Marathon-Induced Cardiac Strain as Model for the Evaluation of Diagnostic microRNAs for Acute Myocardial Infarction
Abstract
1. Introduction
2. Materials and Methods
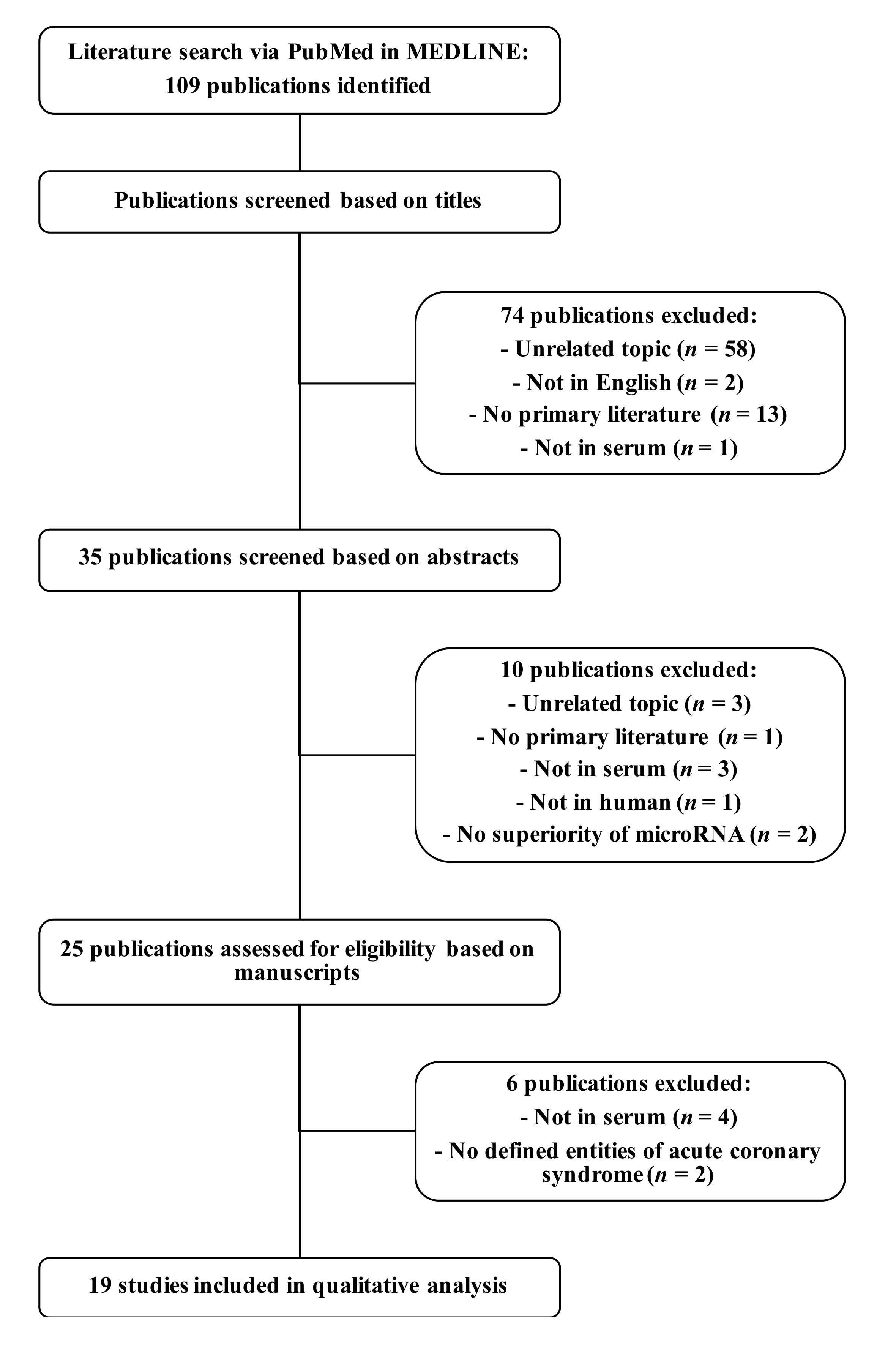
2.1. Comprehensive Review of Potential Diagnostic Serum microRNAs as Biomarkers for Myocardial Infarction
2.2. Participants
2.3. Ethics and Consents
2.4. Sample Preparation for Next-Generation Sequencing
2.5. Next-Generation Sequencing: Data Preparation for Analyzes
2.6. Sample Preparation for Quantitative Real-Time Polymerase Chain Reaction
2.7. Quantitative Real-Time Polymerase Chain Reaction: Data Preparation for Statistical Analyzes
2.8. Statistics
3. Results
3.1. Comprehensive Review of Serum microRNA Biomarkers of Myocardial Infarction
3.2. Patients Undergoing Strenuous Exercise
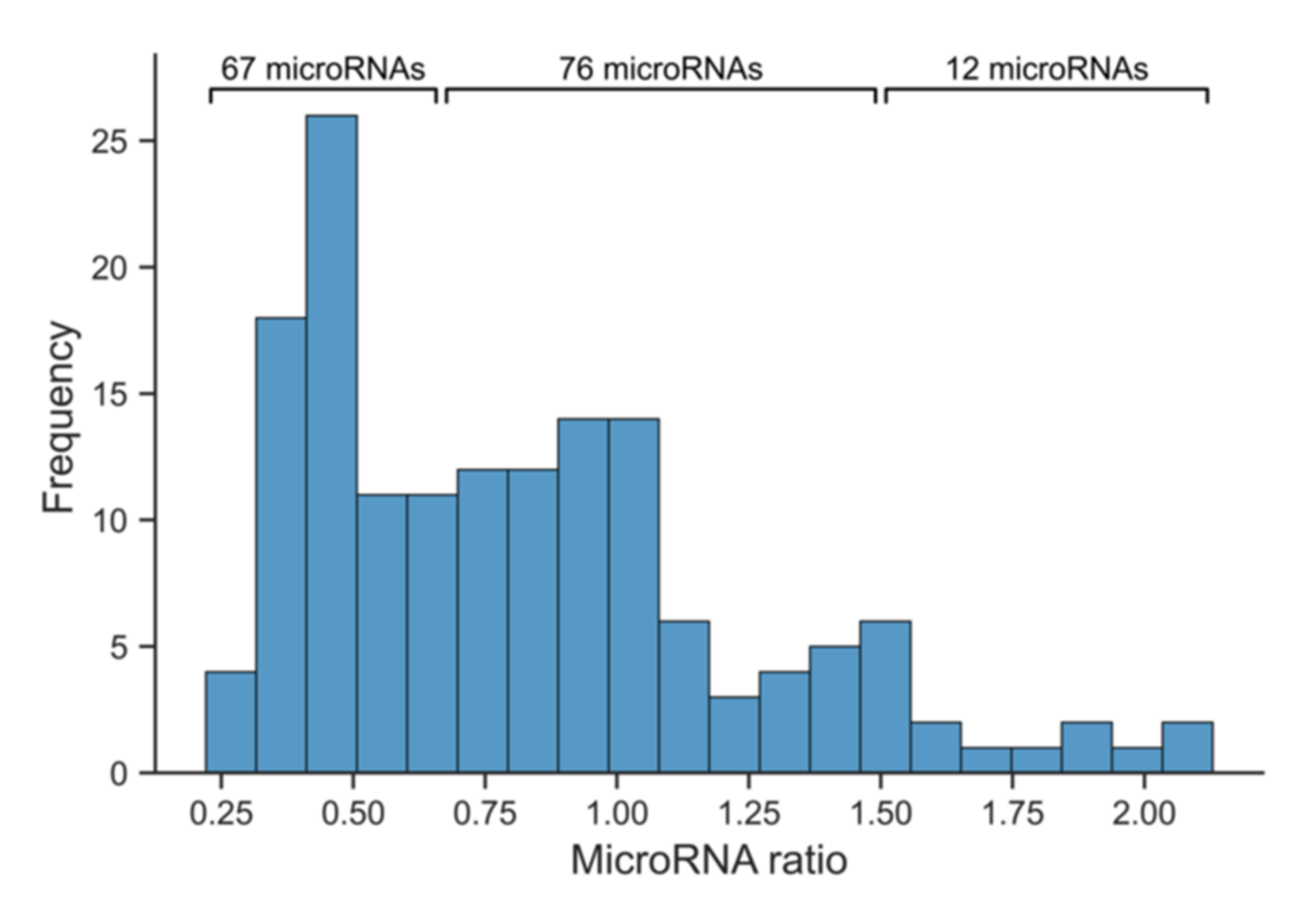
3.3. Next-Generation Sequencing to Identify Abundant and Reliably Measurable Serum microRNAs in Marathon Runners
3.4. Quantitative Real-Time Polymerase Chain Reaction to Further Stratify Suitable Myocardial Infarction Candidate microRNAs
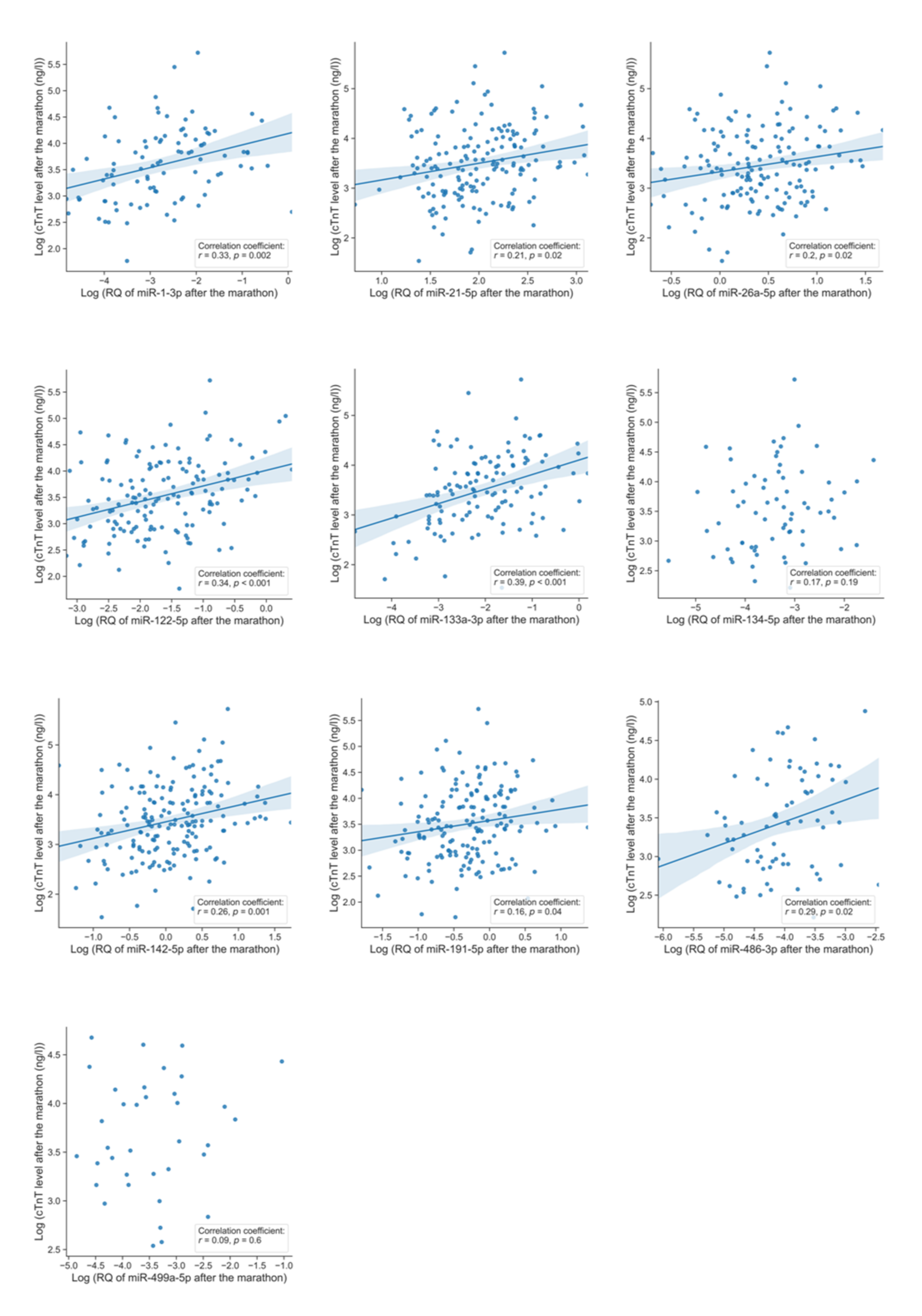
3.5. Co-Release of microRNAs and Cardiac Troponin T
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santos, I.S.; Goulart, A.C.; Brandao, R.M.; Santos, R.C.; Bittencourt, M.S.; Sitnik, D.; Pereira, A.C.; Pastore, C.A.; Samesima, N.; Lotufo, P.A.; et al. One-year Mortality after an Acute Coronary Event and its Clinical Predictors: The ERICO Study. Arquivos Brasileiros de Cardiologia 2015, 105, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.C.; Mateus, P.; Santos, L.; Mateus, C.; Sampaio, F.; Adao, L.; Bettencourt, N.; Ribeiro, J.; Teixeira, M.; Fonseca, C.; et al. Acute coronary syndrome and predictors of quality of life. Revista Portuguesa de Cardiologia 2005, 24, 819–831. [Google Scholar] [PubMed]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D.; Group, E.S.C.S.D. Fourth universal definition of myocardial infarction. Eur. Heart J. 2018, 72, 2231–2264. [Google Scholar] [CrossRef] [PubMed]
- Pickering, J.W.; Greenslade, J.H.; Cullen, L.; Flaws, D.; Parsonage, W.; George, P.; Worster, A.; Kavsak, P.A.; Than, M.P. Validation of presentation and 3 h high-sensitivity troponin to rule-in and rule-out acute myocardial infarction. Heart 2016, 102, 1270–1278. [Google Scholar] [CrossRef]
- Kanjwal, K.; Imran, N.; Grubb, B.; Kanjwal, Y. Troponin elevation in patients with various tachycardias and normal epicardial coronaries. Indian Pacing Electrophysiol. J. 2008, 8, 172–174. [Google Scholar]
- McCarthy, C.P.; Donnellan, E.; Phelan, D.; Griffin, B.P.; Enriquez-Sarano, M.; McEvoy, J.W. High sensitivity troponin and valvular heart disease. Trends Cardiovasc. Med. 2017, 27, 326–333. [Google Scholar] [CrossRef]
- Perna, E.R.; Macin, S.M.; Parras, J.I.; Pantich, R.; Farias, E.F.; Badaracco, J.R.; Jantus, E.; Medina, F.; Brizuela, M. Cardiac troponin T levels are associated with poor short- and long-term prognosis in patients with acute cardiogenic pulmonary edema. Am. Heart J. 2002, 143, 814–820. [Google Scholar] [CrossRef]
