Quantification of Bacteria in Mouth-Rinsing Solution for the Diagnosis of Periodontal Disease
Abstract
1. Introduction
2. Materials and Methods
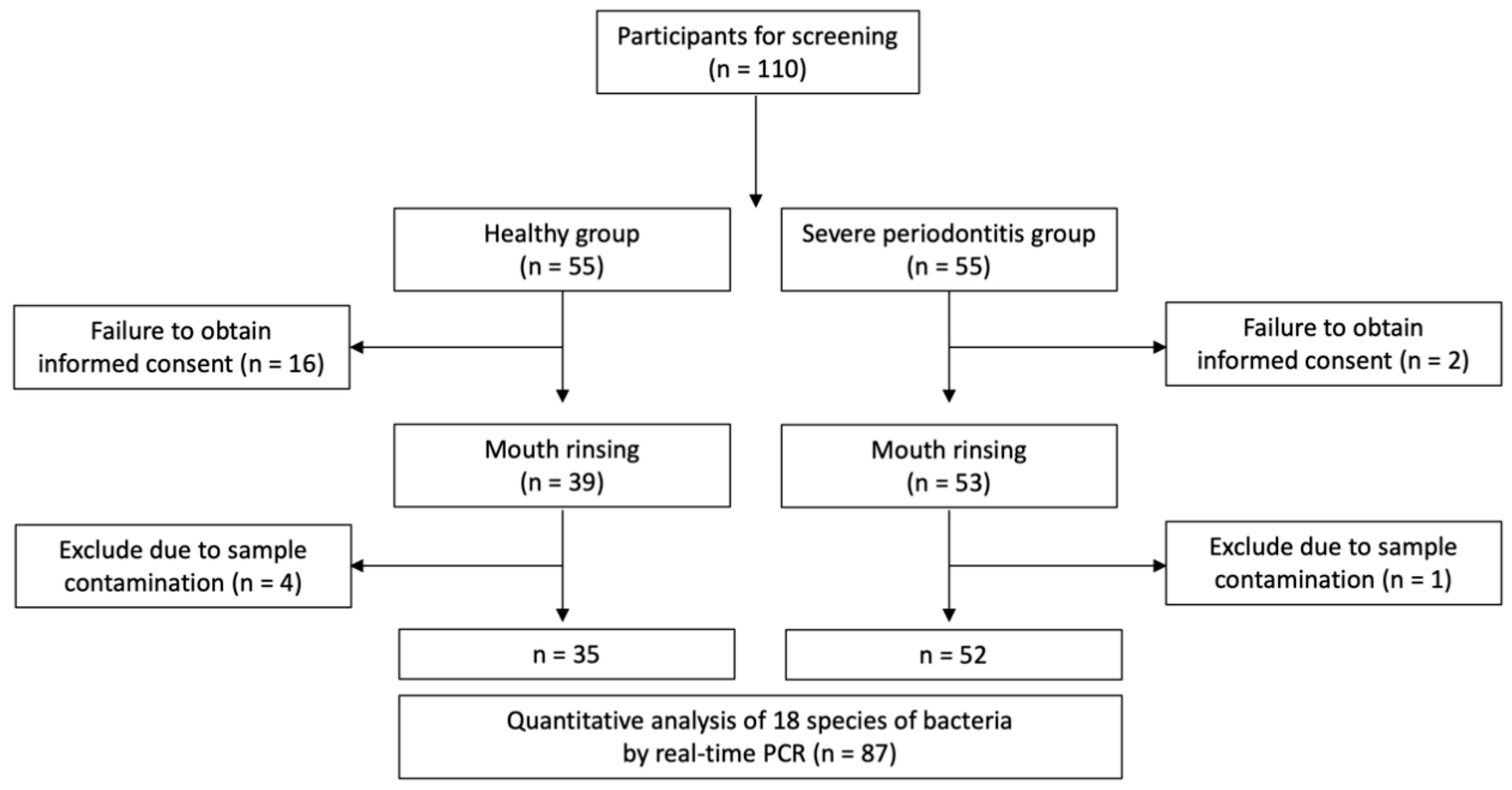
2.1. Patient Selection
2.2. Sample Size Determination
2.3. Periodontal Examination
2.4. Sample Collection and DNA Extraction
2.5. Multiplex Quantitative RT-PCR (qPCR)
2.6. Bacterial Quantification
2.7. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Teanpaisan, R.; Obiechina, N.S.; Pithpornchaiyakul, W.; Pongpaisal, S.; Pisuithanakan, S.; Baelum, V.; Fejerskov, O.; Dahlen, G. Periodontal microbiota and clinical periodontal status in a rural sample in southern Thailand. Eur. J. Oral Sci. 2002, 110, 345–352. [Google Scholar] [CrossRef]
- Pradhan-Palikhe, P.; Mäntylä, P.; Paju, S.; Buhlin, K.; Persson, G.R.; Nieminen, M.S.; Sinisalo, J.; Pussinen, P.J. Subgingival Bacterial Burden in Relation to Clinical and Radiographic Periodontal Parameters. J. Periodontol. 2013, 84, 1809–1817. [Google Scholar] [CrossRef]
- Hyvärinen, K.; Laitinen, S.; Paju, S.; Hakala, A.; Suominen-Taipale, L.; Skurnik, M.; Könönen, E.; Pussinen, P.J. Detection and quantification of five major periodontal pathogens by single copy gene-based real-time PCR. Innate Immun. 2009, 15, 195–204. [Google Scholar] [CrossRef]
- Paju, S.; Pussinen, P.J.; Suominen-Taipale, L.; Hyvönen, M.; Knuuttila, M.; Könönen, E. Detection of Multiple Pathogenic Species in Saliva is Associated with Periodontal Infection in Adults. J. Clin. Microbiol. 2008, 47, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Mineoka, T.; Awano, S.; Rikimaru, T.; Kurata, H.; Yoshida, A.; Ansai, T.; Takehara, T. Site-Specific Development of Periodontal Disease is Associated with Increased Levels of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythiain Subgingival Plaque. J. Periodontol. 2008, 79, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Torrungruang, K.; Jitpakdeebordin, S.; Charatkulangkun, O.; Gleebbua, Y. Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Treponema denticola/Prevotella intermedia Co-Infection are Associated with Severe Periodontitis in a Thai Population. PLoS ONE 2015, 10, e0136646. [Google Scholar] [CrossRef]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary Biomarkers: Toward Future Clinical and Diagnostic Utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.; Testa-Weintraub, L.; Banderas, J.; Haraszthy, G.; Reddy, M.; Levine, M. Sialochemistry: A Diagnostic Tool? Crit. Rev. Oral Biol. Med. 1993, 4, 343–350. [Google Scholar] [CrossRef]
- Liguori, G.; Lucariello, A.; Colella, G.; de Luca, A.; Marinelli, P. Rapid identification of Candida species in oral rinse solutions by PCR. J. Clin. Pathol. 2006, 60, 1035–1039. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Buchner, A.; Lang, A.-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 2009, 41, 1149–1160. [Google Scholar] [CrossRef]
