Putative Biomarkers for Acute Pulmonary Embolism in Exhaled Breath Condensate
Abstract
:1. Introduction
2. Materials and Methods
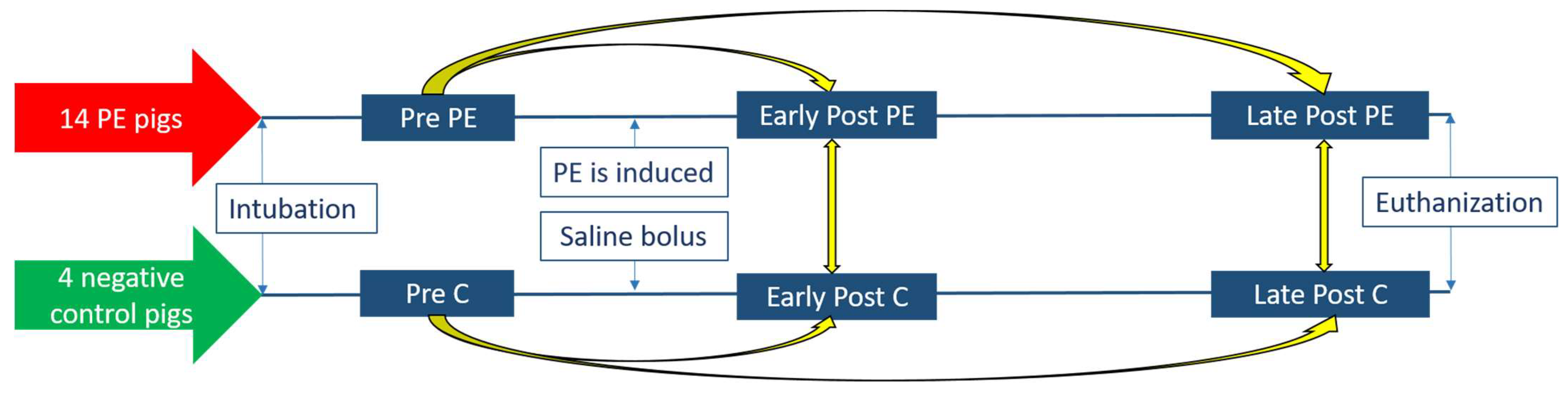
2.1. Study Design
2.2. Research Animals
2.3. Collection and Storage of the Exhaled Breath Condensate
2.4. Preparation of EBC Samples for Mass Spectrometry
2.5. Nano Liquid Chromatography–Tandem Mass Spectrometry (nLC-MS/MS)
2.6. Identification and Label-Free Quantification of the Proteins
2.7. Differential Expression Analysis for Discovery of Putative Markers
2.8. Bioinformatic Analysis
2.9. Adjusted Differential Expression Analysis and Prediction Models
2.10. Statistics Concerning Clinical Parameters and Protein Concentrations
3. Results
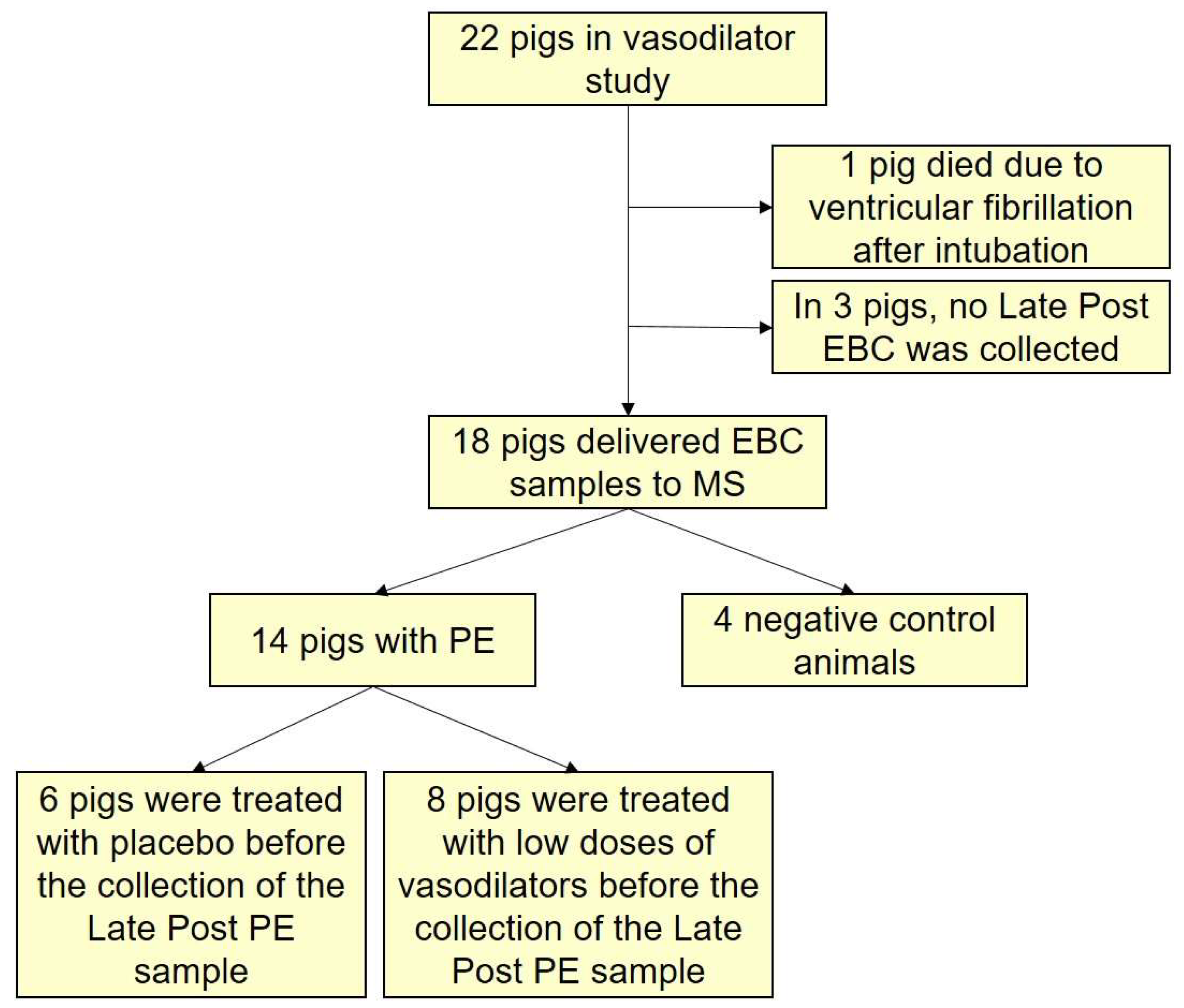
3.1. The Research Animals
3.2. The Proteins in the EBC
3.3. Discovery Based Differential Expression Analysis and Bioinformatics
3.4. Sensitivity Analysis
3.5. Analyses Adjusted for Multiple Comparisons and Prediction Models
4. Discussion
4.1. Study Limitations
4.2. Perspectives
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Konstantinides, S.V.; Meyer, G.; Becattini, C.; Bueno, H.; Geersing, G.J.; Harjola, V.P.; Huisman, M.V.; Humbert, M.; Jennings, C.S.; Jiménez, D.; et al. 2019 ESC Guidelines for the diagnosis and management of acute pulmonary embolism developed in collaboration with the European Respiratory Society (ERS): The Task Force for the diagnosis and management of acute pulmonary embolism of the European Society of Cardiology (ESC). Eur. Heart J. 2020, 41, 543–603. [Google Scholar] [CrossRef]