- Rittoo, D.; Jones, A.; Lecky, B.; Neithercut, D. Elevation of cardiac troponin T, but not cardiac troponin I, in patients with neuromuscular diseases: Implications for the diagnosis of myocardial infarction. J. Am. Coll. Cardiol. 2014, 63, 2411–2420. [Google Scholar] [CrossRef]
- Urhausen, A.; Scharhag, J.; Herrmann, M.; Kindermann, W. Clinical significance of increased cardiac troponins T and I in participants of ultra-endurance events. Am. J. Cardiol. 2004, 94, 696–698. [Google Scholar] [CrossRef]
- Regwan, S.; Hulten, E.A.; Martinho, S.; Slim, J.; Villines, T.C.; Mitchell, J.; Slim, A.M. Marathon Running as a Cause of Troponin Elevation: A Systematic Review and Meta-Analysis. J. Interv. Cardiol. 2010, 23, 443–450. [Google Scholar] [CrossRef]
- Baker, P.; Leckie, T.; Harrington, D.; Richardson, A. Exercise-induced cardiac troponin elevation: An update on the evidence, mechanism and implications. Int. J. Cardiol. Heart Vasc. 2019, 22, 181–186. [Google Scholar] [CrossRef]
- Thygesen, K.; Mair, J.; Katus, H.; Plebani, M.; Venge, P.; Collinson, P.; Lindahl, B.; Giannitsis, E.; Hasin, Y.; Galvani, M.; et al. Recommendations for the use of cardiac troponin measurement in acute cardiac care (dagger). Eur. Heart J. 2010, 31, 2197–2204b. [Google Scholar] [CrossRef]
- Neumayr, G.; Pfister, R.; Mitterbauer, G.; Maurer, A.; Gaenzer, H.; Sturm, W.; Hoertnagl, H. Effect of the “Race Across The Alps” in elite cyclists on plasma cardiac troponins I and T. Am. J. Cardiol. 2002, 89, 484–486. [Google Scholar] [CrossRef]
- Scherr, J.; Braun, S.; Schuster, T.; Hartmann, C.; Moehlenkamp, S.; Wolfarth, B.; Pressler, A.; Halle, M. 72-h kinetics of high-sensitive troponin T and inflammatory markers after marathon. Med. Sci. Sports Exerc. 2011, 43, 1819–1827. [Google Scholar] [CrossRef]
- Giannitsis, E.; Kehayova, T.; Vafaie, M.; Katus, H.A. Combined testing of high-sensitivity troponin T and copeptin on presentation at prespecified cutoffs improves rapid rule-out of non-ST-segment elevation myocardial infarction. Clin. Chem. 2011, 57, 1452–1455. [Google Scholar] [CrossRef] [PubMed]
- Figiel, L.; Kasprzak, J.D.; Peruga, J.; Lipiec, P.; Drozdz, J.; Krzeminska-Pakula, M.; Smigielski, J. Heart-type fatty acid binding protein--a reliable marker of myocardial necrosis in a heterogeneous group of patients with acute coronary syndrome without persistent ST elevation. Kardiologia Polska 2008, 66, 253–259. [Google Scholar]
- Ertekin, B.; Kocak, S.; Dundar, Z.D.; Girisgin, S.; Cander, B.; Gul, M.; Doseyici, S.; Mehmetoglu, I.; Sahin, T.K. Diagnostic value of ischemia-modified albumin in acute coronary syndrome and acute ischemic stroke. Pak. J. Med. Sci. 2013, 29, 1003–1007. [Google Scholar] [CrossRef]
- Meder, B.; Keller, A.; Vogel, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Just, S.; Borries, A.; Rudloff, J.; Leidinger, P.; et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res. Cardiol. 2011, 106, 13–23. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005, 309, 1573–1576. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 2013, 7, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in Breastmilk and the Lactating Breast: Potential Immunoprotectors and Developmental Regulators for the Infant and the Mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [PubMed]
