Genetic Modifying Factors of Cystic Fibrosis Phenotype: A Challenge for Modern Medicine
Abstract
1. Introduction
2. Literature Search Strategies and Data Collection
3. The Role of Genetic Heterogeneity in Cystic Fibrosis and Genotype-Phenotype Correlation
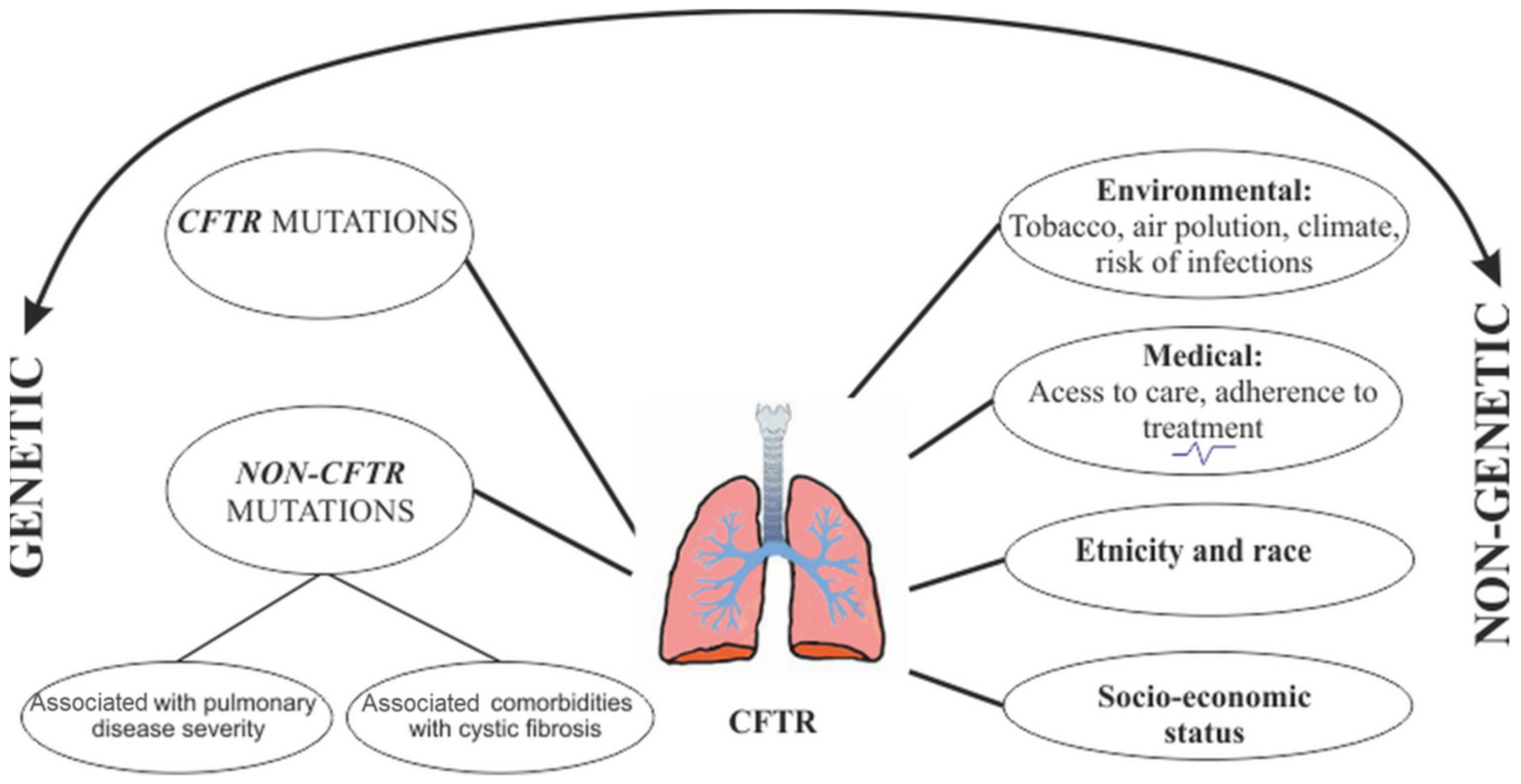
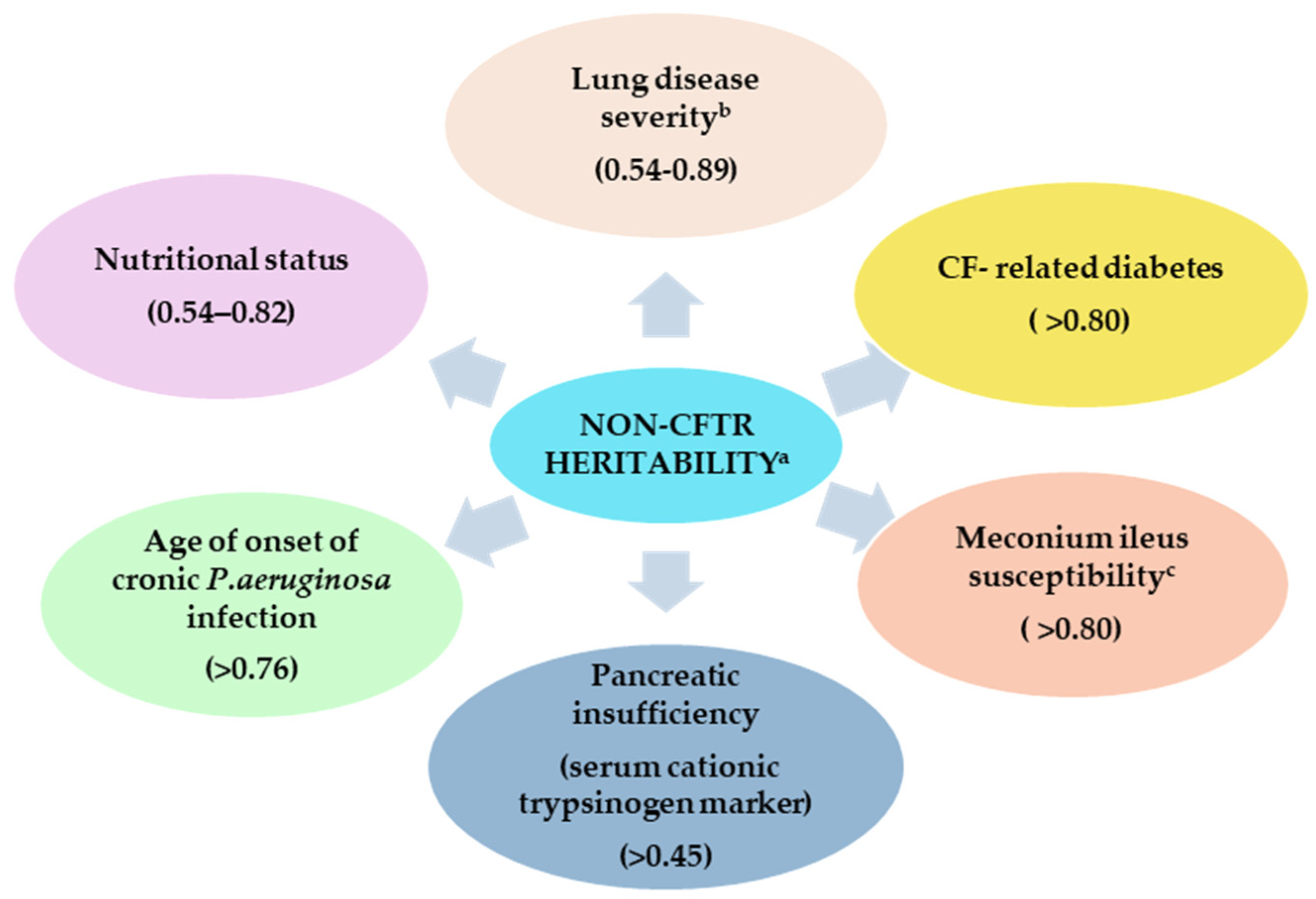
4. The Role of Modifier Genes and Phenotypic Variability in Cystic Fibrosis
4.1. The Concept of Modifier Genes
4.2. The Role of Modifier Factors
5. Candidate Gene Studies
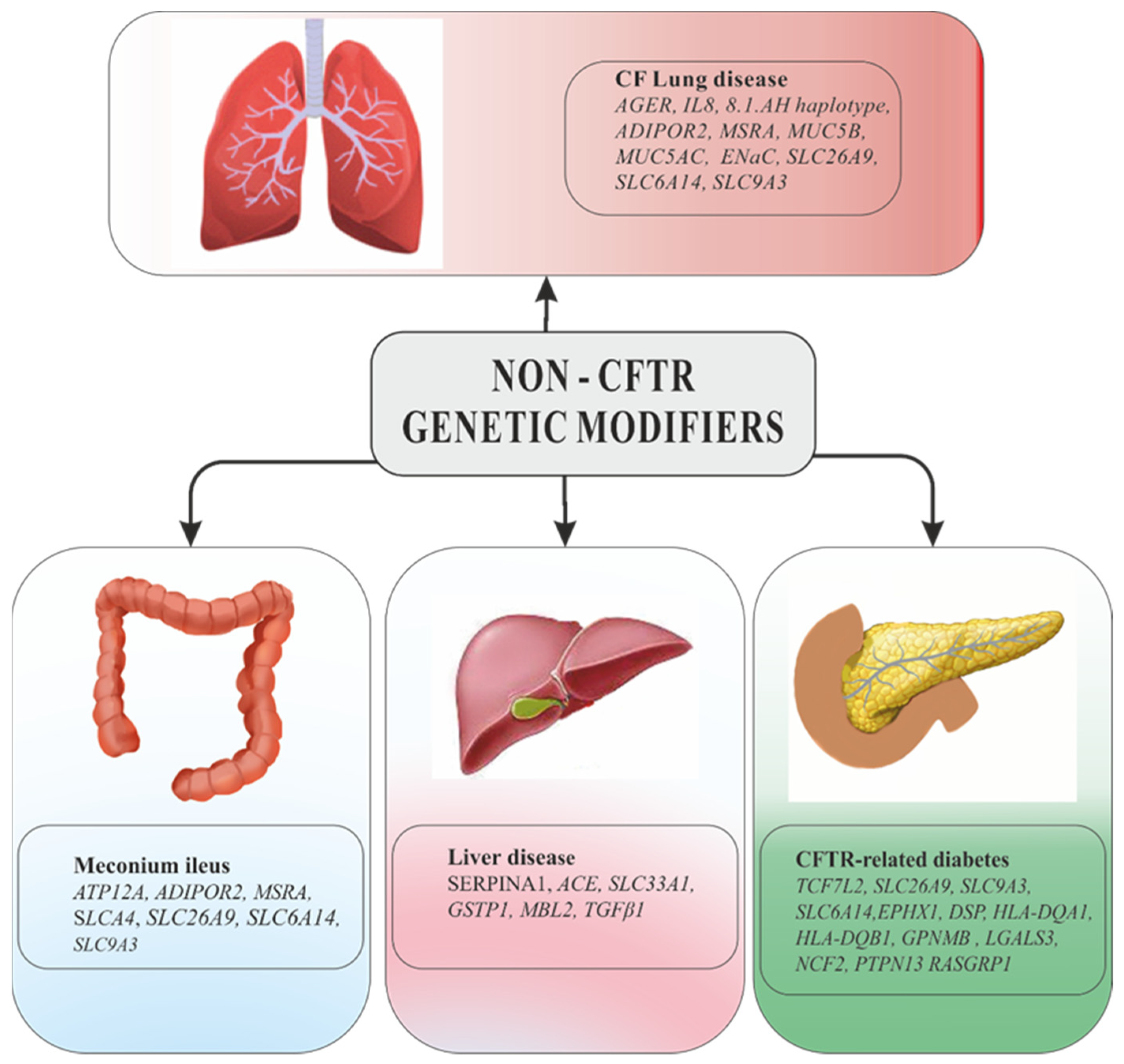
5.1. Modifier Genes of CF Lung Disease
5.1.1. Candidate Genes Related to the Inflammatory Mechanism
- a.
- Cytokines
- b.
- Other Genes Involved in Inflammation
5.1.2. Candidate Genes Related to the Infectious Response
5.1.3. Candidate Genes Involved in Epithelial Tissue Repair Mechanism
Glutathion and Glutathion-S-transferase
Nitric Oxide Synthases (NOS)
5.1.4. Candidate Genes Associated with the Response to Drug Therapy
5.1.5. Candidate Genes Encoding Ion Channels
5.1.6. Genes Encoding Cytoskeletal Proteins
5.2. Modifier Genes Related to Cystic Fibrosis Comorbidities
6. Whole Exome Sequencing (WES) and Genome-Wide Association Studies (GWAS)
6.1. Whole Exome Sequencing (WES)
6.2. Genome-Wide Associations Studies (GWASs)
6.3. Genome-Wide Associations Studies (GWAS) Results for Cystic Fibrosis Lung Disease
6.4. Genome-Wide Associations Studies (GWAS) Results for Cystic Fibrosis-Related Diabets (CFRD)
6.5. Genome-Wide Associations Studies (GWAS) Results for Meconium Ileus
7. CF Lung Disease Sverity: Non-Genetic Modifiers
8. Could Modifier Genes Influence the Response to CFTR Modulators?
9. Genetic Counseling of CF Patients in the Context of the Action of Modifier Genes
10. Discussion
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
References
- Cystic Fybrosis Foundation. Available online: https://www.cff.org/Research/Researcher-Resources/Patient-Registry/Understanding-Changes-in-Life-Expectancy/ (accessed on 19 November 2021).