- Eke, P.I.; Page, R.C.; Wei, L.; Thornton-Evans, G.; Genco, R.J. Update of the Case Definitions for Population-Based Surveillance of Periodontitis. J. Periodontol. 2012, 83, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-H.; Joo, J.-Y.; Lee, Y.J.; Koh, J.-K.; Choi, J.-H.; Shin, Y.; Cho, J.; Park, E.; Kang, J.; Lee, K.; et al. Grading system for periodontitis by analyzing levels of periodontal pathogens in saliva. PLoS ONE 2018, 13, e0200900. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Rodríguez, M.; Córdoba, J.J.; Andrade, M.J. Design of primers and probes for quantitative real-time PCR methods. In PCR Primer Design; Humana Press: New York, NY, USA, 2015; Volume 1275, pp. 31–56. [Google Scholar]
- Boutaga, K.; van Winkelhoff, A.J.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M. Periodontal pathogens: A quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol. Med. Microbiol. 2005, 45, 191–199. [Google Scholar] [CrossRef]
- Elkaïm, R.; Dahan, M.; Kocgozlu, L.; Werner, S.; Kanter, D.; Kretz, J.G.; Tenenbaum, H. Prevalence of periodontal pathogens in subgingival lesions, atherosclerotic plaques and healthy blood vessels: A preliminary study. J. Periodontal Res. 2007, 43, 224–231. [Google Scholar] [CrossRef]
- Boutaga, K.; van Winkelhoff, A.J.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M. Comparison of real-time PCR and culture for detection of Porphyromonas gingivalis in subgingival plaque samples. J. Clin. Microbiol. 2003, 41, 4950–4954. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Yoshida, A.; Saito, T.; Kawada, M.; Nakano, Y. Quantitative Microbiological Study of Subgingival Plaque by Real-Time PCR Shows Correlation between Levels of Tannerella forsythensis and Fusobacterium spp. J. Clin. Microbiol. 2004, 42, 2255–2257. [Google Scholar] [CrossRef]
- Suzuki, N.; Nakano, Y.; Yoshida, A.; Yamashita, Y.; Kiyoura, Y. Real-Time TaqMan PCR for Quantifying Oral Bacteria during Biofilm Formation. J. Clin. Microbiol. 2004, 42, 3827–3830. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Asai, Y.; Jinno, T.; Igarashi, H.; Ohyama, Y.; Ogawa, T. Detection and Quantification of Oral Treponemes in Subgingival Plaque by Real-Time PCR. J. Clin. Microbiol. 2002, 40, 3334–3340. [Google Scholar] [CrossRef]
- Rawashdeh, R.Y.; Malkawi, H.I.; Al-Hiyasat, A.S.; Hammad, M.M. A fast and sensitive molecular detection of Streptococcus mutans and Actinomyces viscosus from dental plaques. Jordan J. Biol. Sci. 2008, 1, 135–139. [Google Scholar]
- Yun, J.-H.; Park, J.-E.; Kim, D.-I.; Lee, S.-I.; Choi, S.-H.; Cho, K.-S.; Lee, D.-S. Identification of putative periodontal pathogens in Korean chronic periodontitis patients. J. Korean Acad. Periodontol. 2008, 38, 143–152. [Google Scholar] [CrossRef][Green Version]
- Yoshida, A.; Suzuki, N.; Nakano, Y.; Kawada, M.; Oho, T.; Koga, T. Development of a 5′ Nuclease-Based Real-Time PCR Assay for Quantitative Detection of Cariogenic Dental Pathogens Streptococcus mutans and Streptococcus sobrinus. J. Clin. Microbiol. 2003, 41, 4438–4441. [Google Scholar] [CrossRef]