- Tomankova, T.; Petrek, M.; Kriegova, E. Involvement of microRNAs in physiological and pathological processes in the lung. Respir. Res. 2010, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar]
- Jia, K.; Shi, P.; Han, X.; Chen, T.; Tang, H.; Wang, J. Diagnostic value of miR-30d-5p and miR-125b-5p in acute myocardial infarction. Mol. Med. Rep. 2016, 14, 184–194. [Google Scholar] [CrossRef]
- Oerlemans, M.I.; Mosterd, A.; Dekker, M.S.; de Vrey, E.A.; van Mil, A.; Pasterkamp, G.; Doevendans, P.A.; Hoes, A.W.; Sluijter, J.P. Early assessment of acute coronary syndromes in the emergency department: The potential diagnostic value of circulating microRNAs. EMBO Mol. Med. 2012, 4, 1176–1185. [Google Scholar] [CrossRef]
- Bai, R.; Yang, Q.; Xi, R.; Li, L.; Shi, D.; Chen, K. miR-941 as a promising biomarker for acute coronary syndrome. BMC Cardiovasc. Disord. 2017, 17, 227. [Google Scholar] [CrossRef]
- Corsten, M.F.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Wagner, D.R.; Staessen, J.A.; Heymans, S.; Schroen, B. Circulating MicroRNA-208b and MicroRNA-499 Reflect Myocardial Damage in Cardiovascular Disease. Circ. Cardiovasc. Gene 2010, 3, 499–506. [Google Scholar] [CrossRef]
- Biener, M.; Giannitsis, E.; Thum, T.; Bar, C.; Costa, A.; Andrzejewski, T.; Stoyanov, K.M.; Vafaie, M.; Meder, B.; Katus, H.A.; et al. Diagnostic value of circulating microRNAs compared to high-sensitivity troponin T for the detection of non-ST-segment elevation myocardial infarction. Eur. Heart J. Acute Cardiovasc. Care 2021, 10, 653–660. [Google Scholar] [CrossRef]
- Goldbergova, M.P.; Lipkova, J.; Fedorko, J.; Sevcikova, J.; Parenica, J.; Spinar, J.; Masarik, M.; Vasku, A. MicroRNAs in pathophysiology of acute myocardial infarction and cardiogenic shock. Bratislavske Lekarske Listy 2018, 119, 341–347. [Google Scholar] [CrossRef]
- Mompeon, A.; Ortega-Paz, L.; Vidal-Gomez, X.; Costa, T.J.; Perez-Cremades, D.; Garcia-Blas, S.; Brugaletta, S.; Sanchis, J.; Sabate, M.; Novella, S.; et al. Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: A systematic and paired comparative analysis. Sci. Rep. 2020, 10, 5373. [Google Scholar] [CrossRef]
- Backes, C.; Fehlmann, T.; Kern, F.; Kehl, T.; Lenhof, H.P.; Meese, E.; Keller, A. miRCarta: A central repository for collecting miRNA candidates. Nucleic Acids Res. 2018, 46, D160–D167. [Google Scholar] [CrossRef]
- Scherr, J.; Nieman, D.C.; Schuster, T.; Habermann, J.; Rank, M.; Braun, S.; Pressler, A.; Wolfarth, B.; Halle, M. Nonalcoholic beer reduces inflammation and incidence of respiratory tract illness. Med. Sci. Sports Exerc. 2012, 44, 18–26. [Google Scholar] [CrossRef]
- Dill, D.B.; Costill, D.L. Calculation of percentage changes in volumes of blood, plasma, and red cells in dehydration. J. Appl. Physiol. 1974, 37, 247–248. [Google Scholar] [CrossRef]
