated across a broad range of pathologies, where their elevated levels in patient plasma are indicative of cellular stress and disease severity [17,18].
For more than two decades, EVs have been studied, and their roles in malaria infection have been shown to include regulating parasite growth, driving malaria pathogenesis, and modulating host immune responses. In experimental rodent malaria models, levels of MPs are high in the plasma of mice with cerebral malaria [19]. iRBC-derived MPs from the plasma of Plasmodium berghei strain ANKA (PbA)-infected mice induce proinflammatory responses in macrophages resulting in systemic inflammation [20]. In addition, the adoptive transfer of MPs from PbA-infected mice leads to localized MPs in cerebral microvessels of PbA-infected recipient mice [19]. The decrease in MP production in knockout mice or with pharmacological inhibition protects these mice from cerebral malaria [21,22]. Similarly, EV levels are increased and associated with disease severity in malaria-infected patients [23,24]. Platelets and RBCs are major EV cell sources in patient plasma [24]. From in vitro experiments, Pf-EVs are internalized by monocytes and macrophages, deliver biomolecular cargos into the cytosol, and elicit innate immune cytokine responses [25,26,27]. Moreover, the internalization of Pf-EVs modulates endothelial cell barriers, including adherence-related proteins that lead to vascular dysfunction and support cytoadherence of iRBCs [28].
Previous studies indicate the role of Pf-EVs in parasite-parasite communication, which mediate the horizontal transfer of nucleic acids to regulate the formation of gametocytes (the sexual stage responsible for malaria transmission) [25,29]. Moreover, Pf-EVs derived from the cultures of low and high parasitemia carry P. falciparum lactate dehydrogenase (PfLDH) in differential levels; only Pf-EVs derived from high parasite density regulate the parasite population by apoptosis induction [30]. This evidence implies the functional mechanism of Pf-EVs in a quorum sensing-like mechanism in which a cell secretes a signaling quorum-sensing molecule to communicate with other cells in response to the cell population density. As well-known studies reported in bacteria, the higher density of bacteria, higher level of the production and accumulation of quorum-sensing molecules. These molecules induce the positive feedback mechanism that allows communication between bacteria to regulate the gene expression involved in virulence, competition, pathogenicity, and resistance and to limit the population size [31]. The quorum sensing-like mechanism in protozoa has been proposed as the mechanism regulating the balance between virulence, survival in the host, and potential for transmission [32,33].
The proteomic profiles of malaria-derived EVs are now being studied. Sources of these EVs include plasma of Plasmodium spp.-infected patients, mice, and humanized mice, as well as from supernatants of Plasmodium spp. cultures [25,27,34]. Results show that these EVs contain both host and parasite proteins. Some have been identified as malaria antigens (MSP1, MSP3, MSP7, MSP9, serine-repeat antigen 1, and heat shock protein (HSP) 70), which might have potential for vaccine development [35]. Some might also be used as biomarkers of infection [34]. However, little information is available on the protein profiles of EVs derived from different P. falciparum strains. We hypothesized that the differences in P. falciparum strain virulence might be related to the EV’s selective and unique cell-specific proteins and thus their different disease outcomes. In this study, the proteomic profiles of EVs derived from different P. falciparum strains and growth stages were compared. We found that most virulence-associated proteins were highly conserved among P. falciparum strains. We further explored the effects of these Pf-EVs on invasion efficiency, growth, and development during intraerythrocytic schizogony.
2. Materials and Methods
2.1. Parasite Culture
Four P. falciparum parasite strains (3D7, NF54, TM01, and TM02) were grown in human RBCs in complete culture media (RPMI 1640 (Gibco; Grand Island, NY, USA) supplemented with 25 mM HEPES, 0.225% NaHCO3, 0.1 mM hypoxanthine, 25 g/mL gentamicin, and 5% AlbuMax II (Gibco; Auckland, New Zealand)) under an atmosphere of 5% O2, 5% CO2, and 90% N2. The culture media were changed daily. Parasite growth was monitored daily by Giemsa-stained thin blood smears. At a certain percentage of parasitemia, cultures were synchronized into the ring stage by resuspension of culture pellets in 5% D-sorbitol solution (Sigma-Aldrich; St. Louis, MO, USA) and incubated at 37 °C for 10 min, followed by centrifugation. The synchronized ring stage parasites were continuously re-cultured at 5% parasitemia and 5% hematocrit under an atmosphere of 5% O2, 5% CO2, and 90% N2 for 24 h. Culture supernatants were collected to isolate EVs of early-stage parasites (ring form to mid-trophozoite). New culture media were added and parasite cultures were continued for 24 h. During parasite growth beyond the mid-trophozoite stage, culture supernatants were collected to isolate EVs of late-stage parasites (mid-trophozoite to early-ring). These culture supernatants were centrifuged at 1500× g for 15 min to remove residual RBCs and cell debris and kept at −20 °C until used for EV isolation. This study was approved by the Siriraj Institutional Review Board of the Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand (COA no Si 634/2019).
2.2. Extracellular Vesicle Isolation
The Pf-EVs were isolated from malarial culture supernatants by multi-step sequential centrifugation. Briefly, the 400 mL of cell-depleted culture supernatant was centrifuged at 13,000× g for 2 min and subsequently passed through a 1.2 µm syringe filter. The filtered culture supernatant was further centrifuged at 21,000× g (Thermo Scientific TM Sorvall RC-6 Plus superspeed centrifuge; Waltham, MA, USA), 4 °C for 70 min to collect the MV pellet. Then, the upper 90% of the supernatant was further passed through a 0.22 µm membrane filter and centrifuged at 110,000× g (Thermo Scientific ™ Sorvall WX80 ultracentrifuge), 4 °C for 90 min to collect the Exo pellet. Finally, these EV pellets were washed with sterile 0.22 µm-filtered phosphate-buffered saline (PBS) and stored at −80 °C until use. The EVs isolated from the supernatants of the cultures with early-stage development were designated as early-stage-derived MV (MV-E) and early-stage-derived Exo (Exo-E), while the EVs isolated from the culture supernatants of late-stage development were called late-stage-derived MV (MV-L) and late-stage-derived Exo (Exo-L), respectively. The control EVs were separated from the culture supernatant of 10 µM A23187-activated uninfected RBCs at 20% hematocrit for 24 h; culture supernatant was collected to isolate A23187-MV and A23187-Exo in the same manner as the Pf-EVs [36].
2.3. Transmission Electron Microscopy (TEM)
The morphology of Pf-EVs was visualized by TEM. First, the isolated Pf-EVs were fixed with 2% paraformaldehyde solution for 30 min. Next, the 200-mesh formvar/carbon-coated copper electron microscope (EM) grid was glow-discharged. The PFA-fixed Pf-EVs solution (3 µL) was loaded on the EM grid and incubated for 15 min. Then, the EM grid was stained with 50 µL of 2% uranyl acetate and incubated for 5–7 min. Finally, the excess solution was removed by filter paper and the EM grid was washed ten times with PBS and distilled water. The grid was imaged using an FEI Tecnai T12 Transmission Electron Microscope (Field Electron and Ion Co., Hillsboro, OR, USA) operated at 80 kV. Images were analyzed by DigitalMicrograph® software (Gatan; Pleasanton, CA, USA).
2.4. Nanoparticle Tracking Analysis (NTA)
To characterize the size and particle concentration of Pf-EVs, the isolated EVs were diluted with sterile 0.22 µm-filtered PBS at the appropriate dilution to obtain the optimum measurement range. The diluted samples were injected into the Nanosight NS300 Analyzer (Malvern Pananalytical; Worcester, UK). Nanosight acquisition and analysis parameters were as follows: camera level: 14–15; detection threshold: 5; syringe pump speed: 50 µL/s; number of videos acquired per sample: 5; video duration: 60 s. Data analysis was performed using NTA software version 3.4 (Malvern Pananalytical; Worcester, UK). The system was washed with PBS between samples.