- Haarman, M.; Knol, J. Quantitative Real-Time PCR Analysis of Fecal Lactobacillus Species in Infants Receiving a Prebiotic Infant Formula. Appl. Environ. Microbiol. 2006, 72, 2359–2365. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Procop, G.W.; Kobayashi, N.; Togawa, D.; Wilson, D.A.; Borden, L.; Krebs, V.; Bauer, T.W. Simultaneous Detection of Staphylococcus aureus and Coagulase-Negative Staphylococci in Positive Blood Cultures by Real-Time PCR with Two Fluorescence Resonance Energy Transfer Probe Sets. J. Clin. Microbiol. 2004, 42, 5739–5744. [Google Scholar] [CrossRef] [PubMed]
- Granger, K.; Rundell, M.S.; Pingle, M.R.; Shatsky, R.; Larone, D.H.; Golightly, L.M.; Barany, F.; Spitzer, E.D. Multiplex PCR-Ligation Detection Reaction Assay for Simultaneous Detection of Drug Resistance and Toxin Genes from Staphylococcus aureus, Enterococcus faecalis, and Enterococcus faecium. J. Clin. Microbiol. 2009, 48, 277–280. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Firth, D. Bias Reduction of Maximum Likelihood Estimates. Biometrika 1993, 80, 27. [Google Scholar] [CrossRef]
- Heinze, G.; Schemper, M. A solution to the problem of separation in logistic regression. Stat. Med. 2002, 21, 2409–2419. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89, S159–S172. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, X.; Yu, T.; Brinkman, B.M.; Zimmermann, B.G.; Palanisamy, V.; Wong, D.T. Characterization of salivary RNA by cDNA library analysis. Arch. Oral Biol. 2007, 52, 30–35. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Baelum, V.; Luan, W.-M.; Madianos, P.N.; Chen, X.; Fejerskov, O.; Dahlén, G. Subgingival Microbiota in Adult Chinese: Prevalence and Relation to Periodontal Disease Progression. J. Periodontol. 1997, 68, 651–666. [Google Scholar] [CrossRef]
- Kuboniwa, M.; Amano, A.; Kimura, K.R.; Sekine, S.; Kato, S.; Yamamoto, Y.; Okahashi, N.; Iida, T.; Shizukuishi, S. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol. Immunol. 2004, 19, 168–176. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Huang, W.; Pan, Z.; Cui, H.; Qi, G.; Zhou, X.; Chen, H. Quantitative analysis of microbiota in saliva, supragingival, and subgingival plaque of Chinese adults with chronic periodontitis. Clin. Oral Investig. 2011, 16, 1579–1588. [Google Scholar] [CrossRef]
- Slots, J. Subgingival microflora and periodontal disease. J. Clin. Periodontol. 1979, 6, 351–382. [Google Scholar] [CrossRef]
- Haubek, D.; Ennibi, O.-K.; Poulsen, K.; Vaeth, M.; Poulsen, S.; Kilian, M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet 2008, 371, 237–242. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Neiderud, A.M.; Disick, E.; Lalla, E.; Miller, G.C.; Dahlén, G. Longitudinal stability of serum immunoglobulin G responses to periodontal bacteria. J. Clin. Periodontol. 2004, 31, 985–990. [Google Scholar] [CrossRef]
- Van Winkelhoff, A.J.; Loos, B.G.; van der Reijden, W.A.; van der Velden, U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J. Clin. Periodontol. 2002, 29, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Palmer, R.J.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell–cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. The Bacterial Etiology of Destructive Periodontal Disease: Current Concepts. J. Periodontol. 1992, 63, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef]