- Fehlmann, T.; Backes, C.; Kahraman, M.; Haas, J.; Ludwig, N.; Posch, A.E.; Wurstle, M.L.; Hubenthal, M.; Franke, A.; Meder, B.; et al. Web-based NGS data analysis using miRMaster: A large-scale meta-analysis of human miRNAs. Nucleic Acids Res. 2017, 45, 8731–8744. [Google Scholar] [CrossRef]
- Rosner, B. Percentage Points for a Generalized Esd Many-Outlier Procedure. Technometrics 1983, 25, 165–172. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Cheng, Y.; Tan, N.; Yang, J.; Liu, X.; Cao, X.; He, P.; Dong, X.; Qin, S.; Zhang, C. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin. Sci. 2010, 119, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Fang, Z.; Jiang, T.; Zhang, Q.; Liu, C.; Zhang, C.; Xiang, Y. Serum microRNAs profile from genome-wide serves as a fingerprint for diagnosis of acute myocardial infarction and angina pectoris. BMC Med. Genom. 2013, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Guo, Z.; Shi, Y.; Zhang, L.; Song, C. Serum Exosomal MicroRNA-21, MicroRNA-126, and PTEN Are Novel Biomarkers for Diagnosis of Acute Coronary Syndrome. Front. Physiol. 2020, 11, 654. [Google Scholar] [CrossRef]
- Gao, C.; Zhao, D.; Wang, J.; Liu, P.; Xu, B. Clinical significance and correlation of microRNA-21 expression and the neutrophil-lymphocyte ratio in patients with acute myocardial infarction. Clinics 2019, 74, e1237. [Google Scholar] [CrossRef]
- Maciejak, A.; Kiliszek, M.; Opolski, G.; Segiet, A.; Matlak, K.; Dobrzycki, S.; Tulacz, D.; Sygitowicz, G.; Burzynska, B.; Gora, M. miR-22-5p revealed as a potential biomarker involved in the acute phase of myocardial infarction via profiling of circulating microRNAs. Mol. Med. Rep. 2016, 14, 2867–2875. [Google Scholar] [CrossRef][Green Version]
- Bukauskas, T.; Mickus, R.; Cereskevicius, D.; Macas, A. Value of Serum miR-23a, miR-30d, and miR-146a Biomarkers in ST-Elevation Myocardial Infarction. Med. Sci. Monit. 2019, 25, 3925–3932. [Google Scholar] [CrossRef]
- Hsu, A.; Chen, S.J.; Chang, Y.S.; Chen, H.C.; Chu, P.H. Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. BioMed Res. Int. 2014, 2014, 418628. [Google Scholar] [CrossRef]
- Dai, Y.; Yan, T.; Gao, Y. Silence of miR-32-5p promotes endothelial cell viability by targeting KLF2 and serves as a diagnostic biomarker of acute myocardial infarction. Diagn. Pathol. 2020, 15, 19. [Google Scholar] [CrossRef]
- Ling, H.; Guo, Z.; Du, S.; Liao, Y.; Li, Y.; Ding, C.; Song, C. Serum exosomal miR-122-5p is a new biomarker for both acute coronary syndrome and underlying coronary artery stenosis. Biomarkers 2020, 25, 539–547. [Google Scholar] [CrossRef]
- Ji, Q.; Jiang, Q.; Yan, W.; Li, X.; Zhang, Y.; Meng, P.; Shao, M.; Chen, L.; Zhu, H.; Tian, N. Expression of circulating microRNAs in patients with ST segment elevation acute myocardial infarction. Minerva Cardioangiol. 2015, 63, 397–402. [Google Scholar]
- Chen, Z.; Pan, X.; Yan, G.; Sheng, Z.; Ma, G. miR-142 is a Sensitive Biomarker for the Diagnosis and Prognosis of Acute Myocardial Infarction. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Guo, X.; Chen, Y.; Lu, Y.; Li, P.; Yu, H.; Diao, F.R.; Tang, W.D.; Hou, P.; Zhao, X.X.; Shi, C.Y. High level of circulating microRNA-142 is associated with acute myocardial infarction and reduced survival. Ir. J. Med. Sci. 2020, 189, 933–937. [Google Scholar] [CrossRef]