- Konstantinides, S.V.; Torbicki, A.; Agnelli, G.; Danchin, N.; Fitzmaurice, D.; Galie, N.; Gibbs, J.S.; Huisman, M.V.; Humbert, M.; Kucher, N.; et al. 2014 ESC guidelines on the diagnosis and management of acute pulmonary embolism. Eur. Heart J. 2014, 35, 3033–3069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Lyon, A.; Saggar, R.; Heaney, L.M.; Aizawa, K.; Cittadini, A.; Mauro, C.; Citro, R.; Limongelli, G.; Ferrara, F.; et al. Editor’s Choice-Biomarkers of acute cardiovascular and pulmonary diseases. Eur. Hear. J. Acute Cardiovasc. Care 2016, 5, 416–433. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Hunt, J.; Barnes, P.J.; Alving, K.; Antczak, A.; Baraldi, E.; Becher, G.; van Beurden, W.J.; Corradi, M.; Dekhuijzen, R.; et al. Exhaled breath condensate: Methodological recommendations and unresolved questions. Eur. Respir. J. 2005, 26, 523–548. [Google Scholar] [CrossRef] [Green Version]
- Beck, O.; Olin, A.C.; Mirgorodskaya, E. Potential of mass spectrometry in developing clinical laboratory biomarkers of nonvolatiles in exhaled breath. Clin. Chem. 2016, 62, 84–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beale, D.J.; Jones, O.A.H.; Karpe, A.V.; Dayalan, S.; Oh, D.Y.; Kouremenos, K.A.; Ahmed, W.; Palombo, E.A. A review of analytical techniques and their application in disease diagnosis in breathomics and salivaomics research. Int. J. Mol. Sci. 2016, 18, 24. [Google Scholar] [CrossRef] [Green Version]
- Harshman, S.; Grigsby, C.; Ott, D. Exhaled breath condensate for proteomic biomarker discovery. Chromatography 2014, 1, 108–119. [Google Scholar] [CrossRef] [Green Version]
- Fumagalli, M.; Ferrari, F.; Luisetti, M.; Stolk, J.; Hiemstra, P.S.; Capuano, D.; Viglio, S.; Fregonese, L.; Cerveri, I.; Corana, F.; et al. Profiling the proteome of exhaled breath condensate in healthy smokers and COPD patients by LC-MS/MS. Int. J. Mol. Sci. 2012, 13, 13894–13910. [Google Scholar] [CrossRef] [Green Version]