- Gallati, S. Disease-modifying genes and monogenic disorders: Experience in cystic fibrosis. Appl. Clin. Genet. 2014, 7, 133–146. [Google Scholar] [CrossRef]
- Genetic Testing for Cystic Fibrosis. National Institutes of Health Consensus Development Conference Statement on genetic testing for cystic fibrosis. Arch. Intern. Med. 1999, 159, 1529–1539. [Google Scholar]
- Eurogappp Project 1999–2000 Public and Professional Policy Committee (PPPC) Population genetic screening programmes: Proposed recommendations of the European Society of Human Genetics. Eur. J. Hum. Genet. 2000, 8, 998–1000. [CrossRef]
- Barrio, R. Management of endocrine disease: Cystic fibrosis-related diabetes: Novel pathogenic insights opening new therapeutic avenues. Eur. J. Endocrinol. 2015, 172, R131–R141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cutting, G.R. Causes of Variation in the Cystic Fibrosis Phenotype. Ann. Nestlé 2006, 64, 111–117. [Google Scholar] [CrossRef]
- Marson, F.A.L. Disease-modifying genetic factors in cystic fibrosis. Curr. Opin. Pulm. Med. 2018, 24, 296–308. [Google Scholar] [CrossRef]
- Guillot, L.; Beucher, J.; Tabary, O.; Le Rouzic, P.; Clement, A.; Corvol, H. Lung disease modifier genes in cystic fibrosis. Int. J. Biochem. Cell Biol. 2014, 52, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Drumm, M.L.; Konstan, M.W.; Schluchter, M.D.; Handler, A.; Pace, R.; Zou, F.; Zariwala, M.; Fargo, D.; Xu, A.; Dunn, J.M.; et al. Gene Modifier Study Group. Genetic modifiers of lung disease in cystic fibrosis. N. Engl. J. Med. 2005, 353, 1443–1453. [Google Scholar] [CrossRef]
- Bonfield, T.L.; Panuska, J.R.; Konstan, M.W.; Hilliard, K.A.; Hilliard, J.B.; Ghnaim, H.; Berger, M. Inflammatory cytokines in cystic fibrosis lungs. Am. J. Respir. Crit. Care Med. 1995, 152, 2111–2118, Erratum in 1996, 154, 1217. [Google Scholar] [CrossRef]
- Arkwright, P.D.; Laurie, S.; Super, M.; Pravica, V.; Schwarz, M.J.; Webb, A.K.; Hutchinson, I.V. TGF-beta 1 genotype and accelerated decline in lung function of patients with cystic fibrosis. Thorax 2000, 55, 459–462. [Google Scholar] [CrossRef][Green Version]
- Celedón, J.C.; Lange, C.; Raby, B.A.; Litonjua, A.A.; Palmer, L.J.; DeMeo, D.L.; Reilly, J.J.; Kwiatkowski, D.J.; Chapman, H.A.; Laird, N.; et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum. Mol. Genet. 2004, 13, 1649–1656. [Google Scholar] [CrossRef]
- Li, H.; Romieu, I.; Wu, H.; Sienra-Monge, J.J.; Ramírez-Aguilar, M.; del Río-Navarro, B.E.; del Lara-Sánchez, I.C.; Kistner, E.O.; Gjessing, H.K.; London, S.J. Genetic polymorphisms in transforming growth factor beta-1 (TGFB1) and childhood asthma and atopy. Hum. Genet. 2007, 121, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Weiler, C.A.; Drumm, M.L. Genetic influences on cystic fibrosis lung disease severity. Front. Pharmacol. 2013, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Collaco, J.M.; Cutting, G.R. Update on gene modifiers in cystic fibrosis. Curr. Opin. Pulm. Med. 2008, 14, 559–566. [Google Scholar] [CrossRef]
- Hillian, A.D.; Londono, D.; Dunn, J.M.; Goddard, K.A.; Pace, R.G.; Knowles, M.R.; Drumm, M.L. CF Gene Modifier Study Group. Modulation of cystic fibrosis lung disease by variants in interleukin-8. Genes Immun. 2008, 9, 501–508. [Google Scholar] [CrossRef][Green Version]
- Corvol, H.; Boelle, P.Y.; Brouard, J.; Knauer, N.; Chadelat, K.; Henrion-Caude, A.; Flamant, C.; Muselet-Charlier, C.; Boule, M.; Fauroux, B.; et al. Genetic variations in inflammatory mediators influence lung disease progression in cystic fibrosis. Pediatr. Pulmonol. 2008, 43, 1224–1232. [Google Scholar] [CrossRef]
- Levy, H.; Murphy, A.; Zou, F.; Gerard, C.; Klanderman, B.; Schuemann, B.; Lazarus, R.; García, K.C.; Celedón, J.C.; Drumm, M.; et al. IL1B polymorphisms modulate cystic fibrosis lung disease. Pediatr. Pulmonol. 2009, 44, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Labenski, H.; Hedtfeld, S.; Becker, T.; Tümmler, B.; Stanke, F. Initial interrogation, confirmation and fine mapping of modifying genes: STAT3, IL1B and IFNGR1 determine cystic fibrosis disease manifestation. Eur. J. Hum. Genet. 2011, 19, 1281–1288. [Google Scholar] [CrossRef]
- Noah, T.L.; Black, H.R.; Cheng, P.W.; Wood, R.E.; Leigh, M.W. Nasal and bronchoalveolar lavage fluid cytokines in early cystic fibrosis. J. Infect. Dis. 1997, 175, 638–647. [Google Scholar] [CrossRef]
- Osika, E.; Cavaillon, J.M.; Chadelat, K.; Boule, M.; Fitting, C.; Tournier, G.; Clement, A. Distinct sputum cytokine profiles in cystic fibrosis and other chronic inflammatory airway disease. Eur. Respir. J. 1999, 14, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Tesse, R.; Cardinale, F.; Santostasi, T.; Polizzi, A.; Mappa, L.; Manca, A.; De Robertis, F.; Silecchia, O.; Armenio, L. Association of interleukin-10 gene haplotypes with Pseudomonas aeruginosa airway colonization in cystic fibrosis. J. Cyst. Fibros. 2008, 7, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Castro-Giner, F.; Kogevinas, M.; Mächler, M.; de Cid, R.; Van Steen, K.; Imboden, M.; Schindler, C.; Berger, W.; Gonzalez, J.R.; Franklin, K.A.; et al. TNFA -308G>A in two international population-based cohorts and risk of asthma. Eur. Respir. J. 2008, 32, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Dominguez, C.N.; Reyes-Lopez, M.A.; Bustamante, A.; Cerda-Flores, R.M.; Villalobos-Torres, M.C.; Gallardo-Blanco, H.L.; Rojas-Martinez, A.; Martinez-Rodriguez, H.G.; Barrera-Saldaña, H.A.; Ortiz-Lopez, R. The tumor necrosis factor α (-308 A/G) polymorphism is associated with cystic fibrosis in Mexican patients. PLoS ONE 2014, 9, e90945. [Google Scholar] [CrossRef]
- Kaluza, W.; Reuss, E.; Grossmann, S.; Hug, R.; Schopf, R.E.; Galle, P.R.; Maerker-Hermann, E.; Hoehler, T. Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism. J. Investig. Dermatol. 2000, 114, 1180–1183. [Google Scholar] [CrossRef]
- Yarden, J.; Radojkovic, D.; De Boeck, K.; Macek, M., Jr.; Zemkova, D.; Vavrova, V.; Vlietinck, R.; Cassiman, J.J.; Cuppens, H. Association of tumour necrosis factor alpha variants with the CF pulmonary phenotype. Thorax 2005, 60, 320–325. [Google Scholar] [CrossRef]