- Wu, S.; Sun, H.; Sun, B. MicroRNA-145 is involved in endothelial cell dysfunction and acts as a promising biomarker of acute coronary syndrome. Eur. J. Med. Res. 2020, 25, 2. [Google Scholar] [CrossRef]
- Chen, Z.; Yan, Y.; Wu, J.; Qi, C.; Liu, J.; Wang, J. Expression level and diagnostic value of exosomal NEAT1/miR-204/MMP-9 in acute ST-segment elevation myocardial infarction. IUBMB Life 2020, 72, 2499–2507. [Google Scholar] [CrossRef]
- Shalaby, S.M.; El-Shal, A.S.; Shoukry, A.; Khedr, M.H.; Abdelraheim, N. Serum miRNA-499 and miRNA-210: A potential role in early diagnosis of acute coronary syndrome. IUBMB Life 2016, 68, 673–682. [Google Scholar] [CrossRef]
- Gao, C.; Qian, H.; Shi, Q.; Zhang, H. MicroRNA-363-3p serves as a diagnostic biomarker of acute myocardial infarction and regulates vascular endothelial injury by targeting KLF2. Cardiovasc. Diagn. Ther. 2020, 10, 421–430. [Google Scholar] [CrossRef]
- Guo, L.L.; Chen, H.H.; Qu, F.C.; Lu, Q.H. Clinical significance of miR-492 in peripheral blood of acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9041–9045. [Google Scholar] [CrossRef]
- Su, J.; Li, J.; Yu, Q.; Wang, J.; Li, X.; Yang, J.; Xu, J.; Liu, Y.; Xu, Z.; Ji, L.; et al. Exosomal miRNAs as potential biomarkers for acute myocardial infarction. IUBMB Life 2020, 72, 384–400. [Google Scholar] [CrossRef]
- Gholikhani-Darbroud, R.; Khaki-Khatibi, F.; Mansouri, F.; Hajahmadipoorrafsanjani, M.; Ghojazadeh, M. Decreased circulatory microRNA-4478 as a specific biomarker for diagnosing non-ST-segment elevation myocardial infarction (NSTEMI) and its association with soluble leptin receptor. Bratislavske Lekarske Listy 2017, 118, 684–690. [Google Scholar] [CrossRef]
- Tanaka, H.; Monahan, K.D.; Seals, D.R. Age-predicted maximal heart rate revisited. J. Am. Coll. Cardiol. 2001, 37, 153–156. [Google Scholar] [CrossRef]
- Soplinska, A.; Zareba, L.; Wicik, Z.; Eyileten, C.; Jakubik, D.; Siller-Matula, J.M.; De Rosa, S.; Malek, L.A.; Postula, M. MicroRNAs as Biomarkers of Systemic Changes in Response to Endurance Exercise-A Comprehensive Review. Diagnostics 2020, 10, 813. [Google Scholar] [CrossRef]
- Aengevaeren, V.L.; Hopman, M.T.E.; Thompson, P.D.; Bakker, E.A.; George, K.P.; Thijssen, D.H.J.; Eijsvogels, T.M.H. Exercise-Induced Cardiac Troponin I Increase and Incident Mortality and Cardiovascular Events. Circulation 2019, 140, 804–814. [Google Scholar] [CrossRef]
- Gioffre, S.; Chiesa, M.; Cardinale, D.M.; Ricci, V.; Vavassori, C.; Cipolla, C.M.; Masson, S.; Sandri, M.T.; Salvatici, M.; Ciceri, F.; et al. Circulating MicroRNAs as Potential Predictors of Anthracycline-Induced Troponin Elevation in Breast Cancer Patients: Diverging Effects of Doxorubicin and Epirubicin. J. Clin. Med. 2020, 9, 1418. [Google Scholar] [CrossRef]
- May, S.M.; Abbott, T.E.F.; Del Arroyo, A.G.; Reyes, A.; Martir, G.; Stephens, R.C.M.; Brealey, D.; Cuthbertson, B.H.; Wijeysundera, D.N.; Pearse, R.M.; et al. MicroRNA signatures of perioperative myocardial injury after elective noncardiac surgery: A prospective observational mechanistic cohort study. Br. J. Anaesth. 2020, 125, 661–671. [Google Scholar] [CrossRef]