- Gade, I.L.; Schultz, J.G.; Cehofski, L.J.; Kjærgaard, B.; Severinsen, M.T.; Rasmussen, B.S.; Vorum, H.; Honoré, B.; Kristensen, S.R. Exhaled breath condensate in acute pulmonary embolism; a porcine study of effect of condensing temperature and feasibility of protein analysis by mass spectrometry. J. Breath Res. 2021, 15, 26005. [Google Scholar] [CrossRef]
- Lacombe, M.; Marie-Desvergne, C.; Combes, F.; Kraut, A.; Bruley, C.; Vandenbrouck, Y.; Mossuz, V.C.; Coute, Y.; Brun, V. Proteomic characterization of human exhaled breath condensate. J. Breath Res. 2017, 12, 021001. [Google Scholar] [CrossRef]
- Schultz, J.; Andersen, A.; Gade, I.L.; Kjaergaard, B.; Nielsen-Kudsk, J.E. Riociguat, sildenafil and inhaled nitric oxide reduces pulmonary vascular resistance and improves right ventricular function in a porcine model of acute pulmonary embolism. Eur. Hear. J. Acute Cardiovasc. Care 2019, 9, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Gaugaz, F.Z. Fast and sensitive total protein and peptide assays for proteomic analysis. Anal. Chem. 2015, 87, 4110–4116. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Project for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Gatto, L.; Lilley, K.S. Msnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 2012, 28, 288–289. [Google Scholar] [CrossRef] [Green Version]
- Gatto, L.; Gibb, S.; Rainer, J. MSnbase, efficient and elegant R-based processing and visualization of raw mass spectrometry data. J. Proteome Res. 2020, 20, 1063–1069. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregori, J.; Sanchez, A.; Villanueva, J. msmsTests: LC-MS/MS Differential Expression Tests, R Package Version 1.18.0; 2013. Available online: https://bioconductor.org/packages/release/bioc/html/msmsTests.html (accessed on 10 September 2021).