2.5. Western Blotting Analysis
The protein concentration of Pf-EVs was determined by Bradford protein assay (Bio-Rad Laboratories; Hercules, CA, USA) according to the manufacturer’s protocol. Isolated Pf-EVs (10 µg) were run on 12% SDS-polyacrylamide electrophoresis, and proteins were transferred to polyvinylidene difluoride membranes. These were blocked with non-fat dry milk in Tris-buffered saline containing 0.1% Tween20 (TBST), and the blots were probed with primary antibodies including β-actin (clone 8H10D10, Cell Signaling Technology®; Danvers, MA, USA), stomatin (clone E-5, Santa Cruz Biotechnology; Dallas, TX, USA), and flotillin-1 (clone C-2, Santa Cruz Biotechnology; Dallas, TX, USA), each diluted 1:400 in blocking buffer in TBST for 1 h. Then, the blots were washed with TBST and incubated with goat anti-mouse, PER-conjugated HRP (ImmunoTools GmbH; Friesoythe, Germany) at a dilution of 1:4000 for 1 h at room temperature. After washing, Clarity ™ Western ECL substrate was added (Bio-Rad Laboratories; Hercules, CA, USA); signal was detected by ImageQuant LAS 4010 (GE Healthcare; Boston, CA, USA).
2.6. Exosomal Protein Preparation
Total protein was isolated using 0.5% SDS solution and measured with Lowry assay using bovine serum albumin as the standard [37]. Protein samples (5 µg) were subjected to in-solution digestion. The samples were completely dissolved in 10 mM ammonium bicarbonate (AMBIC), then disulfide bonds were reduced with 5 mM dithiothreitol (DTT) in 10 mM AMBIC at 60 °C for 1 h; sulfhydryl groups were alkylated using 15 mM Iodoacetamide (IAA) in 10 mM AMBIC at room temperature for 45 min in the dark. For digestion, samples were mixed with 50 ng/µL of sequencing grade trypsin (1:20 ratio) (Promega; Madison, WI, USA) and incubated at 37 °C overnight. Before LC-MS/MS analysis, the digested samples were dried and protonated with 0.1% formic acid before injection into LC-MS/MS.
2.7. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
Tryptic peptide samples were injected in triplicate (5 µL each) into an HCT Ultra Ion Trap LC-MS System (Bruker Daltonics; Hamburg, Germany) which was coupled to a nanoLC system: UltiMate 3000 LC System (Thermo Fisher Scientific; Waltham, MA, USA) as well as an electrospray at the flow rate of 300 ոL/min to a nanocolumn (PepSwift monolithic column 100 mm internal diameter 50 mm). Each injection was spiked with 200 fmol of tryptic digested bovine serum albumin. Mobile phases of solvent A (0.1% formic acid) and solvent B (80% acetonitrile and 0.1% formic acid) were used to elute peptides, employing a linear gradient (10–70%) of solvent B at 0–13 min (the time-point of retention) followed by 90% solvent B at 13–15 min to remove all peptides in the column. A final elution of 10% solvent B at 15–20 min was carried out to remove any remaining salts.
2.8. Bioinformatics and Data Analysis
For protein quantitation, DeCyder MS Differential Analysis software (DeCyder MS, GE Healthcare; Boston, MA, USA) was used. Acquired LC-MS raw data were converted and the PepDetect module was used for automated peptide detection, charge state assignments, and quantitation based on peptide ion signal intensities in MS mode. The analyzed MS/MS data from DeCyder MS were submitted to the Mascot software version 2.2 (Matrix Science; London, UK) and searched against the Uniprot database for protein identification. The interrogation consisted of taxonomy (Homo sapiens or Plasmodium falciparum); enzyme (trypsin); variable modifications (carbamidomethyl, oxidation of methionine residues); mass values (monoisotopic); protein mass (unrestricted); peptide mass tolerance (1.2 Da); fragment mass tolerance (±0.6 Da); peptide charge state (1+, 2+ and 3+); max missed cleavages. Data normalization and quantification of differences in protein abundance between the control and treated samples were performed and visualized using MultiExperiment Viewer (MeV) software version 4.6.1 [38]. Briefly, peptide intensities from the LC-MS analyses were transformed and normalized using a mean central tendency procedure. Using statistical tests of variance of differences (ANOVA), the data sets were assessed for statistically significant amounts of protein (p < 0.05). Then, all differentially expressed proteins were analyzed for intersections among the different sample groups using jvenn [39]. Gene ontology annotation, including biological process, molecular function and cellular component, was performed using the PANTHER Classification System version 17.0 (http://www.pantherdb.org, accessed on 22 March 2022) [40]. The identified proteins were simultaneously submitted to Search Tool for Interacting Chemicals (STITCH) version 5.0 (http://stitch.embl.de, accessed on 5 May 2021) (European Molecular Biology Laboratory; Heidelberg, Germany) to search for linked cellular functions and other interactions between proteins and small molecules [41]. A heatmap analysis was generated using FunRich version 3.1.3 (http://www.funrich.org, accessed on 3 April 2022) [42].
2.9. Invasion and Growth Development Evaluation
Immature schizont parasites (42–44 h) were synchronized with the Percoll gradient solution [43]. In addition, the number of the schizont-infected and newly prepared RBCs were counted with BD TrucountTM tubes (BD Biosciences; San Jose, CA, USA). Highly synchronized schizont-stage parasites (0.5% parasitemia in 1 × 107 total RBCs) were seeded onto 96-well flat-bottom plates; each well contained a final culture volume of 100 µL. Isolated EVs were added into the cultures. Invasion efficiency was measured at 6 and 54 h post-coculturing by staining cultured RBCs with SYRB Green I nucleic acid staining dye; at least 50,000 RBCs were analyzed by BD® LSR II Flow Cytometer (BD Biosciences; San Jose, CA, USA) [44]. Data were acquired using FACSDivaTM software Version 7 (BD Biosciences; San Jose, CA, USA) and analyzed by FlowJo software Version 10.8.1 (Becton, Dickinson and Company; Ashland, OR, USA). The percentage parasitemia was determined by gating on SYRB Green I-positive RBCs. Each experiment was set up in triplicate wells, and the average parasitemia calculated.
2.10. Statistic Analysis
Data were analyzed using GraphPad Prism software version 9.3.1 (GraphPad; San Diego, CA, USA) and presented as mean ± SEM. The two-way ANOVA, followed by Tukey’s post-hoc test, was used to assess differences between more than two data groups. Differences were considered significant when the p-value was < 0.05.
3. Results
3.1. Characterization of Pf-Infected RBC-Derived EVs
The Pf-EVs and control A23187-EVs were isolated from culture supernatants by multi-step centrifugation, and their characteristics were analyzed. According to MISEV2018 criteria, the combination of images of single EVs and single-particle analysis should be performed to test the characteristics of EVs [45]. Thus, TEM and NTA were utilized to determine the characteristics of these Pf-EVs and control A23187-EVs. TEM showed the morphology of isolated Pf-Exo to be a cup-shaped membrane-bound vesicle approximately 100 nm in diameter (representative Pf-Exo; Figure 1A). By NTA, the average diameters of Pf-Exo and Pf-MV were 97.9 ± 8.1 nm and 159.7 ± 27.2 nm, respectively. A representative NTA showed that these sizes were 92.8 ± 2.8 nm and 126.6 ± 8.0 nm (Figure 1B). Western blot analysis was also performed to demonstrate the presence of EV markers in isolated Pf-EVs. β-actin and flotillin-1 were used as markers for cytosolic proteins and stomatin, an integral membrane protein of RBCs, was used as a marker of the plasma membrane of the originated cells. The results showed that both Pf-Exo (left lane) and Pf-MV (right lane) carried these three proteins, but at different levels. A representative Western blot analysis of Pf-MV and Pf-Exo isolated from 3D7 strain is shown (Figure 1C). The TEM, NTA and Western blots confirmed that Pf-EVs were successfully isolated from supernatants of malarial cultures.