- Clauss, S.; Wakili, R.; Hildebrand, B.; Kaab, S.; Hoster, E.; Klier, I.; Martens, E.; Hanley, A.; Hanssen, H.; Halle, M.; et al. MicroRNAs as Biomarkers for Acute Atrial Remodeling in Marathon Runners (The miRathon Study--A Sub-Study of the Munich Marathon Study). PLoS ONE 2016, 11, e0148599. [Google Scholar] [CrossRef]
- Xing, X.; Guo, S.; Zhang, G.; Liu, Y.; Bi, S.; Wang, X.; Lu, Q. miR-26a-5p protects against myocardial ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT signaling pathway. Braz. J. Med. Biol. Res. 2020, 53, e9106. [Google Scholar] [CrossRef]
- Yang, X.; Du, X.; Ma, K.; Li, G.; Liu, Z.; Rong, W.; Miao, H.; Zhu, F.; Cui, Q.; Wu, S.; et al. Circulating miRNAs Related to Long-term Adverse Cardiovascular Events in STEMI Patients: A Nested Case-Control Study. Can. J. Cardiol. 2021, 37, 77–85. [Google Scholar] [CrossRef]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stockel, D.; Meese, E.; Lenhof, H.P.; Keller, A. miRPathDB 2.0: A novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2020, 48, D142–D147. [Google Scholar] [CrossRef]
- Hoekstra, M. MicroRNA-499-5p: A therapeutic target in the context of cardiovascular disease. Ann. Transl. Med. 2016, 4, 539. [Google Scholar] [CrossRef]
- Li, P.; Li, S.Y.; Liu, M.; Ruan, J.W.; Wang, Z.D.; Xie, W.C. Value of the expression of miR-208, miR-494, miR-499 and miR-1303 in early diagnosis of acute myocardial infarction. Life Sci. 2019, 232, 116547. [Google Scholar] [CrossRef] [PubMed]
- Kayvanpour, E.; Gi, W.T.; Sedaghat-Hamedani, F.; Lehmann, D.H.; Frese, K.S.; Haas, J.; Tappu, R.; Samani, O.S.; Nietsch, R.; Kahraman, M.; et al. microRNA neural networks improve diagnosis of acute coronary syndrome (ACS). J. Mol. Cell Cardiol. 2021, 151, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef] [PubMed]

| MicroRNA | Disease * | Direction of Dysregulation | Reference |
|---|---|---|---|
| miR-1-3p | MI | Upregulation | [42,43] |
| miR-21-5p | ACS MI | Upregulation | [44] [45] |
| miR-22-5p | STEMI | Upregulation | [46] |
| miR-23a-3p | STEMI | Downregulation | [47] |
| miR-26a-5p | STEMI | Downregulation | [48] |
| miR-32-5p | MI | Upregulation | [49] |
| miR-122-5p | MI | Upregulation | [50] |
| miR-126-3p | MI STEMI | Upregulation Downregulation | [44] [48] |
| miR-133a-3p | MI | Upregulation | [51] |
| miR-133b | MI STEMI | Upregulation | [51] [46] |
| miR-134-5p | MI | Upregulation | [43] |
| miR-142-5p | MI | Upregulation | [52,53] |
| miR-145-5p | ACS | Downregulation | [54] |
| miR-150-3p | STEMI | Upregulation | [48] |
| miR-186-5p | MI | Upregulation | [43] |
| miR-191-5p | STEMI | Downregulation | [48] |
| miR-204-5p | STEMI | Downregulation | [55] |
| miR-208a-3p | MI | Upregulation | [43] |
| miR-210-3p | NSTEMI | Upregulation | [56] |
| miR-223-3p | MI | Upregulation | [43] |
| miR-363-3p | MI | Upregulation | [57] |
| miR-486-3p | STEMI | Upregulation | [48] |
| miR-492 | MI | Upregulation | [58] |
| miR-499a-5p | MI NSTEMI | Upregulation | [43,51] [56] |