- Genco, R.; Kornman, K.; Williams, R.; Offenbacher, S.; Zambon, J.J.; Ishikawa, I.; Listgarten, M.; Michalowicz, B.; Page, R.; Schenkein, H.; et al. Periodontal Diseases: Pathogenesis and Microbial Factors. J. Am. Dent. Assoc. 1998, 129, 58S–62S. [Google Scholar] [CrossRef]
- Kawada, M.; Yoshida, A.; Suzuki, N.; Nakano, Y.; Saito, T.; Oho, T.; Koga, T. Prevalence of Porphyromonas gingivalis in relation to periodontal status assessed by real-time PCR. Oral Microbiol. Immunol. 2004, 19, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.I.; Gonçalves, R.B. Detection ofTannerella forsythensis(Bacteroides forsythus) and Porphyromonas gingivalisby Polymerase Chain Reaction in Subjects with Different Periodontal Status. J. Periodontol. 2003, 74, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.-H.; Lee, E.; Kim, Y.-S.; Kim, W.-K.; Lee, Y.-K.; Kim, S.-H. Differential expression of microRNAs in the saliva of patients with aggressive periodontitis: A pilot study of potential biomarkers for aggressive periodontitis. J. Periodontal Implant. Sci. 2020, 50, 281–290. [Google Scholar] [CrossRef] [PubMed]
| Bacteria | Target Gene | Primer/ Probe | Sequence (5′-3′) | Ref. | Bacteria | Target Gene | Primer/ Probe | Sequence (5′-3′) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Aggregatibacter actinomycetemcomitans | leukotoxin | Forward | CG**********GA | [15] | Eubacterium nodatum | hypothetical protein | Forward | TG**********GA | [16] |
| Reverse | AT**********CA | Reverse | AA**********AT | ||||||
| Probe | [FAM]GG**********CC[BHQ1] | Probe | [TR]TT**********GG[BHQ2] | ||||||
| Porphyromonas gingivalis | hemagglutinin | Forward | AC**********GC | [17] | Eikenella corrodens | prolineiminopeptidase | Forward | GC**********TG | [16] |
| Reverse | GC**********CT | Reverse | GC**********TT | ||||||
| Probe | [HEX]CG**********GA[BHQ1] | Probe | [Cy5]AC**********AT[BHQ2] | ||||||
| Tannerella forsythia | karilysin protease | Forward | TG**********CC | [18] | Streptococcus mitis | 16S ribosomal RNA | Forward | GT**********CG | [19] |
| Reverse | TT**********CA | Reverse | TA**********AT | ||||||
| Probe | [TR]CC**********GG[BHQ2] | Probe | [FAM]TA**********CC[BHQ1] | ||||||
| Treponema denticola | OpdB | Forward | AG**********AG | [20] | Streptococcus mutans | PTS EII | Forward | CA**********CA | [21] |
| Reverse | GC**********AT | Reverse | TG**********CC | ||||||
| Probe | [Cy5]CG**********TC[BHQ2] | Probe | [HEX]TG**********GG[BHQ1] | ||||||
| Fusobacterium nucleatum | 16S ribosomal RNA | Forward | GG**********TC | [22] | Streptococcus sobrinus | Ftsk | Forward | GG**********CC | [23] |
| Reverse | CT**********GC | Reverse | AC**********GG | ||||||
| Probe | [FAM]AA**********CG[BHQ1] | Probe | [TR]AG**********GC[BHQ2] | ||||||
| Prevotella intermedia | hemagglutinin | Forward | CA**********AC | [15] | Lactobacillus casei | att | Forward | CA**********GT | [24] |
| Reverse | CA**********TC | Reverse | AC**********CC | ||||||
| Probe | [HEX]CC**********AC[BHQ1] | Probe | [Cy5]TG**********GT[BHQ2] | ||||||
| Prevotella nigrescens | gyrase subunit B | Forward | AG**********CT | [16] | Staphylococcus aureus | clumping factor A | Forward | GC**********AA | [25] |