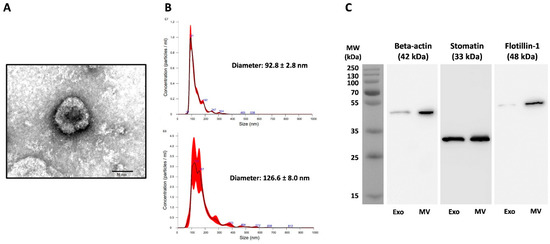
Figure 1.
Characteristics of Pf-EVs isolated from supernatants of P. falciparum cultures. Exo, exosome; MV, microvesicle. (A) Representative TEM of Pf-Exo from P. falciparum 3D7. Image presents negatively stained Pf-Exo, imaged by TEM at amplification of 80,000X; the bar represents 50 nm. (B) Representative NTA results of Pf-Exo (top) and Pf-MV (bottom) from P. falciparum 3D7. (C) Representative Western blot of β-actin, stomatin, and flotillin-1 in Pf-Exo (left lane) and Pf-MV (right lane) from P. falciparum 3D7-infected RBCs.
3.2. Characterization of the Human Proteome in P. falciparum-Infected RBC-Derived EVs
To compare the proteomes found in Pf-EVs derived from different P. falciparum strains, the proteomic profiles of Pf-EVs from the different strains and developmental stages were identified with two databases: the human and P. falciparum protein databases.
Totals of 3717 and 3774 human proteins were found in MV and Exo isolated derived from supernatants of A23187-activated RBCs and P. falciparum-infected RBC cultures, respectively. Among these human proteins, EVs carried numerous RBC-specific proteins including hemoglobin subunit, acetylcholinesterase, excitatory amino acid transporter (EAAT), Band 3 anion transport protein, ankyrin, tropomodulin, CD44, CD47, glutathione S-transferase, thioredoxin reductase, heat shock protein 90-beta (HSP 90), carbonic anhydrase, and 14-3-3 protein [46,47]. Our isolated EVs contained several EV-specific proteins including AHCY, FLNA, ATP1A1, ENO1, EEF2, STOM, YWHAG, ALB, and RAB5A that are classified as Top100 proteins in the ExoCarta database and can be used as EV markers [48]. Thus, the EVs studied here contained both RBC-specific and EV-specific proteins.
When considering the human proteins present only in MV and Exo derived from Pf-infected RBC cultures, there were 150 proteins in Pf-MV and 164 proteins in Pf-Exo. Of these, only three proteins were expressed in both types of EVs. They were ATP-dependent RNA helicase DDX23 (Uniprot accession: Q9BUQ8), protein disulfide-isomerase A4 (Uniprot accession: P13667) and adenylate cyclase type 7 (Uniprot accession: P51828). Thus, there were 147 proteins found exclusively in Pf-MV and 161 proteins found only in Pf-Exo. In analyzing the proteins in Pf-EVs based on P. falciparum stage of development, 139 of the 150 proteins were found in both MV-E and MV-L. Ten proteins were uniquely found in MV-E, only one in MV-L. Among 164 human proteins in the Exo, 157 were found in both Exo-E and Exo-L. Three proteins were found uniquely in Exo-E, and four in Exo-L (Figure 2A). The human proteins detected uniquely in MV-E, MV-L, Exo-E, and Exo-L are listed in Table 1. These data suggested that the human proteins expressed in Pf-EVs were unique for the type of EVs but not the stage of P. falciparum development.
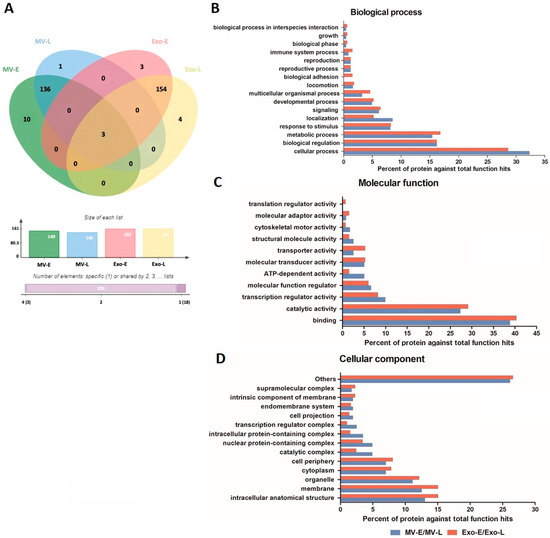
Figure 2.
Human proteins in Pf-EVs of different type and developmental stage (MV-E, MV-L, Exo-E, and Exo-L). (A) Venn diagram showing the proteins differentially expressed in each type of Pf-EVs. The column graph presents the number of proteins in each type of EVs, and the bar graph indicates the number of proteins shared between types of EV. (B–D) GO annotation analyses of the common proteins in MV (MV-E/MV-L) and Exo (Exo-E/Exo-L); (B) biological process, (C) molecular function, and (D) cellular component.

Table 1.
List of human proteins detected only in MV-E, MV-L, Exo-E, and Exo-L.
Next, the common human proteins expressed in Pf-MV and Pf-Exo were analyzed in terms of Gene Ontology (GO) annotation by PANTHER Classification System. The GO classification system revealed that the 138 proteins in Pf-MV (MV-E and MV-L) and 154 in Pf-Exo (Exo-E and Exo-L) could be classified based on functional properties, including those related to biological processes, molecular functions, and cellular components. The biological processes of proteins found commonly in both Pf-MV and Pf-Exo were the cellular process (GO:0009987), biological regulation (GO:0065007), and metabolic process (GO:0008152). The other functional classes (less than 10.0% each) identified were: response to stimulus (GO:0050896), localization (GO:0051179), signaling (GO:0023052), developmental process (GO:0032502), multicellular organismal process (GO:0032501), locomotion (GO:0040011), biological adhesion (GO:0022610), reproductive process (GO:0022414), reproduction (GO:0000003), immune system process (GO:0002376), growth (GO:0040007), biological phase (GO:0044848), and biological process involved in interspecies interaction between organisms (GO:0044419) (Figure 2B). Of molecular functions, the proteins in both Pf-MV and Pf-Exo were associated with binding (GO:0005488), catalytic activity (GO:0003824), transcription regulator activity (GO:0140110), molecular function regulator (GO:0098772), ATP-dependent activity (GO:0140657), molecular transducer activity (GO:0060089), transporter activity (GO:0005215), structural molecule activity (GO:0005198), cytoskeletal motor activity (GO:0003774), molecular adaptor activity (GO:0060090), and translation regulator activity (GO:0045182) (Figure 2C). In addition, the cellular components of proteins in both Pf-MV and Pf-Exo were involved in intracellular anatomical structure (GO:0005622), membrane (GO:0016020), organelle (GO:0043226), cell periphery (GO:0071944), cytoplasm (GO:0005737), catalytic complex (GO:1902494), nuclear protein-containing complex (GO:0140513), and intracellular protein-containing complex (GO:01400535) (Figure 2D).
Comparison of specific proteins carried in each EV type from different P. falciparum strains showed that at least 40% of human proteins were found in EVs from all parasite strains. These common proteins made up 43.6% in MV-E, 45.7% in MV-L, 40.6% in Exo-E, and 42.2% in Exo-L. Less than 10% of these proteins were unique to a parasite strain in each EV group (Figure 3). These data suggested that many human proteins carried in Pf-EVs were conserved among different strains. However, the relative expression levels of these human proteins in Pf-EVs varied with parasite strain and stage. The heatmaps of unique human proteins carried in Pf-MV and Pf-Exo are shown in Figures S1 and S2, respectively.

Figure 3.
Venn diagrams showing the human proteins differentially found in each type of Pf-EVs from 4 P. falciparum strains (3D7, NF54, TM01, and TM02). (A) MV-E, n = 149; (B) MV-L, n = 140; (C) Exo-E, n = 160; (D) Exo-L, n = 161. The column graph presents the number of proteins in each strain of P. falciparum, and the bar graph indicates the cumulative number of proteins shared among the P. falciparum strains.