| miR-1915-3p | MI | Downregulation | [59] |
| miR-3656 ** | MI | Downregulation | [59] |
| miR-4507 | MI | Downregulation | [59] |
| miR-4478 | NSTEMI | Downregulation | [60] |
| Method | NGS | qPCR | |||
|---|---|---|---|---|---|
| Cohort | Low cTnT Cohort (n = 19) | High cTnT Cohort (n = 27) | Low cTnT Cohort n = 31) | High cTnT Cohort (n = 56) | Correlation Cohort (n = 210) |
| Age (yrs) ( ± σ) | 46 ± 5.03 (n = 19) | 48.44 ± 6.69 (n = 27) | 45.19 ± 5.89 (n = 31) | 38.48 ± 11.25 * (n = 56) | 41.63 ± 9.16 (n = 210) |
| Body-Mass-Index (kg/m2) ( ± σ) | 23.18 ± 1.92 (n = 19) | 24.19 ± 2.35 (n = 27) | 23.39 ± 2.31 (n = 31) | 23.25 ± 2.07 (n = 56) | 23.55 ± 2.12 (n = 209) |
| Active smokers (n) | 0 (n = 19) | 0 (n = 27) | 0 (n = 31) | 0 (n = 56) | 6 (n = 210) |
| Maximum heart rate § (bpm) ( ± σ) | 175.8 ± 3.43 (n = 19) | 174.09 ± 4.59 (n = 27) | 176.36 ± 4.06 (n = 31) | 181.06 ± 7.8 (n = 56)* | 178.86 ± 6.4 (n = 210) |
| Running time during marathon (h:min) ( ± σ) | 4:01 ± 0:32 (n = 19) | 3:55 ± 0:32 (n = 27) | 3:53 ± 0:30 (n = 26) | 3:49 ± 0:31 (n = 53) | 3:50 ± 0:30 (n = 196) |
| Mean heart rate during the marathon §§ (bpm) ( ± σ) | 150.94 ± 10.2 (n = 18) | 154.91 ± 10.02 (n = 23) | 153.24 ± 9.09 (n = 25) | 161.44 ± 9.78 * (n = 43) | 156.66 ± 10 (n = 163) |
| Cardiac troponin T before the marathon (ng/L) ( (Q1–Q3)) | 3 (3–3) (n = 19) | 5.75 (3.91–10.13) * (n = 27) | 3 (3–3.18) (n = 31) | 4.15 (3–5.98) * (n = 56) | 3 (3–4.92) (n = 210) |
| Cardiac troponin T after the marathon (ng/L) ( (Q1–Q3)) | 11.41 (6.36–12.72) (n = 19) | 64.34 (58.06–89.81) * (n = 27) | 10.98 (7.22–12.91) (n = 31) | 67.95 (58.55–96.2) * (n = 56) | 31.44 (18.22–53.33) (n = 210) |
| N-terminal pro-brain natriuretic peptide before the marathon (pg/mL) ( (Q1–Q3)) | 31.63 (21.74–54.64) (n = 19) | 37.95 (20.12–55.02) (n = 27) | 28.54 (18.17–38.93) (n = 31) | 21.94 (10.38–37.58) (n = 56) | 24.81 (13.12–42.62) (n = 210) |
| MicroRNA | Correlation Coefficient | p-Value ** | Reliably Measurable in Number of Runners |
|---|---|---|---|
| miR-1-3p | r = 0.33 | p = 0.002 | n = 95 |
| miR-21-5p | r = 0.21 | p = 0.02 | n = 163 |
| miR-26a-5p | r = 0.2 | p = 0.02 | n = 151 |
| miR-122-5p | r = 0.34 | p < 0.001 | n = 147 |
| miR-133a-3p | r = 0.39 | p < 0.001 | n = 120 |
| miR-134-5p | r = 0.17 | p = 0.19 | n = 69 |
| miR-142-5p | r = 0.26 | p = 0.001 | n = 176 |
| miR-191-5p | r = 0.16 | p = 0.04 | n = 167 |
| miR-486-3p | r = 0.29 | p = 0.02 | n = 73 |
| miR-499a-5p | r = 0.09 | p = 0.6 | n = 36 |
| MicroRNA | Fold Change in the Low cTnT Cohort (Median, Q1–Q3) | Fold Change in the High cTnT Cohort (Median, Q1–Q3) | p-Value ** |
|---|---|---|---|
| miR-1-3p | N/A (n = 1) | 2.1, 1.32–4.6 (n = 16) | p = N/A *** |
| miR-21-5p | 1.07, 0.64–1.49 (n = 17) | 1.51, 0.95–2.57 (n = 39) | p = 0.09 |
| miR-26a-5p | 0.6, 0.47–0.82 (n = 17) | 1.11, 0.59–1.77 (n = 33) | p = 0.046 |
| miR-122-5p | 1.13, 0.37–1.61 (n = 12) | 1.19, 0.66–4.31 (n = 31) | p = 0.14 |
| miR-133a-3p | 1.01, 0.61–1.43 (n = 5) | 5.63, 2.86–10.06 (n = 17) | p = 0.03 |
| miR-134-5p | N/A (n = 1) | 2.69, 2.14–3.13 (n = 5) | p = N/A |