| Reverse | GC**********CT | Reverse | GA**********TT | ||||||
| Probe | [TR]GC**********AA[BHQ2] | Probe | [FAM]TG**********CA[BHQ1] | ||||||
| Parvimonas micra | 16S ribosomal RNA | Forward | GA**********AG | [15] | Enterococcus faecalis | gelE-sprE operon | Forward | GA**********TT | [26] |
| Reverse | GG**********CC | Reverse | CG**********AC | ||||||
| Probe | [FAM]GG**********CA[BHQ1] | Probe | [HEX]GC**********GA[BHQ1] | ||||||
| Campylobacter rectus | GroEL | Forward | AA**********GG | [16] | Actinomyces viscosus | nanH | Forward | GC**********CG | [21] |
| Reverse | TC**********GA | Reverse | GA**********CA | ||||||
| Probe | [HEX]GG**********GT[BHQ1] | Probe | [TR]GA**********AA[BHQ2] |
| Characteristic | Healthy Group (n = 35) | Severe Periodontitis Group (n = 52) | |
|---|---|---|---|
| Age (Years, mean ± SD) | 39.0 ± 17.9 | 56.2 ± 15.2 | |
| Sex | Male | 29 (83%) | 44 (85%) |
| Female | 6 (17%) | 8 (15%) | |
| Smoking | Non-smokers | 31 (89%) | 46 (88%) |
| Current smokers | 4 (11%) | 6 (12%) | |
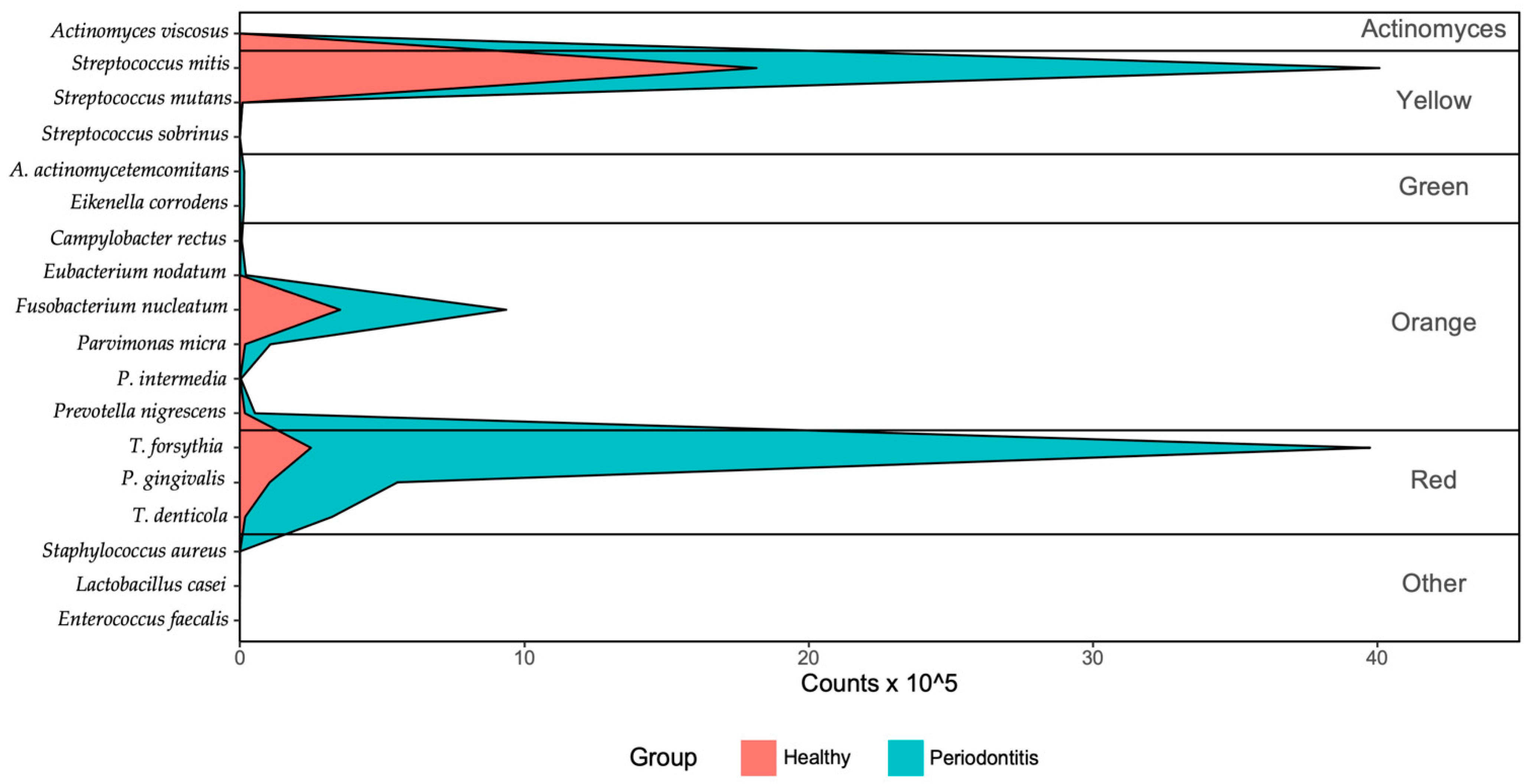
| Bacteria | Healthy Group (n = 35) | Severe Periodontitis Group (n = 52) |
|---|---|---|
| Aggregatibacter actinomycetemcomitans | ||
| Prevalence, n (%) a | 1 (2.9) | 7 (13.5) |
| Median bacterial cells proportion (%) (IQR) | 0.46 (0.46–0.46) | 0.75 (0.46–1.07) |
| Porphyromonas gingivalis | ||
| Prevalence, n (%) | 16 (45.7) | 47 (90.4) |
| Median bacterial cells proportion (%) (IQR) | 3.43 (1.74–5.86) | 3.83 (2.27–8.28) |
| Tannerella forsythia | ||
| Prevalence, n (%) | 26 (74.3) | 38 (73.1) |
| Median bacterial cells proportion (%) (IQR) | 3.07 (0.65–7.74) | 26.07 (4.49–50.82) |
| Treponema denticola | ||
| Prevalence, n (%) a | 15 (42.9) | 36 (69.2) |
| Median bacterial cells proportion (%) (IQR) | 0.59 (0.18–2.34) | 2.91 (1.36–5.39) |
| Fusobacterium nucleatum | ||
| Prevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells proportion (%) (IQR) | 18.73 (13.31–23.15) | 12.89 (7.23–20.02) |
| Prevotella intermedia | ||
| Prevalence, n (%) | 8 (22.9) | 15 (28.8) |
| Median bacterial cells proportion (%) (IQR) | 0.22 (0.05–0.46) | 0.11 (0.05–0.19) |
| Prevotella nigrescens | ||
| Prevalence, n (%) a | 28 (80.0) | 43 (82.7) |
| Median bacterial cells proportion (%) (IQR) | 0.73 (0.4–1.94) | 0.47 (0.14–1.23) |
| Parvimonas micra | ||
| Prevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells proportion (%) (IQR) | 0.5 (0.29–0.82) | 0.99 (0.45–1.81) |
| Campylobacter rectus | ||
| Prevalence, n (%) | 28 (80.0) | 45 (86.5) |
| Median bacterial cells proportion (%) (IQR) | 0.11 (0.06–0.18) | 0.08 (0.04–0.13) |
| Eubacterium nodatum | ||
| Prevalence, n (%) | 3 (8.6) | 14 (26.9) |
| Median bacterial cells proportion (%) (IQR) | 0.21 (0.12–0.27) | 0.71 (0.3–1.34) |
| Eikenella corrodens | ||
| Prevalence, n (%) | 4 (11.4) | 15 (28.8) |
| Median bacterial cells proportion (%) (IQR) | 0.07 (0.04–0.45) | 0.28 (0.15–0.71) |
| Streptococcus mitis | ||
| Prevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells proportion (%) (IQR) | 73.72 (63.61–79.49) | 59.13 (37.87–70.34) |