3.3. Characterization of the P. falciparum Proteome in Pf-EVs
In contrast to human protein profiles, a Venn diagram analysis showed that the P. falciparum proteins in Pf-EVs can be divided into three subgroups: (i) proteins common to all Pf-EVs types (MV-E, MV-L, Exo-E, and Exo-L; 128 proteins), (ii) proteins expressed only in Pf-MV (MV-E and MV-L; 32 proteins), and (iii) proteins expressed only in Pf-Exo (Exo-E and Exo-L; 26 proteins) (Figure 4A). The P. falciparum proteins in each group were further grouped in terms of GO classification using PANTHER Classification System. The biological processes analysis showed that most identified proteins were involved in cellular processes (GO:0009987), followed by the metabolic processes (GO:0008152), biological regulation (GO:0065007), and response to stimulus (GO:0050896) (Figure 4B). The molecular functions analysis showed that identified proteins were mainly involved in binding (GO:0005488) and catalytic activity (GO:0003824) (Figure 4C). The cellular components analysis showed that identified proteins in Pf-EVs were involved in the protein-containing complex (GO:0032991), intracellular anatomical structure (GO:0005622), cytoplasm (GO:0005737), organelle (GO:0043226), and membrane (GO:0016020) (Figure 4D).
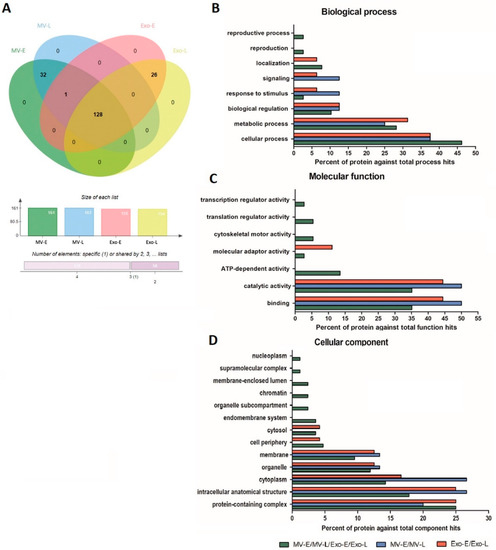
Figure 4.
P. falciparum proteins in different types of Pf-EVs and stages of development (MV-E, MV-L, Exo-E, and Exo-L). (A) Venn diagram showed the proteins differentially expressed in each type of Pf-EVs. The column graph presents the number of proteins in each type of EVs, and the bar graph indicates the number of proteins shared between the type of EVs. (B–D) GO annotation analysis of the proteins common to EVs (MV-E/MV-L/Exo-E/Exo-L), MV (MV-E/MV-L), and Exo (Exo-E/Exo-L); (B) biological process, (C) molecular function, and (D) cellular component.
To explore potential functional interactions among the P. falciparum proteins in Pf-EVs, a network was built based on the STITCH 5 online database. An additional ten most-related proteins have expanded the network based on high-throughput text-mining [49]. Potential functional interactions among the P. falciparum proteins were found only in MVs (MV-E and MV-L), with 31 of 32 proteins in Pf-MV matching proteins in the database. Functional enrichment of the KEGG pathway identified the mismatch repair (pathway ID 03430, FDR = 0.00129), glutathione metabolism (pathway ID 00480, FDR = 0.0045), DNA replication (pathway ID 03030, FDR = 0.0045), nucleotide excision repair (pathway ID 03420, FDR = 0.0045), glycolysis/gluconeogenesis (pathway ID 00010, FDR = 0.0159), purine metabolism (pathway ID 00230, FDR = 0.0169), and metabolic pathways (pathway ID 01100, FDR = 0.0356). In Pf-Exo, 24 of 26 proteins matched with proteins in the database, and the functional enrichment of the KEGG pathway identified ribosome (pathway ID 03010, FDR = 2.72 × 10−26)
In further evaluation of the protein diversity in EVs belonging to each P. falciparum strain, 71.4%, 72%, 65.1%, and 62.3% of proteins shared by all four strains were found in MV-E, MV-L, Exo-E, and Exo-L, respectively (Figure 5). These data suggested that the P. falciparum proteome in Pf-EVs was highly conserved across strains and that these percentages were higher than those belonging to the human proteome. Heatmap analyses comparing levels of P. falciparum proteins in each Pf-EVs type from the four P. falciparum strains are shown in Figure S3.
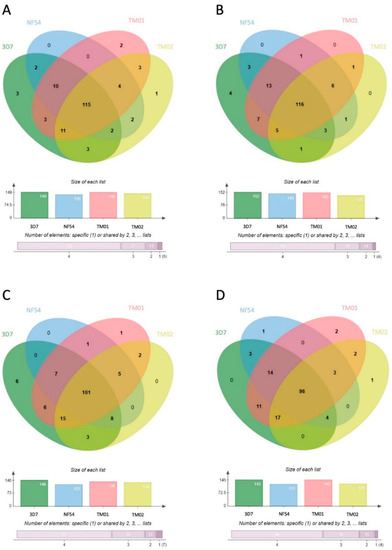
Figure 5.
Venn diagrams showing the P. falciparum proteins differentially expressed in each type of Pf-EVs from 4 P. falciparum strains (3D7, NF54, TM01, and TM02). (A) MV-E, n = 161; (B) MV-L, n = 161; (C) Exo-E, n = 155; (D) Exo-L, n = 154. The column graphs present the number of proteins from each strain of P. falciparum, and the bar graphs indicate the numbers of proteins shared by P. falciparum strains.
3.4. Pf-EVs Contain Parasite-Derived Proteins That Are Associated with Parasite Invasion
Among P. falciparum proteins found in Pf-EVs, many were major virulence proteins which play an essential role in RBC invasion and erythrocytic schizogony, such as AMA-1, reticulocyte binding protein, MSP-1, and EBA-175. Potential invasion-associated proteins found in Pf-EVs are listed in Table 2. Furthermore, a heatmap analysis helped distinguish these proteins by Pf-EV type and parasite strain (Figure 6). These data persuaded us to explore the relationship of these Pf-EVs to invasion efficiency and parasite development during the intraerythrocytic schizogony of each P. falciparum strain.

Table 2.
List of invasion-associated P. falciparum proteins detected in Pf-EVs.

Figure 6.
Heatmap of differential expression levels of invasion associated P. falciparum proteins detected in Pf-EVs. Side bar indicates differential protein expression between each sample (blue: decrease, red: increase). Rows: proteins; columns: samples.
3.5. Pf-EVs in Invasion Efficiency and Growth Development
The following experiment explored whether the Pf-EVs carrying invasion-associated P. falciparum proteins affected the merozoite RBC invasion process during the parasite’s life cycle. We added either MV (A23187-MV, Pf-MV-E and Pf-MV-L) or Exo (A23187-Exo, Pf-Exo-E and Pf-Exo-L) at 1 × 109 particles into cultures containing 1 × 107 RBCs. The cultures began with 0.5% parasitemia, and after two erythrocytic cycles were re-measured.
The first invasion cycle was determined at 6 h post co-incubation or after schizont rupture. The co-incubation then was continued over 48 h for second schizont rupture and was determined percentage of parasitemia as the second invasion cycle. The percent change of parasitemia was calculated relatively to control which was referred to the condition without the addition of EVs. In the first invasion cycle post-coincubation, the percentages of changes in parasitemia of control EVs (A23187-MV and A23187-Exo) from four strains were varied from 90.58 ± 4.38 to 102.04 ± 2.55, while those of Pf-MV coculture ranged from 80.15 ± 5.51 to 104.20 ± 2.38 and those of Pf-Exo coculture ranged from 83.90 ± 1.67 to 102.33 ± 2.54. Thus, the data seem to suggest that the effect of Pf-EVs on the invasion process was not clearly demonstrated at this time point. By contrast, in comparison with A23187-MV in the second invasion cycle, we observed a slight reduction in percent change in parasitemia with Pf-MV derived from NF54, TM01, and TM02 strains; Pf-Exo did not alter the culture parasitemia (Figure 7A).