- Laki, J.; Laki, I.; Németh, K.; Ujhelyi, R.; Bede, O.; Endreffy, E.; Bolbás, K.; Gyurkovits, K.; Csiszér, E.; Sólyom, E.; et al. The 8.1 ancestral MHC haplotype is associated with delayed onset of colonization in cystic fibrosis. Int. Immunol. 2006, 18, 585–590. [Google Scholar] [CrossRef]
- Corvol, H.; Beucher, J.; Boëlle, P.Y.; Busson, P.F.; Muselet-Charlier, C.; Clement, A.; Ratjen, F.; Grasemann, H.; Laki, J.; Palmer, C.N.; et al. Ancestral haplotype 8.1 and lung disease severity in European cystic fibrosis patients. J. Cyst. Fibros. 2012, 11, 63–67. [Google Scholar] [CrossRef]
- Aron, Y.; Bienvenu, T.; Hubert, D.; Dusser, D.; Dall’Ava, J.; Polla, B.S. HLA-DR polymorphism in allergic bronchopulmonary aspergillosis. J. Allergy Clin. Immunol. 1999, 104, 891–892. [Google Scholar] [CrossRef]
- O’Neal, W.K.; Gallins, P.; Pace, R.G.; Dang, H.; Wolf, W.E.; Jones, L.C.; Guo, X.; Zhou, Y.H.; Madar, V.; Huang, J.; et al. Gene expression in transformed lymphocytes reveals variation in endomembrane and HLA pathways modifying cystic fibrosis pulmonary phenotypes. Am. J. Hum. Genet. 2015, 96, 318–328. [Google Scholar] [CrossRef]
- Viel, M.; Hubert, D.; Burgel, P.R.; Génin, E.; Honoré, I.; Martinez, B.; Gaitch, N.; Chapron, J.; Kanaan, R.; Dusser, D.; et al. DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Clin. Respir. J. 2016, 10, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Rahaghi, F.F. Alpha-1 antitrypsin deficiency research and emerging treatment strategies: What’s down the road? Ther. Adv. Chronic. Dis. 2021, 29, 12. [Google Scholar] [CrossRef]
- King, M.A.; Stone, J.A.; Diaz, P.T.; Mueller, C.F.; Becker, W.J.; Gadek, J.E. Alpha 1-antitrypsin deficiency: Evaluation of bronchiectasis with CT. Radiology 1996, 199, 137–141. [Google Scholar] [CrossRef]
- Morgan, K.; Scobie, G.; Marsters, P.; Kalsheker, N.A. Mutation in an alpha1-antitrypsinenhancer results in an interleukin-6 deficient acute-phase response due toloss of cooperativity between transcription factors. Biochim. Biophys. Acta 1997, 1362, 67–76. [Google Scholar] [CrossRef][Green Version]
- Frangolias, D.D.; Ruan, J.; Wilcox, P.J.; Davidson, A.G.; Wong, L.T.; Berthiaume, Y.; Hennessey, R.; Freitag, A.; Pedder, L.; Corey, M.; et al. Alpha 1-antitrypsin deficiency alleles in cystic fibrosis lung disease. Am. J. Respir. Cell Mol. Biol. 2003, 29, 390–396. [Google Scholar] [CrossRef]
- Beucher, J.; Boëlle, P.Y.; Busson, P.F.; Muselet-Charlier, C.; Clement, A.; Corvol, H.; French, C.F. Modifier Gene Study Investigators. AGER -429T/C is associated with an increased lung disease severity in cystic fibrosis. PLoS ONE 2012, 7, e41913. [Google Scholar] [CrossRef] [PubMed]
- Adamali, H.; Armstrong, M.E.; McLaughlin, A.M.; Cooke, G.; McKone, E.; Costello, C.M.; Gallagher, C.G.; Leng, L.; Baugh, J.A.; Fingerle-Rowson, G.; et al. Macrophage migration inhibitory factor enzymatic activity, lung inflammation, and cystic fibrosis. Am. J. Respir. Crit. Care Med. 2012, 186, 162–169. [Google Scholar] [CrossRef]
- Plant, B.J.; Gallagher, C.G.; Bucala, R.; Baugh, J.A.; Chappell, S.; Morgan, L.; O’Connor, C.M.; Morgan, K.; Donnelly, S.C. Cystic fibrosis, disease severity, and a macrophage migration inhibitory factor polymorphism. Am. J. Respir. Crit. Care Med. 2005, 172, 1412–1415. [Google Scholar] [CrossRef] [PubMed]
- Seibold, M.A.; Wise, A.L.; Speer, M.C.; Steele, M.P.; Brown, K.K.; Loyd, J.E.; Fingerlin, T.E.; Zhang, W.; Gudmundsson, G.; Groshong, S.D.; et al. A Common MUC5B Promoter Polymorphism and Pulmonary Fibrosis. N. Engl. J. Med. 2011, 364, 1503–1512. [Google Scholar] [CrossRef]
- Guo, X.; Pace, R.G.; Stonebraker, J.R.; Commander, C.W.; Dang, A.T.; Drumm, M.L.; Harris, A.; Zou, F.; Swallow, D.M.; Wright, F.A.; et al. Mucin variable number tandem repeat polymorphisms and severity of cystic fibrosis lung disease: Significant association with MUC5AC. PLoS ONE 2011, 6, e25452. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Soave, D.; Miller, M.R.; Keenan, K.; Lin, F.; Gong, J.; Chiang, T.; Stephenson, A.L.; Durie, P.; Rommens, J.; et al. Unraveling the complex genetic model for cystic fibrosis: Pleiotropic effects of modifier genes on early cystic fibrosis-related morbidities. Hum. Genet. 2014, 133, 151–161. [Google Scholar] [CrossRef]
- Garred, P.; Larsen, F.; Seyfarth, J.; Fujita, R.; Madsen, H.O. Mannose-binding lectin and its genetic variants. Genes Immun. 2006, 7, 85–94. [Google Scholar] [CrossRef]
- Chalmers, J.D.; Fleming, G.B.; Hill, A.T.; Kilpatrick, D.C. Impact of mannose-binding lectin insufficiency on the course of cystic fibrosis: A review and meta-analysis. Glycobiology 2011, 21, 271–282. [Google Scholar] [CrossRef]
- Blohmke, C.J.; Park, J.; Hirschfeld, A.F.; Victor, R.E.; Schneiderman, J.; Stefanowicz, D.; Chilvers, M.A.; Durie, P.R.; Corey, M.; Zielenski, J.; et al. TLR5 as an anti-inflammatory target and modifier gene in cystic fibrosis. J. Immunol. 2010, 185, 7731–7738. [Google Scholar] [CrossRef]
- Vencken, S.F.; Greene, C.M. Toll-like Receptors in Cystic Fibrosis: Impact of Dysfunctional microRNA on Innate Immune Responses in the Cystic Fibrosis Lung. J. Innate Immun. 2016, 8, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.C.; Laing, I.A.; Zhang, G.; Brennan, S.; Winfield, K.; Sly, P.D.; Stick, S.M.; Goldblatt, J.; LeSouef, P.N. CD14 C-159T and early infection with Pseudomonas aeruginosa in children with cystic fibrosis. Respir. Res. 2005, 6, 63. [Google Scholar] [CrossRef] [PubMed]
- Alexis, N.; Eldridge, M.; Reed, W.; Bromberg, P.; Peden, D.B. CD14-dependent airway neutrophil response to inhaled LPS: Role of atopy. J. Allergy Clin. Immunol. 2001, 107, 31–35. [Google Scholar] [CrossRef]
- Faria, E.J.; Faria, I.C.; Ribeiro, J.D.; Ribeiro, A.F.; Hessel, G.; Bertuzzo, C.S. Association of MBL2, TGF-beta1 and CD14 gene polymorphisms with lung disease severity in cystic fibrosis. J. Bras. Pneumol. 2009, 35, 334–342. [Google Scholar] [CrossRef]