| Streptococcus mutans | ||
| Prevalence, n (%) | 23 (65.7) | 35 (67.3) |
| Median bacterial cells proportion (%) (IQR) | 0.03 (0.02–0.1) | 0.03 (0.01–0.15) |
| Streptococcus sobrinus | ||
| Prevalence, n (%) | 1 (2.9) | 5 (9.6) |
| Median bacterial cells proportion (%) (IQR) | 0.06 (0.06–0.06) | 0 (0–0.01) |
| Lactobacillus casei | ||
| Prevalence, n (%) | 6 (17.1) | 18 (34.6) |
| Median bacterial cells proportion (%) (IQR) | 0.01 (0–0.04) | 0 (0–0.01) |
| Staphylococcus aureus | ||
| Prevalence, n (%) | 15 (42.9) | 4 (7.7) |
| Median bacterial cells proportion (%) (IQR) | 0.02 (0.01–0.14) | 0.03 (0–0.07) |
| Enterococcus faecalis | ||
| Prevalence, n (%) | 0 (0.0) | 0 (0.0) |
| Median bacterial cells proportion (%) (IQR) | NA (NA–NA) | NA (NA–NA) |
| Actinomyces viscosus | ||
| Prevalence, n (%) | 0 (0.0) | 0 (0.0) |
| Median bacterial cells proportion (%) (IQR) | NA (NA–NA) | NA (NA–NA) |
| Total number of cellsPrevalence, n (%) | 35 (100.0) | 52 (100.0) |
| Median bacterial cells (IQR) | 36,126,518 (16,199,034–92,716,204) | 108524910 (69,243,624.5–177,393,988.25) |
| Aa | Pg | Tf | Td | Fn | Pi | Pn | Pm | Cr | En | Ec | Sm | Smu | Ss | Lc | Sa | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa | 0.96 * | −0.49 | 0.3 | 0.38 | 0.98 * | 0.91 * | 0.57 | 0.07 | −0.26 | ||||||||
| Pg | −0.04 | 0.25 | −0.01 | −0.06 | 0.16 | 0.23 | −0.02 | 0.57 * | −0.3 | −0.34 * | 0.16 | 0.5 | −0.05 | −0.27 | −0.06 | ||
| Tf | 0.19 | −0.56 * | −0.21 | −0.28 * | 0.03 | −0.02 | 0.1 | −0.37 | −0.87 * | −0.22 | −0.54 | −0.2 | −0.21 | 0.14 | |||
| Td | −0.17 | 0.53 * | 0.05 | 0.61 * | −0.08 | 0.8 * | −0.24 | −0.42 * | −0.02 | −0.17 | 0.34 | −0.4 | −0.07 | ||||
| Fn | 0.21 | 0.25 * | −0.15 | 0.18 | −0.17 | 0.07 | 0.25 * | 0.02 | 0.9 * | 0.52 * | 0.02 | −0.13 | |||||
| Pi | 0.53 * | 0.14 | 0.51 * | 0.51 | 0.09 | −0.09 | 0.43 | 0.32 | |||||||||
| Pn | 0.15 | 0.23 | −0.16 | −0.13 | 0.07 | −0.18 | 0.75 | −0.03 | −0.32 | −0.21 | |||||||
| Pm | 0.18 | 0.13 | −0.27 | −0.23 * | 0 | 0.17 | −0.02 | −0.23 | −0.02 | ||||||||
| Cr | −0.31 | −0.24 | 0.03 | −0.15 | 0.88 * | 0.06 | −0.24 | −0.16 | |||||||||
| En | −0.13 | 0.35 | 0.3 | 0.42 | |||||||||||||
| Ec | 0.24 | 0.19 | −0.17 | −0.16 | |||||||||||||
| Sm | 0.12 | 0.33 | 0.04 | 0.23 | −0.03 | ||||||||||||
| Smu | −0.12 | −0.13 | −0.15 | −0.09 | |||||||||||||
| Ss | −0.62 | ||||||||||||||||
| Lc | −0.16 | ||||||||||||||||
| Sa | −0.21 | ||||||||||||||||
| Total |
| Aa | Pg | Tf | Td | Fn | Pi | Pn | Pm | Cr | En | Ec | Sm | Smu | Ss | Lc | Sa | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa | 0.96 * | −0.81 | 0.24 | 0.55 | 0.98 * | 0.92 * | 0.56 | 0.18 | −0.34 | ||||||||
| Pg | −0.03 | 0.22 | 0.02 | −0.04 | 0.12 | 0.22 | 0 | 0.57 * | −0.32 | −0.3 * | 0.14 | 0.71 | −0.05 | −0.07 | |||
| Tf | 0.11 | −0.67 * | −0.13 | −0.34 | −0.12 | −0.13 | 0.05 | −0.49 | −0.88 * | −0.29 | 0.11 | −0.31 | 0.05 | ||||
| Td | −0.12 | 0.55 | 0.07 | 0.62 * | −0.09 | 0.78 * | −0.34 | −0.33 * | −0.08 | −0.17 | 0.4 | −0.16 | |||||
| Fn | 0.26 | 0.33 * | −0.1 | 0.24 | −0.12 | 0.28 | 0.43 * | 0.38 * | 0.33 | 0.19 | 0.01 | ||||||