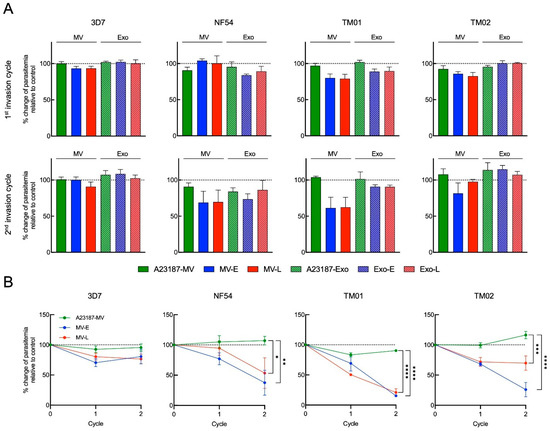
Figure 7.
The effect of MV-E, MV-L, Exo-E and Exo-L treatment in P. falciparum invasion. (A) The percent change of parasitemia in the presence of 1 × 109 EVs relative to control culturing without EVs in P. falciparum 3D7, NF54, TM01 and TM02, as indicated, at 6 h post-co-culture (top row, first invasion cycle) and 54 h post-co-culture (bottom row, second invasion cycle). (B) Line graphs represent percent change of parasitemia in the presence of 5 × 109 Pf-MV relative to control culturing without MV over two invasion cycles of 3D7, NF54, TM01 and TM02, as indicated. Percent change of parasitemia at second invasion cycle were compared between control culturing without MV and MV treatment group using a two-way ANOVA, followed by Tukey’s post-hoc test (*: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
To clarify the effect of Pf-MV on parasite invasion and growth, we increased the number of Pf-MV particles five-fold added to the cultures. As a result, there were short-term effects on P. falciparum development. However, when the co-cultures were observed through the second invasion cycle, significant reductions in parasite populations in cultures were seen with both MV-E and MV-L derived from NF54, TM01, and TM02 strains, but not from the 3D7 strain (Figure 7B). This finding suggested that EVs, especially Pf-MV, secreted from iRBCs moderate parasite growth of many P. falciparum strains.
4. Discussion
Like many other pathogens, EVs secreted from Plasmodium malaria play a role in disease complications. Indeed, elevated levels of circulating EVs have been detected in plasma of malaria-infected patients and correlated with severity of the disease [12,21]. Recently, RBC EVs derived from P. falciparum culture supernatants were widely investigated. In vitro studies demonstrated the role of RBC EVs in intercellular communication between parasite populations. Parasite genetic material is transferred via RBC EVs and internalized into iRBCs, influencing the transmission stage of parasites [25,29]. Obviously, parasite growth in vitro has limited maximum growth capacity. RBC EVs are reported to have a key role in the control of parasite density [30]. However, the way in which EVs impact in vitro cultures may reveal interesting mechanisms. For example, the priming of naïve RBCs with EVs supports parasite growth by altering membrane stiffness [50]. Thus, the involvement of EVs in intercellular parasite communication (parasite–parasite and host–parasite) is complex and needs to be better elucidated [51,52,53].
EVs play a key role in intercellular communication by transferring bioactive molecules such as proteins and genetic materials from parasites to host cells and parasites to parasites, thereby modifying the properties of recipient cells [25,27,34,50,54,55,56]. EVs in plasma have several cellular origins. EVs in supernatants of P. falciparum-RBC cultures are less numerous than in plasma of malaria patients. In this study, Pf-EVs were derived from the cultures of four P. falciparum strains, 3D7, NF54, TM01, and TM02. P. falciparum NF54 is a strain isolated from a patient with uncomplicated malaria in the Netherlands, but the origin of the parasite is thought to be in Africa [57,58]. P. falciparum 3D7 is one of the most studied strains of P. falciparum in research laboratories and is a clone from P. falciparum strain NF54. Although the NF54 and 3D7 are assumed to be genetically identical, phenotypic differences when grown in culture are reported [59]. The P. falciparum strains TM01 and TM02 were culture-adapted isolates which collected from uncomplicated and severe falciparum malaria patients in Thailand, respectively. In this study, EVs were termed as MV or Exo based on differential centrifugation at 21,000× g and 110,000× g, respectively. The average diameter of MV was 159.7 ± 27.2 nm, and the average diameter of Exo was 97.9 ± 8.1 nm. We admit that our separation protocol may not be sufficient to isolate Pf-MV and Pf-Exo as an overlap of the size ranges of both EVs was observed.
To determine protein content during distinct stages of development, parasites were synchronized and RBC culture supernatants were harvested after 24 and 48 h to acquire early and late stages, respectively. From the comparative proteomic profiling, both host and parasite proteins were identified in EVs. Among human proteins, the majority involved RBC membrane and cytosolic proteins such as hemoglobin, band 3, band 7 (stomatin), spectrin, CD47, CD55, acetylcholinesterase, and carbonic anhydrase, which are known to be enriched in RBC EVs [60,61,62]. When considering the human proteins found only in Pf-EVs (not in A23187-EVs), there were 150 proteins in Pf-MV and 164 proteins in Pf-Exo. Of these, only three proteins were expressed in both types of EVs. Considering the parasite protein profiles in early- and late-stage EVs, less than ten proteins were unique to one stage or the other (Table 1). Thus, human proteins expressed in Pf-EVs were unique for the type of EVs but not the stage of parasite and were common across strains. In contrast to human proteins, P. falciparum proteins were most often common to all Pf-EVs types (n = 128 proteins), but 32 were found only in Pf-MV and 26 only in Pf-Exo. Moreover, 62–72% of proteins were associated with all four strains.
Interestingly, P. falciparum protein profiles revealed several invasion-associated proteins in Pf-EVs. For example, MSP-1 (an essential protein in the parasite’s initial contact), proteins involved in the apical orientation step (EBA-175, PfRh1, PfRh2a, PfRh2b, and PfRh3), and AMA1 (a crucial protein in tight junction formation) were all found. Moreover, these parasite proteins are common in RBC EVs derived from P. falciparum culture supernatants [25,27,30,50,56,63]. Other interesting parasite proteins that were identified by proteomics include two proteins linked to the iRBC surface membrane, RESA and KAHRP. While these P. falciparum proteins were present in Pf-EVs from all four strains, the relative expression levels were observed. For instance, myosin-A, GBP-130, and AMA1 were less frequent in Pf-MV from 3D7 in comparison with other strains.
Focusing on the influence of EVs on parasite growth development, there have been interesting approaches to study this aspect. Mantel et al. profiled EV products and identified abundant RBC lipid raft proteins, RBC membrane proteins and others involved in parasite invasion of RBCs [25]. Their approach was to treat ring stage parasites with RBC EVs and investigate parasitemia after 48 h. However, they did not observe significant alterations in growth rates. In contrast to this finding, we showed that Pf-EVs, in particular, Pf-MVs from all strains, had more effect in terms of attenuating parasite invasion efficiency than did Pf-Exo, and the effects of Pf-MVs were significant only at high dose. Our study is in line with a report by Correa et al., who modified the approach by adding EVs isolated from high parasitemia cultures into those with mature trophozoites; they observed significant reductions in growth after 24 h [30]. Moreover, they identified Pf-LDH as an inducer of apoptosis and parasite density control. Recently, Dekel et al. modified the experiment again by priming naïve uninfected-RBCs (uRBCs) with Pf-EVs for 18 h and then introduced them to late-stage parasites [50]. After 12 h, they found an increase in parasite levels in the EV pretreatment cultures. They identified 20S proteasome as a key feature in altering RBC membrane stiffness and so facilitating parasite growth. In consistent with this report, we found both human and parasite proteins except that we did not find proteasomes of parasite origin. These discrepancies in the protein contents could be partially ascribed to: (1) Different approach in treating uRBCs with Pf-EVs. In our study, uRBCs were exposed to both Pf-EVs and parasites at the same time during malaria cultures without pretreatment with Pf-EVs. (2) Different stages of Pf-EVs that were used to treat the uRBCs. Unlike Dekel et al., who used late-stage Pf-EVs [50], our experiment used both early- and late stage-derived EVs. It is reasoned that EVs at different stages of parasite development exploit distinct selective sorting mechanisms for proteins and their mRNAs. Cumulative evidence suggests the existence of regulators that control the sorting of protein/mRNA cargo into EVs and their release by utilizing parasite own protein network [26,29,50,64]. While further study is needed to understand the complex molecular nature of such differences in proteomes, we suggest that such differences might be associated with distinct cargo sorting at each stage of parasite development which might have a crucial role in parasite–host communication.