- Ricciardolo, F.L.M. Multiple roles of nitric oxide in the airways. Thorax 2003, 58, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Dötsch, J.; Puls, J.; Klimek, T.; Rascher, W. Reduction of neuronal and inducible nitric oxide synthase gene expression in patients with cystic fibrosis. Eur. Arch. Otorhinolaryngol. 2002, 259, 222–226. [Google Scholar] [CrossRef]
- Grasemann, H.; Knauer, N.; Büscher, R.; Hübner, K.; Drazen, J.M.; Ratjen, F. Airway Nitric Oxide Levels in Cystic Fibrosis Patients Are Related to a Polymorphism in the Neuronal Nitric Oxide Synthase Gene. Am. J. Respir. Crit. Care Med. 2000, 162, 2172–2176. [Google Scholar] [CrossRef]
- Texereau, J.; Marullo, S.; Hubert, D.; Coste, J.; Dusser, D.J.; Dall’Ava-Santucci, J.; Dinh-Xuan, A.T. Nitric oxide synthase 1 as a potential modifier gene of decline in lung function in patients with cystic fibrosis. Thorax 2004, 59, 156–158. [Google Scholar] [CrossRef]
- Hart, M.A.; Konstan, M.W.; Darrah, R.J.; Schluchter, M.D.; Xue, L.; Londono, U.; Goddard, K.A.; Drumm, M.L.; Storfer-Isser, A. Beta 2 adrenergic receptor polymorphisms in cystic fibrosis. Pediatr. Pulmonol. 2005, 39, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Marson, F.A.; Bertuzzo, C.S.; Ribeiro, A.F.; Ribeiro, J.D. Polymorphisms in ADRB2 gene can modulate the response to bronchodilators and the severity of cystic fibrosis. BMC Pulm. Med. 2012, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- de Paiva, A.C.; Marson, F.A.; Ribeiro, J.D.; Bertuzzo, C.S. Asthma: Gln27Glu and Arg16Gly polymorphisms of the beta2-adrenergic receptor gene as risk factors. Allergy Asthma Clin. Immunol. 2014, 10, 8. [Google Scholar] [CrossRef]
- Corvol, H.; Nathan, N.; Charlier, C.; Chadelat, K.; Le Rouzic, P.; Tabary, O.; Fauroux, B.; Henrion-Caude, A.; Feingold, J.; Boelle, P.Y.; et al. Glucocorticoid receptor gene polymorphisms associated with progression of lung disease in young patients with cystic fibrosis. Respir. Res. 2007, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Norek, A.; Bienvenu, T.; Scheinert, S.; Kusmierek, E.; Chrzescijanska, E.; Sapiejka, E.; Swierczynska, B.; Sands, D.; Derichs, N. The role of CFTR/ENaC genotype in cystic fibrosis-like phenotypes. Eur. Respir. J. 2014, 44, 1959. [Google Scholar]
- Stanke, F.; Becker, T.; Cuppens, H.; Kumar, V.; Cassiman, J.J.; Jansen, S.; Radojkovic, D.; Siebert, B.; Yarden, J.; Ussery, D.W.; et al. The TNFalpha receptor TNFRSF1A and genes encoding the amiloride-sensitive sodium channel ENaC as modulators in cystic fibrosis. Hum. Genet. 2006, 119, 331–343. [Google Scholar] [CrossRef]
- Viel, M.; Leroy, C.; Hubert, D.; Fajac, I.; Bienvenu, T. ENaCbeta and gamma genes as modifier genes in cystic fibrosis. J. Cyst. Fibros. 2008, 7, 23–29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dorfman, R.; Taylor, C.; Lin, F.; Sun, L.; Sandford, A.; Paré, P.; Berthiaume, Y.; Corey, M.; Durie, P.; Zielenski, J.; et al. Modulatory effect of the SLC9A3 gene on susceptibility to infections and pulmonary function in children with cystic fibrosis. Pediatr. Pulmonol. 2011, 46, 385–392. [Google Scholar] [CrossRef]
- Stanke, F.; Hedtfeld, S.; Becker, T.; Tümmler, B. An association study on contrasting cystic fibrosis endophenotypes recognizes KRT8 but not KRT18 as a modifier of cystic fibrosis disease severity and CFTR mediated residual chloride secretion. BMC Med. Genet. 2011, 12, 62. [Google Scholar] [CrossRef]
- Somayaji, R.; Ramos, K.J.; Kapnadak, S.G.; Aitken, M.L.; Goss, C.H. Common clinical features of CF (respiratory disease and exocrine pancreatic insufficiency). Presse Med. 2017, 46, e109–e124. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Schwarzenberg, S.J. Pancreatic insufficiency in Cystic Fibrosis. J. Cyst. Fibros. 2017, 16, S70–S78. [Google Scholar] [CrossRef]
- Blackman, S.M.; Commander, C.W.; Watson, C.; Arcara, K.M.; Strug, L.J.; Stonebraker, J.R.; Wright, F.A.; Rommens, J.M.; Sun, L.; Pace, R.G.; et al. Genetic modifiers of cystic fibrosis-related diabetes. Diabetes 2013, 62, 3627–3635. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, R.; Li, W.; Sun, L.; Lin, F.; Wang, Y.; Sandford, A.; Paré, P.D.; McKay, K.; Kayserova, H.; Piskackova, T.; et al. Modifier gene study of meconium ileus in cystic fibrosis: Statistical considerations and gene mapping results. Hum. Genet. 2009, 126, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Henderson, L.B.; Doshi, V.K.; Blackman, S.M.; Naughton, K.M.; Pace, R.G.; Moskovitz, J.; Knowles, M.R.; Durie, P.R.; Drumm, M.L.; Cutting, G.R. Variation in MSRA modifies risk of neonatal intestinal obstruction in cystic fibrosis. PLoS Genet. 2012, 8, e1002580. [Google Scholar] [CrossRef] [PubMed]
- Strug, L.J.; Gonska, T.; He, G.; Keenan, K.; Ip, W.; Boëlle, P.-Y.; Lin, F.; Panjwani, N.; Gong, J.; Li, W.; et al. Cystic fibrosis gene modifier SLC26A9 modulates airway response to CFTR-directed therapeutics. Hum. Mol. Genet. 2016, 25, 4590–4600. [Google Scholar] [CrossRef]
- Aksit, M.A.; Pace, R.G.; Vecchio-Pagán, B.; Ling, H.; Rommens, J.M.; Boelle, P.Y.; Guillot, L.; Raraigh, K.S.; Pugh, E.; Zhang, P.; et al. Genetic Modifiers of Cystic Fibrosis-Related Diabetes Have Extensive Overlap With Type 2 Diabetes and Related Traits. J. Clin. Endocrinol. Metab. 2020, 105, 1401–1415. [Google Scholar] [CrossRef] [PubMed]
- Sepahzad, A.; Morris-Rosendahl, D.J.; Davies, J.C. Cystic Fibrosis Lung Disease Modifiers and Their Relevance in the New Era of Precision Medicine. Genes 2021, 12, 562. [Google Scholar] [CrossRef]
- Sun, L.; Rommens, J.M.; Corvol, H.; Li, W.; Li, X.; Chiang, T.A.; Lin, F.; Dorfman, R.; Busson, P.F.; Parekh, R.V.; et al. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat. Genet. 2012, 44, 562–569. [Google Scholar] [CrossRef]
- Gong, J.; Wang, F.; Xiao, B.; Panjwani, N.; Lin, F.; Keenan, K.; Avolio, J.; Esmaeili, M.; Zhang, L.; He, G.; et al. Genetic association and transcriptome integration identify contributing genes and tissues at cystic fibrosis modifier loci. PLoS Genet. 2019, 15, e1008007. [Google Scholar] [CrossRef]