| Pi | 0.75 * | 0.21 | 0.56 * | 0.58 | 0.15 | −0.16 | 0.36 | ||||||||||
| Pn | 0.23 | 0.4 * | −0.22 | −0.14 | 0.15 | −0.15 | −0.26 | −0.17 | −0.23 | ||||||||
| Pm | 0.22 | −0.09 | −0.32 | −0.11 | −0.08 | −0.37 | 0.11 | −0.12 | |||||||||
| Cr | −0.32 | −0.22 | 0.12 | −0.18 | 0.95 * | 0.22 | −0.14 | ||||||||||
| En | −0.13 | 0.31 | 0.3 | 0.45 | |||||||||||||
| Ec | 0.2 | 0.18 | −0.23 | −0.24 | |||||||||||||
| Sm | 0.12 | −0.2 | 0.24 | 0.04 | |||||||||||||
| Smu | 0.01 | −0.23 | |||||||||||||||
| Ss | −0.49 | ||||||||||||||||
| Lc | −0.12 | ||||||||||||||||
| Sa | |||||||||||||||||
| Total |
| Aa | Pg | Tf | Td | Fn | Pi | Pn | Pm | Cr | En | Ec | Sm | Smu | Ss | Lc | Sa | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa | 0.96 * | −0.81 | 0.24 | 0.55 | 0.98 * | 0.92 * | 0.56 | 0.18 | −0.34 | ||||||||
| Pg | −0.03 | 0.22 | 0.02 | −0.04 | 0.12 | 0.22 | 0 | 0.57 * | −0.32 | −0.3 * | 0.14 | 0.71 | −0.05 | −0.07 | |||
| Tf | 0.11 | −0.67 * | −0.13 | −0.34 | −0.12 | −0.13 | 0.05 | −0.49 | −0.88 * | −0.29 | 0.11 | −0.31 | 0.05 | ||||
| Td | −0.12 | 0.55 | 0.07 | 0.62 * | −0.09 | 0.78 * | −0.34 | −0.33 * | −0.08 | −0.17 | 0.4 | −0.16 | |||||
| Fn | 0.26 | 0.33 * | −0.1 | 0.24 | −0.12 | 0.28 | 0.43 * | 0.38 * | 0.33 | 0.19 | 0.01 | ||||||
| Pi | 0.75 * | 0.21 | 0.56 * | 0.58 | 0.15 | −0.16 | 0.36 | ||||||||||
| Pn | 0.23 | 0.4 * | −0.22 | −0.14 | 0.15 | −0.15 | −0.26 | −0.17 | −0.23 | ||||||||
| Pm | 0.22 | −0.09 | −0.32 | −0.11 | −0.08 | −0.37 | 0.11 | −0.12 | |||||||||
| Cr | −0.32 | −0.22 | 0.12 | −0.18 | 0.95 * | 0.22 | −0.14 | ||||||||||
| En | −0.13 | 0.31 | 0.3 | 0.45 | |||||||||||||
| Ec | 0.2 | 0.18 | −0.23 | −0.24 | |||||||||||||
| Sm | 0.12 | −0.2 | 0.24 | 0.04 | |||||||||||||
| Smu | 0.01 | −0.23 | |||||||||||||||
| Ss | −0.49 | ||||||||||||||||
| Lc | −0.12 | ||||||||||||||||
| Sa | |||||||||||||||||
| Total |
| Levels | No. of Subjects | No. (%) with Severe Periodontitis | Crude OR (95% CI) | p Value | Adjusted OR (95% CI) | p Value | |
|---|---|---|---|---|---|---|---|
| Aa | |||||||
| 0 | 79 | 45 (57.0) | 1 | 1 | |||
| 1 | 4 | 3 (75.0) | 1.8 (0.3–18.9) | 0.557 | 13.7 (0.9–389.2) | 0.059 | |
| 2 | 4 | 4 (100.0) | 6.8 (0.7–915.8) | 0.111 | 1.6 (0.1–232.1) | 0.750 | |
| Pg | |||||||
| 0 | 24 | 5 (20.8) | 1 | - | 1 | - | |
| 1 | 31 | 23 (74.2) | 10.9 (3.1–39.0) | <0.001 | 3.3 (0.4–26.6) | 0.271 | |
| 2 | 32 | 24 (75.0) | 11.4 (3.2–40.6) | <0.001 | 1.1 (0.1–8.7) | 0.942 | |
| Tf | |||||||
| 0 | 23 | 14 (60.9) | 1 | - | 1 | - | |
| 1 | 32 | 13 (40.6) | 0.4 (0.1–1.3) | 0.141 | 0.1 (0.0–1.1) | 0.059 | |
| 2 | 32 | 25 (78.1) | 2.3 (0.7–7.5) | 0.169 | 3.7 (0.3–48.1) | 0.319 | |
| Td | |||||||
| 0 | 36 | 16 (44.4) | 1 | - | 1 | - | |
| 1 | 25 | 14 (56.0) | 1.6 (0.6–4.4) | 0.375 | 5.3 (0.6–44.8) | 0.129 | |
| 2 | 26 | 22 (84.6) | 6.9 (2.0–24.0) | 0.002 | 7.3 (1.1–47.4) | 0.035 | |
| Fn | |||||||
| 1 | 43 | 30 (69.8) | 1 | - | 1 | - | |
| 2 | 44 | 22 (50.0) | 0.4 (0.2–1.0) | 0.062 | 1.0 (0.2–4.3) | 0.969 | |
| Pi | |||||||
| 0 | 64 | 37 (57.8) | 1 | - | 1 | - | |
| 1 | 11 | 7 (63.6) | 1.3 (0.3–4.8) | 0.717 | 0.7 (0.1–7.6) | 0.762 | |