Because certain P. falciparum strains are considered more virulent than others, and parasites causing severe malaria have greater invasion potential with less selective and multiplied more at high parasitemia than those of uncomplicated malaria [50,65,66]. An analogy with this notion would suggest that EVs from virulent and non-virulent malaria parasites may have different multiplication rates and invasiveness when invading host RBCs. In an effort to better understand the effect of Pf-EVs on the invasion efficiency and growth development during intraerythrocytic schizogony of each P. falciparum strain, in this study, we determined the effects of EVs both immediately after schizogony and then again after a second invasion cycle. Surprisingly, Pf-MVs from all strains had more of an effect attenuating parasite invasion efficiency than Pf-Exo. However, the effects of Pf-MVs were significant only at a high dose. Interestingly, no significant difference in parasite growth was observed when EVs isolated from uncomplicated (TM01) and severe (TM02) falciparum malaria were used to treat uRBCs. It should be emphasized here that the parasite growth in our study has limitations that must be considered; since our approach used an in vitro model, it was limited by the lack of a host immune response since RBCs were the only source of EVs. These limitations might explain the similar findings among strains that differed in virulence. Though the protein contents present in Pf-EVs were highly conserved among all four P. falciparum strains, the effects of Pf-MV were markedly different in culture-adapted isolates versus laboratory reference strains. Long-term P. falciparum laboratory strains, such as 3D7, have been adapted to easily maintain the culture, whereas more recently acquired P. falciparum isolates might be more susceptible to intercellular communication. However, as the effect of EVs in invasion activities is from the whole EVs activity, the results cannot indicate the exact potential essential proteins in the process. In addition, because we did not include the drug-resistant P. falciparum strains in the study, questions about the differences in EVs from these strains still need further investigation.
This finding provides some knowledge to better understand the role of Pf-EVs in regulating malaria parasite growth. Furthermore, as Pf-EVs act as cargo carrying the quorum-sensing molecules with invasion inhibitory activity, identifying potential quorum-sensing molecules in Pf-EVs might benefit the new therapeutic approach and management during malaria infection. For example, administering the current antimalarial drugs combined with synthetic quorum-sensing molecules or bioengineered EVs carrying quorum-sensing molecules, these should synergize with each other in controlling the parasitemia in drug-resistance malaria strain. In addition, the EVs or quorum-sensing molecules might be used as a biomarker for cell-free diagnosis in the endemic malaria area, in which the mutant P. falciparum parasites lack histidine-rich protein and cannot be detected with the present rapid diagnostic tests.
5. Conclusions
This work investigated protein cargos in Pf-EVs generated from four strains of P. falciparum using LC-MS/MS analysis. Their protein profiles revealed human proteins related to RBC EV markers and highly conserved P. falciparum proteins associated with the RBC invasion process. Unexpectedly, Pf-MVs had the effect of attenuating, rather than supporting, parasite invasion efficiency. Our evidence supports the involvement of Pf-EVs in growth-density regulation via a quorum sensing-like mechanism, a process observed across strains of P. falciparum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcm11144250/s1, Figure S1: Heatmap of unique human proteins carried in Pf-MV; Figure S2: Heatmap of unique human proteins carried in Pf-Exo; Figure S3: Heatmap analysis of differential expression levels of P. falciparum protein in each type of Pf-EVs from 4 P. falciparum strains
Author Contributions
Conceptualization, K.P., K.C. and L.K.; methodology, K.P., L.K., S.R., N.P., K.L., A.A., N.S. and K.S.; software, S.R. and N.P.; formal analysis, S.V., L.K., S.R., K.L., A.A. and N.S.; investigation, S.V., L.K., S.R., K.L., A.A., N.S., N.P. and K.S.; resources, K.P., K.C.; writing original draft preparation, L.K. and S.V.; writing—review and editing, L.K., K.P., S.V., S.R., K.C., N.P., K.L., A.A., N.S. and K.S.; supervision, K.P.; project administration, K.S. and L.K.; funding acquisition, K.P., K.C. and L.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Thailand Research Fund (TRF); Distinguished Research Professor Grant, Grant number TRF-DPG5980001; TRF-Royal Golden Jubilee Ph.D. Program, Grant number PHD/0027/2557 and TRF and the Office of the Higher Education Commission; Research Grant for New Scholar, Grant number MRG6080184. S.V. is supported by a fellowship from the TRF, The Royal Golden Jubilee Ph.D. Program (PHD/0027/2557). L.K. is supported by Chalermprakiat grant, Faculty of Medicine Siriraj Hospital, Mahidol University.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Siriraj Institutional Review Board, the Human Research Protection Unit of the Faculty of Medicine Siriraj Hospital, Mahidol University (COA no. Si 634/2019; approved 13 September 2019).
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to the donors who provided blood for use in malarial cultures. Study equipment was supported by the Research Department, Faculty of Medicine Siriraj Hospital, Mahidol University.