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Crofford, O.B.; Mallard, R.E.; Winton, R.E.; Rogers, N.L.; Jackson, J.C.; Keller, U. Acetone in breath and blood. Trans. Am. Clin. Climatol. Assoc. 1977, 88, 128–139. [Google Scholar]
- Fens, N.; Douma, R.A.; Sterk, P.J.; Kamphuisen, P.W. Breathomics as a diagnostic tool for pulmonary embolism. Am. J. Respir. Crit. Care Med. 2011, 183, A2678. [Google Scholar]
- Jr, F.S.C.; Dweik, R.A. Cardiovascular biomarkers in exhaled breath. Prog. Cardiovasc. Dis. 2012, 55, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Muccilli, V.; Saletti, R.; Cunsolo, V.; Ho, J.; Gili, E.; Conte, E.; Sichili, S.; Vancheri, C.; Foti, S. Protein profile of exhaled breath condensate determined by high resolution mass spectrometry. J. Pharm. Biomed. Anal. 2015, 105, 134–149. [Google Scholar] [CrossRef]
- Reinhold, P.; Knobloch, H. Exhaled breath condensate: Lessons learned from veterinary medicine. J. Breath Res. 2010, 4, 017001. [Google Scholar] [CrossRef] [PubMed]
- Czebe, K.; Barta, I.; Antus, B.; Valyon, M.; Horváth, I.; Kullmann, T. Influence of condensing equipment and temperature on exhaled breath condensate pH, total protein and leukotriene concentrations. Respir. Med. 2008, 102, 720–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garey, K.W.; Neuhauser, M.M.; Robbins, R.A.; Danziger, L.H.; Rubinstein, I. Markers of inflammation in exhaled breath condensate of young healthy smokers. Chest 2004, 125, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Kroegel, C.; Reissig, A. Principle mechanisms underlying venous thromboembolism: Epidemiology, risk factors, pathophysiology and pathogenesis. Respiration 2003, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, S.R. Mechanisms of cell damage and enzyme release. Dan. Med. Bull. 1994, 41, 423–433. [Google Scholar]
- Memon, A.A.; Sundquist, K.; PirouziFard, M.; Elf, J.L.; Strandberg, K.; Svensson, P.J.; Sundquist, J.; Zöller, B. Identification of novel diagnostic biomarkers for deep venous thrombosis. Br. J. Haematol. 2018, 181, 378–385. [Google Scholar] [CrossRef]
- Insenser, M.; Montes-Nieto, R.; Martínez-García, M.Á.; Durań, E.F.; Santiuste, C.; Goḿez, V.; Kline, J.A.; Escobar-Morreale, H.F.; Jimeńez, D. Identification of reduced circulating haptoglobin concentration as a biomarker of the severity of pulmonary embolism: A nontargeted proteomic study. PLoS ONE 2014, 9, e100902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Zur Mühlen, C.; Koeck, T.; Schiffer, E.; Sackmann, C.; Zürbig, P.; Hilgendorf, I.; Reinöhl, J.; Rivera, J.; Zirlik, A.; Hehrlein, C.; et al. Urine proteome analysis as a discovery tool in patients with deep vein thrombosis and pulmonary embolism. PROTEOMICS Clin. Appl. 2016, 10, 574–584. [Google Scholar] [CrossRef]