- Trouvé, P.; Génin, E.; Férec, C. In silico search for modifier genes associated with pancreatic and liver disease in Cystic Fibrosis. PLoS ONE 2017, 12, e0173822. [Google Scholar] [CrossRef] [PubMed]
- Sofia, V.M.; Surace, C.; Terlizzi, V.; Da Sacco, L.; Alghisi, F.; Angiolillo, A.; Braggion, C.; Cirilli, N.; Colombo, C.; Di Lullo, A.; et al. Trans-heterozygosity for mutations enhances the risk of recurrent/chronic pancreatitis in patients with Cystic Fibrosis. Mol. Med. 2018, 24, 38. [Google Scholar] [CrossRef] [PubMed]
- Blackman, S.M.; Hsu, S.; Vanscoy, L.L.; Collaco, J.M.; Ritter, S.E.; Naughton, K.; Cutting, G.R. Genetic modifiers play a substantial role in diabetes complicating cystic fibrosis. J. Clin. Endocrinol. Metab. 2009, 94, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Iafusco, F.; Maione, G.; Rosanio, F.M.; Mozzillo, E.; Franzese, A.; Tinto, N. Cystic Fibrosis-Related Diabetes (CFRD): Overview of Associated Genetic Factors. Diagnostics 2021, 11, 572. [Google Scholar] [CrossRef]
- Jin, T. Current Understanding on Role of the Wnt Signaling Pathway Effector TCF7L2 in Glucose Homeostasis. Endocr. Rev. 2016, 37, 254–277. [Google Scholar] [CrossRef]
- Cho, D.H.; Lee, H.J.; Kim, H.J.; Hong, S.H.; Pyo, J.O.; Cho, C.; Jung, Y.-K. Suppression of hypoxic cell death by APIP-induced sustained activation of AKT and ERK1. Oncogene 2007, 26, 2809–2814. [Google Scholar] [CrossRef]
- Fossum, S.L.; Mutolo, M.J.; Tugores, A.; Ghosh, S.; Randell, S.H.; Jones, L.C.; Leir, S.H.; Harris, A. Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. J. Biol. Chem. 2017, 292, 10938–10949. [Google Scholar] [CrossRef]
- Cutting, G.R. Modifier genes in Mendelian disorders: The example of cystic fibrosis. Ann. N. Y. Acad. Sci. 2010, 1214, 57–69. [Google Scholar] [CrossRef]
- Knowles, M.R.; Drumm, M. The influence of genetics on cystic fibrosis phenotypes. Cold. Spring. Harb. Perspect. Med. 2012, 2, a009548. [Google Scholar] [CrossRef]
- Welsh, M.J.; Ramsey, B.W.; Accurso, F.J.; Cutting, G.R. Cystic fibrosis. In The Metabolic and Molecular Bases of Inherited Disease, 8th ed.; Scriver, C.R., Beaudet, A.L., Valle, D., Sly, W.S., Eds.; McGraw-Hill, Inc.: New York, NY, USA, 2001; pp. 5121–5188. [Google Scholar]
- Fanen, P.; Wohlhuter-Haddad, A.; Hinzpeter, A. Genetics of cystic fibrosis: CFTR mutation classifications toward genotype-based CF therapies. Int. J. Biochem. Cell Biol. 2014, 52, 94–102. [Google Scholar] [CrossRef]
- Pranke, I.; Golec, A.; Hinzpeter, A.; Edelman, A.; Sermet-Gaudelus, I. Emerging Therapeutic Approaches for Cystic Fibrosis. From Gene Editing to Personalized Medicine. Front. Pharmacol. 2019, 10, 121. [Google Scholar] [CrossRef]
- Boyle, M.P.; De Boeck, K. A new era in the treatment of cystic fibrosis: Correction of the underlying CFTR defect. Lancet Respir. Med. 2013, 1, 158–163, Erratum in 2013, 1, 101. [Google Scholar] [CrossRef]
- O’Neal, W.K.; Knowles, M.R. Cystic Fibrosis Disease Modifiers: Complex Genetics Defines the Phenotypic Diversity in a Monogenic Disease. Annu. Rev. Genom. Hum. Genet. 2018, 19, 201–222. [Google Scholar] [CrossRef]
- Hull, J.; Thomson, A.H. Contribution of genetic factors other than CFTR to disease severity in cystic fibrosis. Thorax 1998, 53, 1018–1021. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mekus, F.; Ballmann, M.; Bronsveld, I.; Bijman, J.; Veeze, H.; Tümmler, B. Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res. 2000, 3, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Vanscoy, L.L.; Blackman, S.M.; Collaco, J.M.; Bowers, A.; Lai, T.; Naughton, K.; Algire, M.; McWilliams, R.; Beck, S.; Hoover-Fong, J.; et al. Heritability of lung disease severity in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2007, 175, 1036–1043. [Google Scholar] [CrossRef]
- Blackman, S.M.; Deering-Brose, R.; McWilliams, R.; Naughton, K.; Coleman, B.; Lai, T.; Algire, M.; Beck, S.; Hoover-Fong, J.; Hamosh, A.; et al. Relative contribution of genetic and nongenetic modifiers to intestinal obstruction in cystic fibrosis. Gastroenterology 2006, 131, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Sontag, M.K.; Corey, M.; Hokanson, J.E.; Marshall, J.A.; Sommer, S.S.; Zerbe, G.O.; Accurso, F.J. Genetic and physiologic correlates of longitudinal immunoreactive trypsinogen decline in infants with cystic fibrosis identified through newborn screening. J. Pediatr. 2006, 149, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Bradley, G.M.; Blackman, S.M.; Watson, C.P.; Doshi, V.K.; Cutting, G.R. Genetic modifiers of nutritional status in cystic fibrosis. Am. J. Clin. Nutr. 2012, 96, 1299–1308. [Google Scholar] [CrossRef]
- Green, D.M.; Collaco, J.M.; McDougal, K.E.; Naughton, K.M.; Blackman, S.M.; Cutting, G.R. Heritability of respiratory infection with Pseudomonas aeruginosa in cystic fibrosis. J. Pediatr. 2012, 161, 290–295. [Google Scholar] [CrossRef]
- Mekus, F.; Laabs, U.; Veeze, H.; Tümmler, B. Genes in the vicinity of CFTR modulate the cystic fibrosis phenotype in highly concordant or discordant F508del homozygous sib pairs. Hum. Genet. 2003, 112, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Naren, A.P. Macromolecular complexes of cystic fibrosis transmembrane conductance regulator and its interacting partners. Pharmacol. Ther. 2005, 108, 208–223. [Google Scholar] [CrossRef]
- Guggino, W.B.; Stanton, B.A. New insights into cystic fibrosis: Molecular switches that regulate CFTR. Nat. Rev. Mol. Cell Biol. 2006, 7, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Hancock, D.B.; Artigas, M.S.; Gharib, S.A.; Henry, A.; Manichaikul, A.; Ramasamy, A.; Loth, D.W.; Imboden, M.; Koch, B.; McArdle, W.L.; et al. Genome-Wide Joint Meta-Analysis of SNP and SNP-by-Smoking Interaction Identifies Novel Loci for Pulmonary Function. PLoS Genet. 2012, 8, e1003098. [Google Scholar] [CrossRef]
- Corvol, H.; Blackman, S.M.; Boëlle, P.-Y.; Gallins, P.J.; Pace, R.G.; Stonebraker, J.R.; Accurso, F.J.; Clement, A.; Collaco, J.M.; Dang, H.; et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 2015, 6, 8382. [Google Scholar] [CrossRef] [PubMed]