Conflicts of Interest
The authors declare that they have no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- WHO. World Malaria Report 2021; WHO Press: Geneva, Switzerland, 2021. [Google Scholar]
- Volz, J.C.; Yap, A.; Sisquella, X.; Thompson, J.K.; Lim, N.T.; Whitehead, L.W.; Chen, L.; Lampe, M.; Tham, W.H.; Wilson, D.; et al. Essential role of the PfRh5/PfRipr/CyRPA complex during Plasmodium falciparum invasion of erythrocytes. Cell Host Microbe 2016, 20, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Mayer, D.C.; Singh, S.; Reid, M.; Miller, L.H. Domain III of Plasmodium falciparum apical membrane antigen 1 binds to the erythrocyte membrane protein Kx. Proc. Natl. Acad. Sci. USA 2005, 102, 5552–5557. [Google Scholar] [CrossRef] [PubMed]
- Treeck, M.; Zacherl, S.; Herrmann, S.; Cabrera, A.; Kono, M.; Struck, N.S.; Engelberg, K.; Haase, S.; Frischknecht, F.; Miura, K.; et al. Functional analysis of the leading malaria vaccine candidate AMA-1 reveals an essential role for the cytoplasmic domain in the invasion process. PLoS Pathog. 2009, 5, e1000322. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chu, R.; Yin, Y.; Yu, H.; Xu, Q.; Yang, B.; Sun, Y.; Song, J.; Wang, Q.; Xu, J.; et al. Glycosylphosphatidylinositol-anchored micronemal antigen (GAMA) interacts with the band 3 receptor to promote erythrocyte invasion by malaria parasites. J. Biol. Chem. 2022, 298, 101765. [Google Scholar] [CrossRef] [PubMed]
- Schlott, A.C.; Knuepfer, E.; Green, J.L.; Hobson, P.; Borg, A.J.; Morales-Sanfrutos, J.; Perrin, A.J.; Maclachlan, C.; Collinson, L.M.; Snijders, A.P.; et al. Inhibition of protein N-myristoylation blocks Plasmodium falciparum intraerythrocytic development, egress and invasion. PLoS Biol. 2021, 19, e3001408. [Google Scholar] [CrossRef]
- Robert-Paganin, J.; Robblee, J.P.; Auguin, D.; Blake, T.C.A.; Bookwalter, C.S.; Krementsova, E.B.; Moussaoui, D.; Previs, M.J.; Jousset, G.; Baum, J.; et al. Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism. Nat. Commun. 2019, 10, 3286. [Google Scholar] [CrossRef]
- Patarroyo, M.A.; Molina-Franky, J.; Gomez, M.; Arevalo-Pinzon, G.; Patarroyo, M.E. Hotspots in Plasmodium and RBC Receptor-Ligand Interactions: Key Pieces for Inhibiting Malarial Parasite Invasion. Int. J. Mol. Sci. 2020, 21, 4729. [Google Scholar] [CrossRef]
- Cowman, A.F.; Tonkin, C.J.; Tham, W.H.; Duraisingh, M.T. The Molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe 2017, 22, 232–245. [Google Scholar] [CrossRef]
- Lopaticki, S.; Maier, A.G.; Thompson, J.; Wilson, D.W.; Tham, W.H.; Triglia, T.; Gout, A.; Speed, T.P.; Beeson, J.G.; Healer, J.; et al. Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect. Immun. 2011, 79, 1107–1117. [Google Scholar] [CrossRef]
- Williams, A.R.; Douglas, A.D.; Miura, K.; Illingworth, J.J.; Choudhary, P.; Murungi, L.M.; Furze, J.M.; Diouf, A.; Miotto, O.; Crosnier, C.; et al. Enhancing blockade of Plasmodium falciparum erythrocyte invasion: Assessing combinations of antibodies against PfRH5 and other merozoite antigens. PLoS Pathog. 2012, 8, e1002991. [Google Scholar] [CrossRef]
- Nantakomol, D.; Dondorp, A.M.; Krudsood, S.; Udomsangpetch, R.; Pattanapanyasat, K.; Combes, V.; Grau, G.E.; White, N.J.; Viriyavejakul, P.; Day, N.P.; et al. Circulating red cell-derived microparticles in human malaria. J. Infect. Dis. 2011, 203, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Slomka, A.; Urban, S.K.; Lukacs-Kornek, V.; Zekanowska, E.; Kornek, M. Large extracellular vesicles: Have we found the holy grail of inflammation? Front. Immunol. 2018, 9, 2723. [Google Scholar] [CrossRef]
- Hezel, M.E.V.; Nieuwland, R.; Bruggen, R.V.; Juffermans, N.P. The Ability of Extracellular vesicles to induce a pro-inflammatory host response. Int. J. Mol. Sci. 2017, 18, 1285. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xie, F.; Wang, L.; Zhang, L.; Zhang, S.; Fang, M.; Zhou, F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell Mol. Immunol. 2020, 17, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Julich, H.; Willms, A.; Lukacs-Kornek, V.; Kornek, M. Extracellular vesicle profiling and their use as potential disease specific biomarker. Front. Immunol. 2014, 5, 413. [Google Scholar] [CrossRef]
- El-Assaad, F.; Wheway, J.; Hunt, N.H.; Grau, G.E.; Combes, V. Production, fate and pathogenicity of plasma microparticles in murine cerebral malaria. PLoS Pathog. 2014, 10, e1003839. [Google Scholar] [CrossRef]
- Couper, K.N.; Barnes, T.; Hafalla, J.C.; Combes, V.; Ryffel, B.; Secher, T.; Grau, G.E.; Riley, E.M.; de Souza, J.B. Parasite-derived plasma microparticles contribute significantly to malaria infection-induced inflammation through potent macrophage stimulation. PLoS Pathog. 2010, 6, e1000744. [Google Scholar] [CrossRef]
- Combes, V.; Coltel, N.; Alibert, M.; van Eck, M.; Raymond, C.; Juhan-Vague, I.; Grau, G.E.; Chimini, G. ABCA1 gene deletion protects against cerebral malaria: Potential pathogenic role of microparticles in neuropathology. Am. J. Pathol. 2005, 166, 295–302. [Google Scholar] [CrossRef]
- Penet, M.F.; Abou-Hamdan, M.; Coltel, N.; Cornille, E.; Grau, G.E.; de Reggi, M.; Gharib, B. Protection against cerebral malaria by the low-molecular-weight thiol pantethine. Proc. Natl. Acad. Sci. USA 2008, 105, 1321–1326. [Google Scholar] [CrossRef] [PubMed]
- Campos, F.M.; Franklin, B.S.; Teixeira-Carvalho, A.; Filho, A.L.; de Paula, S.C.; Fontes, C.J.; Brito, C.F.; Carvalho, L.H. Augmented plasma microparticles during acute Plasmodium vivax infection. Malar. J. 2010, 9, 327. [Google Scholar] [CrossRef] [PubMed]
- Pankoui Mfonkeu, J.B.; Gouado, I.; Fotso Kuate, H.; Zambou, O.; Amvam Zollo, P.H.; Grau, G.E.; Combes, V. Elevated cell-specific microparticles are a biological marker for cerebral dysfunctions in human severe malaria. PLoS ONE 2010, 5, e13415. [Google Scholar] [CrossRef] [PubMed]
- Mantel, P.Y.; Hoang, A.N.; Goldowitz, I.; Potashnikova, D.; Hamza, B.; Vorobjev, I.; Ghiran, I.; Toner, M.; Irimia, D.; Ivanov, A.R.; et al. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 2013, 13, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Sisquella, X.; Ofir-Birin, Y.; Pimentel, M.A.; Cheng, L.; Abou Karam, P.; Sampaio, N.G.; Penington, J.S.; Connolly, D.; Giladi, T.; Scicluna, B.J.; et al. Malaria parasite DNA-harbouring vesicles activate cytosolic immune sensors. Nat. Commun. 2017, 8, 1985. [Google Scholar] [CrossRef]
- Sampaio, N.G.; Emery, S.J.; Garnham, A.L.; Tan, Q.Y.; Sisquella, X.; Pimentel, M.A.; Jex, A.R.; Regev-Rudzki, N.; Schofield, L.; Eriksson, E.M. Extracellular vesicles from early stage Plasmodium falciparum-infected red blood cells contain PfEMP1 and induce transcriptional changes in human monocytes. Cell Microbiol. 2018, 20, e12822. [Google Scholar] [CrossRef]
- Mantel, P.Y.; Hjelmqvist, D.; Walch, M.; Kharoubi-Hess, S.; Nilsson, S.; Ravel, D.; Ribeiro, M.; Gruring, C.; Ma, S.; Padmanabhan, P.; et al. Infected erythrocyte-derived extracellular vesicles alter vascular function via regulatory Ago2-miRNA complexes in malaria. Nat. Commun. 2016, 7, 12727. [Google Scholar] [CrossRef]
- Regev-Rudzki, N.; Wilson, D.W.; Carvalho, T.G.; Sisquella, X.; Coleman, B.M.; Rug, M.; Bursac, D.; Angrisano, F.; Gee, M.; Hill, A.F.; et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 2013, 153, 1120–1133. [Google Scholar] [CrossRef]