| miR-142-5p | 0.64, 0.45–1.08 (n = 18) | 1.72, 0.83–2.73 (n = 40) | p = 0.01 |
| miR-191-5p | 0.91, 0.63–1.08 (n = 15) | 0.96, 0.76–2.28 (n = 40) | p = 0.21 |
| miR-486-3p | N/A (n = 4) | N/A (n = 4) | p = N/A |
| miR-499a-5p | N/A (n = 0) | N/A (n = 1) | p = N/A |
| MicroRNA | Direction of Dysregulation in Patients with MI * | Direction of Dysregulation in Marathon Runners with cTnT Rise from qPCR ** |
|---|---|---|
| miR-1-3p | Upregulation | N/A |
| miR-21-5p | Upregulation | No significant difference of Dysregulation *** |
| miR-22-5p | Upregulation | N/A |
| miR-23a-3p | Downregulation | N/A |
| miR-26a-5p | Downregulation | Upregulation |
| miR-32-5p | Upregulation | N/A |
| miR-122-5p | Upregulation | No significant difference of dysregulation |
| miR-126-3p | Upregulation and downregulation | N/A |
| miR-133a-3p | Upregulation | Upregulation |
| miR-133b | Upregulation | N/A |
| miR-134-5p | Upregulation | N/A |
| miR-142-5p | Upregulation | Upregulation |
| miR-145-5p | Downregulation | N/A |
| miR-150-3p | Upregulation | N/A |
| miR-186-5p | Upregulation | N/A |
| miR-191-5p | Downregulation | No significant difference of dysregulation |
| miR-204-5p | Downregulation | N/A |
| miR-208a-3p | Upregulation | N/A |
| miR-210-3p | Upregulation | N/A |
| miR-223-3p | Upregulation | N/A |
| miR-363-3p | Upregulation | N/A |
| miR-486-3p | Upregulation | N/A |
| miR-492 | Upregulation | N/A |
| miR-499a-5p | Upregulation | N/A |
| miR-1915-3p | Downregulation | N/A |
| miR-4507 | Downregulation | N/A |
| miR-4478 | Downregulation | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirvani Samani, O.; Scherr, J.; Kayvanpour, E.; Haas, J.; Lehmann, D.H.; Gi, W.-T.; Frese, K.S.; Nietsch, R.; Fehlmann, T.; Sandke, S.; et al. Marathon-Induced Cardiac Strain as Model for the Evaluation of Diagnostic microRNAs for Acute Myocardial Infarction. J. Clin. Med. 2022, 11, 5. https://doi.org/10.3390/jcm11010005
Shirvani Samani O, Scherr J, Kayvanpour E, Haas J, Lehmann DH, Gi W-T, Frese KS, Nietsch R, Fehlmann T, Sandke S, et al. Marathon-Induced Cardiac Strain as Model for the Evaluation of Diagnostic microRNAs for Acute Myocardial Infarction. Journal of Clinical Medicine. 2022; 11(1):5. https://doi.org/10.3390/jcm11010005
Chicago/Turabian StyleShirvani Samani, Omid, Johannes Scherr, Elham Kayvanpour, Jan Haas, David H. Lehmann, Weng-Tein Gi, Karen S. Frese, Rouven Nietsch, Tobias Fehlmann, Steffi Sandke, and et al. 2022. "Marathon-Induced Cardiac Strain as Model for the Evaluation of Diagnostic microRNAs for Acute Myocardial Infarction" Journal of Clinical Medicine 11, no. 1: 5. https://doi.org/10.3390/jcm11010005
APA StyleShirvani Samani, O., Scherr, J., Kayvanpour, E., Haas, J., Lehmann, D. H., Gi, W.-T., Frese, K. S., Nietsch, R., Fehlmann, T., Sandke, S., Weis, T., Keller, A., Katus, H. A., Halle, M., Frey, N., Meder, B., & Sedaghat-Hamedani, F. (2022). Marathon-Induced Cardiac Strain as Model for the Evaluation of Diagnostic microRNAs for Acute Myocardial Infarction. Journal of Clinical Medicine, 11(1), 5. https://doi.org/10.3390/jcm11010005