| 2 | 12 | 8 (66.7) | 1.5 (0.4–5.3) | 0.568 | 0.5 (0.1–3.4) | 0.504 | |
| Pn | |||||||
| 0 | 16 | 9 (56.2) | 1 | - | 1 | - | |
| 1 | 35 | 25 (71.4) | 1.9 (0.6–6.7) | 0.289 | 120.4 (5.3–2725.4) | 0.002 | |
| 2 | 36 | 18 (50.0) | 0.8 (0.2–2.5) | 0.677 | 22.5 (2.0–260.6) | 0.012 | |
| Pm | |||||||
| 1 | 43 | 19 (44.2) | 1 | - | 1 | - | |
| 2 | 44 | 33 (75.0) | 3.8 (1.5–9.4) | 0.004 | 4.4 (1.0–20.1) | 0.057 | |
| Cr | |||||||
| 0 | 14 | 7 (50.0) | 1 | - | 1 | - | |
| 1 | 36 | 25 (69.4) | 2.3 (0.6–8.1) | 0.203 | 3.5 (0.5–27.3) | 0.226 | |
| 2 | 37 | 20 (54.1) | 1.2 (0.3–4.0) | 0.795 | 1.6 (0.2–11.0) | 0.628 | |
| En | |||||||
| 0 | 70 | 38 (54.3) | 1 | - | 1 | - | |
| 1 | 8 | 5 (62.5) | 1.3 (0.3–6.1) | 0.694 | 0.3 (0.1–2.0) | 0.227 | |
| 2 | 9 | 9 (100.0) | 16.0 (1.9–2097.0) | 0.006 | 4.3 (0.4–609.5) | 0.280 | |
| Ec | |||||||
| 0 | 68 | 37 (54.4) | 1 | - | 1 | - | |
| 1 | 9 | 6 (66.7) | 1.7 (0.4–7.3) | 0.490 | 2.6 (0.2–30.9) | 0.441 | |
| 2 | 10 | 9 (90.0) | 7.5 (0.9–62.8) | 0.061 | 13.6 (0.5–380.9) | 0.124 | |
| Sm | |||||||
| 1 | 43 | 33 (76.7) | 1 | - | 1 | - | |
| 2 | 44 | 19 (43.2) | 0.2 (0.1–0.6) | 0.001 | 0.1 (0.0–0.8) | 0.024 | |
| Smu | |||||||
| 0 | 29 | 17 (58.6) | 1 | - | 1 | - | |
| 1 | 29 | 18 (62.1) | 1.2 (0.4–3.3) | 0.788 | 2.5 (0.4–18.3) | 0.354 | |
| 2 | 29 | 17 (58.6) | 1.0 (0.4–2.8) | 1 | 0.4 (0.1–2.5) | 0.316 | |
| Ss | |||||||
| 0 | 81 | 47 (58.0) | 1 | 1 | |||
| 1 | 3 | 3 (100.0) | 5.1 (0.5–692.1) | 0.205 | 1.6 (0.1–244.0) | 0.755 | |
| 2 | 3 | 2 (66.7) | 1.2 (0.2–13.7) | 0.856 | 1.4 (0.1–193.5) | 0.873 | |
| Lc | |||||||
| 0 | 63 | 34 (54.0) | 1 | - | 1 | - | |
| 1 | 12 | 11 (91.7) | 9.4 (1.1–77.0) | 0.037 | 1.4 (0.1–14.1) | 0.795 | |
| 2 | 12 | 7 (58.3) | 1.2 (0.3– 4.2) | 0.780 | 0.3 (0.0–1.9) | 0.192 | |
| Sa | |||||||
| 0 | 68 | 48 (70.6) | 1 | - | 1 | - | |
| 1 | 9 | 2 (22.2) | 0.1 (0.0–0.6) | 0.011 | 0.2 (0.0–2.0) | 0.162 | |
| 2 | 10 | 2 (20.0) | 0.1 (0.0–0.5) | 0.006 | 0.4 (0.0–7.4) | 0.525 | |
| Total | |||||||
| 1 | 43 | 18 (41.9) | 1 | - | 1 | - | |
| 2 | 44 | 34 (77.3) | 4.7 (1.9–12.0) | 0.001 | 1.4 (0.3–5.8) | 0.673 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-H.; Oh, J.-W.; Lee, Y.; Yun, J.-H.; Choi, S.-H.; Lee, D.-W. Quantification of Bacteria in Mouth-Rinsing Solution for the Diagnosis of Periodontal Disease. J. Clin. Med. 2021, 10, 891. https://doi.org/10.3390/jcm10040891
Kim J-H, Oh J-W, Lee Y, Yun J-H, Choi S-H, Lee D-W. Quantification of Bacteria in Mouth-Rinsing Solution for the Diagnosis of Periodontal Disease. Journal of Clinical Medicine. 2021; 10(4):891. https://doi.org/10.3390/jcm10040891
Chicago/Turabian StyleKim, Jeong-Hwa, Jae-Woon Oh, Young Lee, Jeong-Ho Yun, Seong-Ho Choi, and Dong-Woon Lee. 2021. "Quantification of Bacteria in Mouth-Rinsing Solution for the Diagnosis of Periodontal Disease" Journal of Clinical Medicine 10, no. 4: 891. https://doi.org/10.3390/jcm10040891
APA StyleKim, J.-H., Oh, J.-W., Lee, Y., Yun, J.-H., Choi, S.-H., & Lee, D.-W. (2021). Quantification of Bacteria in Mouth-Rinsing Solution for the Diagnosis of Periodontal Disease. Journal of Clinical Medicine, 10(4), 891. https://doi.org/10.3390/jcm10040891