- Correa, R.; Coronado, L.; Caballero, Z.; Faral-Tello, P.; Robello, C.; Spadafora, C. Extracellular vesicles carrying lactate dehydrogenase induce suicide in increased population density of Plasmodium falciparum in vitro. Sci. Rep. 2019, 9, 5042. [Google Scholar] [CrossRef]
- Pena, R.T.; Blasco, L.; Ambroa, A.; Gonzalez-Pedrajo, B.; Fernandez-Garcia, L.; Lopez, M.; Bleriot, I.; Bou, G.; Garcia-Contreras, R.; Wood, T.K.; et al. Relationship between quorum sensing and secretion systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef]
- Briggs, E.M.; Rojas, F.; McCulloch, R.; Matthews, K.R.; Otto, T.D. Single-cell transcriptomic analysis of bloodstream Trypanosoma brucei reconstructs cell cycle progression and developmental quorum sensing. Nat. Commun. 2021, 12, 5268. [Google Scholar] [CrossRef] [PubMed]
- Tettey, M.D.; Rojas, F.; Matthews, K.R. Extracellular release of two peptidases dominates generation of the trypanosome quorum-sensing signal. Nat. Commun. 2022, 13, 3322. [Google Scholar] [CrossRef] [PubMed]
- Gualdron-Lopez, M.; Flannery, E.L.; Kangwanrangsan, N.; Chuenchob, V.; Fernandez-Orth, D.; Segui-Barber, J.; Royo, F.; Falcon-Perez, J.M.; Fernandez-Becerra, C.; Lacerda, M.V.G.; et al. Characterization of Plasmodium vivax proteins in plasma-derived exosomes from malaria-infected liver-chimeric humanized mice. Front. Microbiol. 2018, 9, 1271. [Google Scholar] [CrossRef]
- Aparici-Herraiz, I.; Gualdron-Lopez, M.; Castro-Cavadia, C.J.; Carmona-Fonseca, J.; Yasnot, M.F.; Fernandez-Becerra, C.; Del Portillo, H.A. Antigen discovery in circulating extracellular vesicles from Plasmodium vivax patients. Front. Cell Infect. Microbiol. 2021, 11, 811390. [Google Scholar] [CrossRef]
- Salzer, U.; Hinterdorfer, P.; Hunger, U.; Borken, C.; Prohaska, R. Ca(++)-dependent vesicle release from erythrocytes involves stomatin-specific lipid rafts, synexin (annexin VII), and sorcin. Blood J. Am. Soc. Hematol. 2002, 99, 2569–2577. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef] [PubMed]
- Rivadeneira, E.M.; Wasserman, M.; Espinal, C.T. Separation and concentration of schizonts of Plasmodium falciparum by Percoll gradients. J. Protozool. 1983, 30, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Vimonpatranon, S.; Chotivanich, K.; Sukapirom, K.; Lertjuthaporn, S.; Khowawisetsut, L.; Pattanapanyasat, K. Enumeration of the invasion efficiency of Plasmodium falciparum in vitro in four different red blood cell populations using a three-color flow cytometry-based method. Cytom. Part A 2019, 95, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Bryk, A.H.; Wisniewski, J.R. Quantitative Analysis of Human Red Blood Cell Proteome. J. Proteome Res. 2017, 16, 2752–2761. [Google Scholar] [CrossRef]
- Ravenhill, B.J.; Kanjee, U.; Ahouidi, A.; Nobre, L.; Williamson, J.; Goldberg, J.M.; Antrobus, R.; Dieye, T.; Duraisingh, M.T.; Weekes, M.P. Quantitative comparative analysis of human erythrocyte surface proteins between individuals from two genetically distinct populations. Commun. Biol. 2019, 2, 350. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Kuhn, M.; von Mering, C.; Campillos, M.; Jensen, L.J.; Bork, P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36, D684–D688. [Google Scholar] [CrossRef]
- Dekel, E.; Yaffe, D.; Rosenhek-Goldian, I.; Ben-Nissan, G.; Ofir-Birin, Y.; Morandi, M.I.; Ziv, T.; Sisquella, X.; Pimentel, M.A.; Nebl, T.; et al. 20S proteasomes secreted by the malaria parasite promote its growth. Nat. Commun. 2021, 12, 1172. [Google Scholar] [CrossRef]
- Yoon, Y.J.; Kim, O.Y.; Gho, Y.S. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014, 47, 531–539. [Google Scholar] [CrossRef]
- Opadokun, T.; Rohrbach, P. Extracellular vesicles in malaria: An agglomeration of two decades of research. Malar. J. 2021, 20, 442. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, N.G.; Cheng, L.; Eriksson, E.M. The role of extracellular vesicles in malaria biology and pathogenesis. Malar. J. 2017, 16, 245. [Google Scholar] [CrossRef] [PubMed]
- Antwi-Baffour, S.; Adjei, J.K.; Agyemang-Yeboah, F.; Annani-Akollor, M.; Kyeremeh, R.; Asare, G.A.; Gyan, B. Proteomic analysis of microparticles isolated from malaria positive blood samples. Proteome Sci. 2016, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Martin-Jaular, L.; Nakayasu, E.S.; Ferrer, M.; Almeida, I.C.; Del Portillo, H.A. Exosomes from Plasmodium yoelii-infected reticulocytes protect mice from lethal infections. PLoS ONE 2011, 6, e26588. [Google Scholar] [CrossRef] [PubMed]
- Abdi, A.; Yu, L.; Goulding, D.; Rono, M.K.; Bejon, P.; Choudhary, J.; Rayner, J. Proteomic analysis of extracellular vesicles from a Plasmodium falciparum Kenyan clinical isolate defines a core parasite secretome. Wellcome Open Res. 2017, 2, 50. [Google Scholar] [CrossRef]
- Ponnudurai, T.; Leeuwenberg, A.D.; Meuwissen, J.H. Chloroquine sensitivity of isolates of Plasmodium falciparum adapted to in vitro culture. Trop. Geogr. Med. 1981, 33, 50–54. [Google Scholar]
- Bryant, J.M.; Baumgarten, S.; Lorthiois, A.; Scheidig-Benatar, C.; Claes, A.; Scherf, A. De Novo Genome Assembly of a Plasmodium falciparum NF54 clone using single-molecule real-time sequencing. Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Delves, M.J.; Straschil, U.; Ruecker, A.; Miguel-Blanco, C.; Marques, S.; Dufour, A.C.; Baum, J.; Sinden, R.E. Routine in vitro culture of P. falciparum gametocytes to evaluate novel transmission-blocking interventions. Nat. Protoc. 2016, 11, 1668–1680. [Google Scholar] [CrossRef]
- Prudent, M.; Delobel, J.; Hubner, A.; Benay, C.; Lion, N.; Tissot, J.D. Proteomics of stored red blood cell membrane and storage-induced microvesicles reveals the association of flotillin-2 with band 3 complexes. Front. Physiol. 2018, 9, 421. [Google Scholar] [CrossRef]
- Chiangjong, W.; Netsirisawan, P.; Hongeng, S.; Chutipongtanate, S. Red blood cell extracellular vesicle-based drug delivery: Challenges and opportunities. Front. Med. 2021, 8, 761362. [Google Scholar] [CrossRef]
- Thangaraju, K.; Neerukonda, S.N.; Katneni, U.; Buehler, P.W. Extracellular vesicles from red blood cells and their evolving roles in health, coagulopathy and therapy. Int. J. Mol. Sci. 2020, 22, 153. [Google Scholar] [CrossRef] [PubMed]
- Tiberti, N.; Latham, S.L.; Bush, S.; Cohen, A.; Opoka, R.O.; John, C.C.; Juillard, A.; Grau, G.E.; Combes, V. Exploring experimental cerebral malaria pathogenesis through the characterisation of host-derived plasma microparticle protein content. Sci. Rep. 2016, 6, 37871. [Google Scholar] [CrossRef] [PubMed]
- Avalos-Padilla, Y.; Georgiev, V.N.; Lantero, E.; Pujals, S.; Verhoef, R.; Borgheti-Cardoso, L.N.; Albertazzi, L.; Dimova, R.; Fernandez-Busquets, X. The ESCRT-III machinery participates in the production of extracellular vesicles and protein export during Plasmodium falciparum infection. PLoS Pathog. 2021, 17, e1009455. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.A.; Silamut, K.; Chotivanich, K.; Pukrittayakamee, S.; White, N.J. Red cell selectivity in malaria: A study of multiple-infected erythrocytes. Trans. R. Soc. Trop. Med. Hyg. 1999, 93, 165–168. [Google Scholar] [CrossRef][Green Version]
- Chotivanich, K.; Udomsangpetch, R.; Simpson, J.A.; Newton, P.; Pukrittayakamee, S.; Looareesuwan, S.; White, N.J. Parasite multiplication potential and the severity of Falciparum malaria. J. Infect. Dis. 2000, 181, 1206–1209. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).