- Wright, F.A.; Strug, L.J.; Doshi, V.K.; Commander, C.W.; Blackman, S.M.; Sun, L.; Berthiaume, Y.; Cutler, D.; Cojocaru, A.; Collaco, J.M.; et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat. Genet. 2011, 43, 539–546. [Google Scholar] [CrossRef]
- Castellani, C.; Assael, B.M. Cystic fibrosis: A clinical view. Cell. Mol. Life Sci. 2017, 74, 129–140. [Google Scholar] [CrossRef]
- Klimova, B.; Kuca, K.; Novotny, M.; Maresova, P. Cystic Fibrosis Revisited—A Review Study. Med. Chem. 2017, 13, 102–109. [Google Scholar] [CrossRef]
- Hamosh, A.; Corey, M. Correlation between genotype and phenotype in patients with cystic fibrosis. The Cystic Fibrosis Genotype-Phenotype Consortium. N. Engl. J. Med. 1993, 329, 1308–1313. [Google Scholar] [CrossRef]
- Ramsey, K.; Ratjen, F.; Latzin, P. Elucidating progression of early cystic fibrosis lung disease. Eur. Respir. J. 2017, 50, 1701916. [Google Scholar] [CrossRef]
- Ghigo, A.; Prono, G.; Riccardi, E.; De Rose, V. Dysfunctional Inflammation in Cystic Fibrosis Airways: From Mechanisms to Novel Therapeutic Approaches. Int. J. Mol. Sci. 2021, 22, 1952. [Google Scholar] [CrossRef] [PubMed]
- Stahl, M.; Steinke, E.; Mall, M.A. Quantification of Phenotypic Variability of Lung Disease in Children with Cystic Fibrosis. Genes 2021, 12, 803. [Google Scholar] [CrossRef] [PubMed]
- Konstan, M.W.; Wagener, J.S.; VanDevanter, D.R.; Pasta, D.J.; Millar, S.J.; Morgan, W.J. Scientific Advisory Group and the Investigators and Coordinators of the Epidemiologic Study of Cystic Fibrosis. Comparison of FEV1 reference equations for evaluating a cystic fibrosis therapeutic intervention. Pediatr. Pulmonol. 2017, 52, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Conese, M.; Di Gioia, S. Pathophysiology of Lung Disease and Wound Repair in Cystic Fibrosis. Pathophysiology 2021, 28, 155–188. [Google Scholar] [CrossRef]
- Hudson, V.M. Rethinking cystic fibrosis pathology: The critical role of abnormal reduced glutathione (GSH) transport caused by CFTR mutation. Free Radic. Biol. Med. 2001, 30, 1440–1461. [Google Scholar] [CrossRef]
- Flamant, C.; Henrion-Caude, A.; Boëlle, P.Y.; Brémont, F.; Brouard, J.; Delaisi, B.; Duhamel, J.F.; Marguet, C.; Roussey, M.; Miesch, M.C.; et al. Glutathione-S-transferase M1, M3, P1 and T1 polymorphisms and severity of lung disease in children with cystic fibrosis. Pharmacogenetics 2004, 14, 295–301. [Google Scholar] [CrossRef]
- Baldan, A.; Lo Presti, A.R.; Belpinati, F.; Castellani, C.; Bettin, M.D.; Xumerle, L.; Pignatti, P.R.; Malerba, G.; Bombieri, C. IFRD1 gene polymorphisms are associated with nasal polyposis in cystic fibrosis patients. Rhinology 2015, 53, 359–364. [Google Scholar] [CrossRef]
- Castaldo, A.; Cernera, G.; Iacotucci, P.; Cimbalo, C.; Gelzo, M.; Comegna, M.; Di Lullo, A.M.; Tosco, A.; Carnovale, V.; Raia, V.; et al. TAS2R38 is a novel modifier gene in patients with cystic fibrosis. Sci. Rep. 2020, 10, 5806. [Google Scholar] [CrossRef]
- Boyle, M.P. Strategies for identifying modifier genes in cystic fibrosis. Proc. Am. Thorac. Soc. 2007, 4, 52–57. [Google Scholar] [CrossRef]
- Wright, J.M.; Merlo, C.A.; Reynolds, J.B.; Zeitlin, P.L.; Garcia, J.G.; Guggino, W.B.; Boyle, M.P. Respiratory epithelial gene expression in patients with mild and severe cystic fibrosis lung disease. Am. J. Respir. Cell. Mol. Biol. 2006, 35, 327–336. [Google Scholar] [CrossRef]
- Emond, M.J.; Louie, T.; Emerson, J.; Zhao, W.; Mathias, R.A.; Knowles, M.R.; Wright, F.A.; Rieder, M.J.; Tabor, H.K.; Nickerson, D.A.; et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat. Genet. 2012, 44, 886–889. [Google Scholar] [CrossRef]
- Emond, M.J.; Louie, T.; Emerson, J.; Chong, J.X.; Mathias, R.A.; Knowles, M.R.; Rider, M.J.; Tabor, H.K.; Nickerson, D.A.; Barnes, K.C.; et al. Exome Sequencing of Phenotypic Extremes Identifies CAV2 and TMC6 as Interacting Modifiers of Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. PLoS Genet. 2015, 11, e1005273, Erratum in 2015, 11, e1005424. [Google Scholar] [CrossRef]
- Kontakioti, E.; Domvri, K.; Papakosta, D.; Daniilidis, M. HLA and asthma phenotypes/endotypes: A review. Hum. Immunol. 2014, 75, 930–939. [Google Scholar] [CrossRef]
- Dupuis, A.; Keenan, K.; Ooi, C.Y.; Dorfman, R.; Sontag, M.K.; Naehrlich, L.; Castellani, C.; Strug, L.J.; Rommens, J.M.; Gonska, T. Prevalence of meconium ileus marks the severity of mutations of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene. Genet. Med. 2016, 18, 333–340. [Google Scholar] [CrossRef]
- Rozmahel, R.; Wilschanski, M.; Matin, A.; Plyte, S.; Oliver, M.; Auerbach, W.; Moore, A.; Forstner, J.; Durie, P.; Nadeau, J.; et al. Modulation of disease severity in cystic fibrosis transmembrane conductance regulator deficient mice by a secondary genetic factor. Nat. Genet. 1996, 12, 280–287, Erratum in: 1996, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Zielenski, J.; Corey, M.; Rozmahel, R.; Markiewicz, D.; Aznarez, I.; Casals, T.; Larriba, S.; Mercier, B.; Cutting, G.R.; Krebsova, A.; et al. Detection of a cystic fibrosis modifier locus for meconium ileus on human chromosome 19q13. Nat. Genet. 1999, 22, 128–129. [Google Scholar] [CrossRef] [PubMed]
- Zielenski, J.; Dorfman, R.; Markeiwiz, D.; Corey, M.; Ng, P.; Mak, W.; Durie, P.; Tsui, L.C. Tagging SNP analyses of the CFM1 locus in CF patients with and without meconium ileus. Pediatr. Pulmonol. 2005, S28, 168. [Google Scholar]
- Kopp, B.T.; Ortega-García, J.A.; Sadreameli, S.C.; Wellmerling, J.; Cormet-Boyaka, E.; Thompson, R.; McGrath-Morrow, S.; Groner, J.A. The Impact of Secondhand Smoke Exposure on Children with Cystic Fibrosis: A Review. Int. J. Environ. Res. Public Health 2016, 13, 1003. [Google Scholar] [CrossRef]
- Oates, G.R.; Baker, E.; Rowe, S.M.; Gutierrez, H.H.; Schechter, M.S.; Morgan, W.; Harris, W.T. Tobacco smoke exposure and socioeconomic factors are independent predictors of pulmonary decline in pediatric cystic fibrosis. J. Cyst. Fibros. 2020, 19, 783–790. [Google Scholar] [CrossRef]