- Muñoz, R.; Santamaría, E.; Rubio, I.; Ausín, K.; Ostolaza, A.; Labarga, A.; Roldán, M.; Zandio, B.; Mayor, S.; Bermejo, R.; et al. Mass spectrometry-based proteomic profiling of thrombotic material obtained by endovascular thrombectomy in patients with ischemic stroke. Int. J. Mol. Sci. 2018, 19, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moxon, J.V.; Padula, M.P.; Clancy, P.; Emeto, T.I.; Herbert, B.R.; Norman, P.E.; Golledge, J. Proteomic analysis of intra-arterial thrombus secretions reveals a negative association of clusterin and thrombospondin-1 with abdominal aortic aneurysm. Atherosclerosis 2011, 219, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Eagan, T.M.; Damås, J.K.; Ueland, T.; Voll-Aanerud, M.; Mollnes, T.E.; Hardie, J.A.; Bakke, P.S.; Aukrust, P. Neutrophil gelatinase-associated lipocalin: A biomarker in COPD. Chest 2010, 138, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, G.; Li, J.; Zeng, Y.; Wu, Y.; Yan, X. Neutrophil gelatinase-associated lipocalin predicts myocardial dysfunction and mortality in severe sepsis and septic shock. Int. J. Cardiol. 2017, 227, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Barnes, P.J.; Loukides, S.; Sterk, P.J.; Hogman, M.; Olin, A.C.; Amann, A.; Antus, B.; Baraldi, E.; Bikov, A.; et al. A European Respiratory Society technical standard: Exhaled biomarkers in lung disease. Eur. Respir. J. 2017, 49, 1600965. [Google Scholar] [CrossRef] [Green Version]



| PE Animals (n = 14) | Negative Controls (n = 4) | |||||
|---|---|---|---|---|---|---|
| Before PE (Baseline) | Early Post PE | p-Value | Baseline C | Early Post C | p-Value | |
| Heart rate (beats/min) | 62.3 ± 2.9 | 70.9 ± 5.0 | 0.09 | 59.3±5.4 | 57.5±5.2 | 0.57 |
| Pulmonary vascular resistance (dyn·s/cm5) | 99.1 ± 4.6 | 332.8 ± 18.1 | <0.01 | 139.7 ± 36.1 | 164.4 ± 42.8 | 0.06 |
| Mean pulmonary arterial pressure (mmHg) | 18.3 ± 0.5 | 35.0 ± 1.1 | <0.01 | 20.0 ± 2.4 | 21.8 ± 1.5 | 0.19 |
| Mean Arterial blood pressure (mmHg) | 82.9 ± 3.0 | 84.3 ± 4.0 | 0.49 | 79.8 ± 7.4 | 75.8 ± 5.5 | 0.32 |
| Cardiac Output (L/min) | 5.67 ± 0.22 | 6.26 ± 0.24 | <0.01 | 5.23 ± 0.12 | 5.48 ± 0.13 | 0.37 |
| RV/LV end systolic | 0.64 ± 0.04 | 1.29 ± 0.05 | <0.01 | 0.63 ± 0.03 | 0.61 ± 0.02 | 0.73 |
| End tidal-CO2 (kPa) | 5.1 ± 0.1 | 4.9 ± 0.1 | 0.06 | 5.0 ± 0.1 | 5.0 ± 0.3 | 0.77 |
| pH | 7.50 ± 0.01 | 7.23 ± 0.21 | 0.23 | 7.54 ± 0.01 | 7.55 ± 0.02 | 0.65 |
| PaCO2 (kPa) | 4.58 ± 0.07 | 5.40 ± 0.17 | <0.01 | 4.55 ± 0.15 | 4.68 ± 0.12 | 0.57 |
| PaO2 (kPa) | 16.60 ± 0.80 | 12.82 ± 0.84 | <0.01 | 18.05 ± 0.32 | 17.05 ± 0.34 | 0.06 |
| Rectal temperature | 38.7 ± 0.1 | 39.0 ± 0.2 | 0.08 | 39.3 ± 0.5 | 39.2 ± 0.3 | 0.8 |
| Weight (kg) | 57.3 ± 0.9 | - | - | 56.9 ± 0.4 | - | - |
| Majority Protein IDs | Protein Name | Gene Name | Early Post PE vs. Pre PE | Late Post PE vs. Pre PE | Early Post PE vs. Early Post C | |||
|---|---|---|---|---|---|---|---|---|
| Positive Fold Changes (i.e., Higher Amount after PE) | Log Fold Change | p-Value | Log Fold Change | p-Value | Log Fold Change | p-Value | ||
| P01876 | Immunoglobulin heavy constant alpha 1 | IGHA1 | 4.58 | <0.001 | 4.03 | <0.001 | ||
| P31025 | Lipocalin-1 | LCN1 | 2.51 | 0.005 | ||||
| P61626 | Lysozyme C | LYZ | 3.29 | 0.019 | ||||
| Negative fold changes (ie lower amount after PE) | ||||||||