- Yen, E.H.; Quinton, H.; Borowitz, D. Better nutritional status in early childhood is associated with improved clinical outcomes and survival in patients with cystic fibrosis. J. Pediatr. 2013, 16, 530–535.e1. [Google Scholar] [CrossRef]
- Varannai, O.; Gede, N.; Juhász, M.F.; Szakács, Z.; Dembrovszky, F.; Németh, D.; Hegyi, P.; Párniczky, A. Therapeutic Approach of Chronic Pseudomonas Infection in Cystic Fibrosis-A Network Meta-Analysis. Antibiotics 2021, 10, 936. [Google Scholar] [CrossRef]
- Bass, R.; Brownell, J.N.; Stallings, V.A. The Impact of Highly Effective CFTR Modulators on Growth and Nutrition Status. Nutrients 2021, 13, 2907. [Google Scholar] [CrossRef]
- Terlizzi, V.; Colangelo, C.; Marsicovetere, G.; D’Andria, M.; Francalanci, M.; Innocenti, D.; Masi, E.; Avarello, A.; Taccetti, G.; Amato, F.; et al. Effectiveness of Elexacaftor/Tezacaftor/Ivacaftor Therapy in Three Subjects with the Cystic Fibrosis Genotype Phe508del/Unknown and Advanced Lung Disease. Genes 2021, 12, 1178. [Google Scholar] [CrossRef]
- Duckers, J.; Lesher, B.; Thorat, T.; Lucas, E.; McGarry, L.J.; Chandarana, K.; De Iorio, F. Real-World Outcomes of Ivacaftor Treatment in People with Cystic Fibrosis: A Systematic Review. J. Clin. Med. 2021, 10, 1527. [Google Scholar] [CrossRef] [PubMed]
- Munce, D.; Lim, M.; Akong, K. Persistent recovery of pancreatic function in patients with cystic fibrosis after ivacaftor. Pediatr. Pulmonol. 2020, 55, 3381–3383. [Google Scholar] [CrossRef]
- Corvol, H.; Mésinèle, J.; Douksieh, I.H.; Strug, L.J.; Boëlle, P.Y.; Guillot, L. SLC26A9 Gene Is Associated with Lung Function Response to Ivacaftor in Patients with Cystic Fibrosis. Front. Pharmacol. 2018, 9, 828. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.; Harris, W.T.; Rowe, S.M.; Rutland, S.B.; Oates, G.R. Tobacco smoke exposure limits the therapeutic benefit of tezacaftor/ivacaftor in pediatric patients with cystic fibrosis. J. Cyst. Fibros. 2021, 20, 612–661. [Google Scholar] [CrossRef]
- Stanton, B.A.; Coutermarsh, B.; Barnaby, R.; Hogan, D. Pseudomonas aeruginosa Reduces VX-809 Stimulated F508del-CFTR Chloride Secretion by Airway Epithelial Cells. PLoS ONE 2015, 10, e0127742. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]

| Method CGA/ GWAS | Gene | CF Lung Disease | P. aeruginosa Infection | MI | PI | CFRD | CFLD | Study/Author References |
|---|---|---|---|---|---|---|---|---|
| CGA | TGFβ1 | + | - | - | - | + | [8,9,10,11,12,13,14,15] | |
| CGA | IL8 | + | + | [16,17] | ||||
| CGA | IL1B | + | + | [18,19] | ||||
| CGA | IL10 | + | - | [17,20,21,22] | ||||
| CGA | TNFα | + | + | [23,24,25,26] | ||||
| CGA | 8.1 AH | + | + | [27,28,29] | ||||
| GWAS | HLA II | + | + | [30,31] | ||||
| CGA | SERPINA1 | + | + | [2,8,9,32,33,34,35] | ||||
| CGA | AGER | + | [8,36] | |||||
| CGA | MIF | + | + | [37,38] | ||||
| CGA /GWAS | MUC5B MUC4 MUC 20 | + | [39,40,41] | |||||
| CGA | MBL2 | + | + | + | [2,42,43] | |||
| CGA | CD14 | + | [44,45,46,47,48] | |||||
| CGA | NOS | + | - | [49,50,51,52] | ||||
| GA | ADRB2 | + | + | + | [53,54,55] | |||
| CGA | GR | + | [56] | |||||
| CGA | EnaC SCNN1B, SCNN1G TNFRSF1A | + | [57,58,59,60] | |||||
| CGA | KRT8 KRT19 | + | [61] | |||||
| GWAS | ADIPOR2 | + | [7,62,63] | |||||
| GWAS | SLCA4 | + | [60,62] | |||||
| GWAS | MSRA | + | [60,62,64,65,66] | |||||
| GWAS | SLC26A9 | + | + | [64,67,68,69] | ||||
| CGA/ GWAS | SLC9A3 | + | + | + | [41,60] | |||
| GWAS | SLC6A14 | + | + | + | [2,41,69,70,71] | |||
| CGA | EPHX1 GPNMB DSP | + | [7,72] | |||||
| CGA/ GWAS | TCF7L2 | + | [64,68,73,74,75,76] | |||||
| CGA/ GWAS | IGF2BP2 CDKN2A/B CDKAL1 | + | [64] | |||||
| GWAS | EHF | + | [77,78] | |||||
| GWAS | ATP12A PRSS1 | + | [71,73] |
| Class of Mutation | Class I | Class II | Class III | Class IV | Class V | Class VI |
|---|---|---|---|---|---|---|
| Severity | Severe | Severe | Severe | Mild | Mild | Mild |
| Type | Nonsense/ Frame-shift | Missense; amino acid deletion | Missense | Missense | Missense splicing defect | Missense |
| Frequent mutation | G542X, R553X, R1162X, W1282X | G85E, I507del, F508del, N1303K | S549R, G551D, G1349D | R117H, R347P, R334W, R1070W | A455E 3272-26A > G | 4326del TC, Gln1412X, 4279insA |
| CFTR defect | No CFTR synthesis | CFTR trafficking and processing defect | Abnormal channel function, block in regulation; defecting gaiting regulation | Abnormal channel function, decreased conductance | Reduced synthesis of CFTR protein | Decreased protein stability |
| Potential therapy | Read-throug agents (Ataluren, amynoglicosydes) | Correctors (+Potentiators) Lumacaftor (+Ivacaftor) | Potentiators (Ivacaftor) | Potentiators (Ivacaftor) | Splicing modulators amplifiers | Stabilizers HGF (hepatocyte growth factor) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butnariu, L.I.; Țarcă, E.; Cojocaru, E.; Rusu, C.; Moisă, Ș.M.; Leon Constantin, M.-M.; Gorduza, E.V.; Trandafir, L.M. Genetic Modifying Factors of Cystic Fibrosis Phenotype: A Challenge for Modern Medicine. J. Clin. Med. 2021, 10, 5821. https://doi.org/10.3390/jcm10245821
Butnariu LI, Țarcă E, Cojocaru E, Rusu C, Moisă ȘM, Leon Constantin M-M, Gorduza EV, Trandafir LM. Genetic Modifying Factors of Cystic Fibrosis Phenotype: A Challenge for Modern Medicine. Journal of Clinical Medicine. 2021; 10(24):5821. https://doi.org/10.3390/jcm10245821
Chicago/Turabian StyleButnariu, Lăcrămioara Ionela, Elena Țarcă, Elena Cojocaru, Cristina Rusu, Ștefana Maria Moisă, Maria-Magdalena Leon Constantin, Eusebiu Vlad Gorduza, and Laura Mihaela Trandafir. 2021. "Genetic Modifying Factors of Cystic Fibrosis Phenotype: A Challenge for Modern Medicine" Journal of Clinical Medicine 10, no. 24: 5821. https://doi.org/10.3390/jcm10245821
APA StyleButnariu, L. I., Țarcă, E., Cojocaru, E., Rusu, C., Moisă, Ș. M., Leon Constantin, M.-M., Gorduza, E. V., & Trandafir, L. M. (2021). Genetic Modifying Factors of Cystic Fibrosis Phenotype: A Challenge for Modern Medicine. Journal of Clinical Medicine, 10(24), 5821. https://doi.org/10.3390/jcm10245821