| P08835 | Albumin | ALB | −4.54 | 0.0134 | ||||
| P55072 | Transitional endoplasmic reticulum ATPase | VCP | −1.82 | 0.025 | ||||
| P19971 | Thymidine phosphorylase | TYMP | −2.76 | 0.003 | ||||
| Q2XVP4 | Tubulin alpha-1B chain | TUBA1B | −2.71 | 0.003 | −2.70 | 0.007 | ||
| A8K2U0 | Alpha-2-macroglobulin-like protein 1 | A2ML1 | −2.05 | 0.003 | ||||
| P58107 | Epiplakin | EPPK1 | −2.59 | 0.003 | ||||
| P48594 | Serpin B4 | SERPINB4 | −1.90 | 0.003 | ||||
| P68371 | Tubulin beta-4B chain | TUBB4B | −1.98 | 0.013 | ||||
| P35579 | Myosin-9 | MYH9 | −2.34 | 0.002 | ||||
| Q5VTE0 | Putative elongation factor 1-alpha-like 3 | EEF1A1P5 | −1.85 | 0.019 | ||||
| P13639 | Elongation factor 2 | EEF2 | −2.05 | 0.010 | ||||
| P01857 | Ig gamma-1 chain C region | IGHG1 | −2.06 | 0.006 | −2.30 | 0.019 | ||
| P47929 | Galectin-7 | LGALS7 | −1.98 | 0.004 | ||||
| P06733 | Alpha-enolase | ENO1 | −1.22 | 0.032 | ||||
| P60174 | Triosephosphate isomerase | TPI1 | −1.81 | 0.002 | ||||
| P00558 | Phosphoglycerate kinase 1 | PGK1 | −1.70 | 0.016 | ||||
| P05387 | 60S acidic ribosomal protein P2 | RPLP2 | −1.24 | 0.019 | ||||
| P06702 | Protein S100-A9 | S100A9 | −2.16 | 0.032 | ||||
| P14618 | Pyruvat kinase | PKM | −1.67 | 0.032 | ||||
| P80188 | Neutrophil gelatinase-associated lipocalin | LCN2 | −1.33 | 0.035 | −2.10 | 0.003 | ||
| P06576 | ATP synthase subunit beta, mitochondrial | ATP5B | −1.78 | 0.035 | ||||
| P01024 | Complement C3 | C3 | −1.27 | 0.048 | ||||
| P09211 | Glutathione S-transferase P | GSTP1 | −1.19 | 0.062 | ||||
| Q09666 | Neuroblast differentiation-associated protein AHNAK | AHNAK | −1.59 | 0.062 | ||||
| P29508 | Serpin B3 | SERPINB3 | −1.15 | 0.063 | ||||
| P31151 | Protein S100-A7 | S100A7 | −1.16 | 0.077 | ||||
| O02705 | Heat shock protein HSP 90-alpha | HSP90AA1 | −1.20 | 0.085 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gade, I.L.; Schultz, J.G.; Brøndum, R.F.; Kjærgaard, B.; Nielsen-Kudsk, J.E.; Andersen, A.; Kristensen, S.R.; Honoré, B. Putative Biomarkers for Acute Pulmonary Embolism in Exhaled Breath Condensate. J. Clin. Med. 2021, 10, 5165. https://doi.org/10.3390/jcm10215165
Gade IL, Schultz JG, Brøndum RF, Kjærgaard B, Nielsen-Kudsk JE, Andersen A, Kristensen SR, Honoré B. Putative Biomarkers for Acute Pulmonary Embolism in Exhaled Breath Condensate. Journal of Clinical Medicine. 2021; 10(21):5165. https://doi.org/10.3390/jcm10215165
Chicago/Turabian StyleGade, Inger Lise, Jacob Gammelgaard Schultz, Rasmus Froberg Brøndum, Benedict Kjærgaard, Jens Erik Nielsen-Kudsk, Asger Andersen, Søren Risom Kristensen, and Bent Honoré. 2021. "Putative Biomarkers for Acute Pulmonary Embolism in Exhaled Breath Condensate" Journal of Clinical Medicine 10, no. 21: 5165. https://doi.org/10.3390/jcm10215165
APA StyleGade, I. L., Schultz, J. G., Brøndum, R. F., Kjærgaard, B., Nielsen-Kudsk, J. E., Andersen, A., Kristensen, S. R., & Honoré, B. (2021). Putative Biomarkers for Acute Pulmonary Embolism in Exhaled Breath Condensate. Journal of Clinical Medicine, 10(21), 5165. https://doi.org/10.3390/jcm10215165







