Potential Muscle-Related Biomarkers in Predicting Curve Progression to the Surgical Threshold in Adolescent Idiopathic Scoliosis—A Pilot Proteomic Study Comparing Four Non-Progressive vs. Four Progressive Patients vs. A Control Cohort
Abstract
1. Introduction
2. Materials and Methods
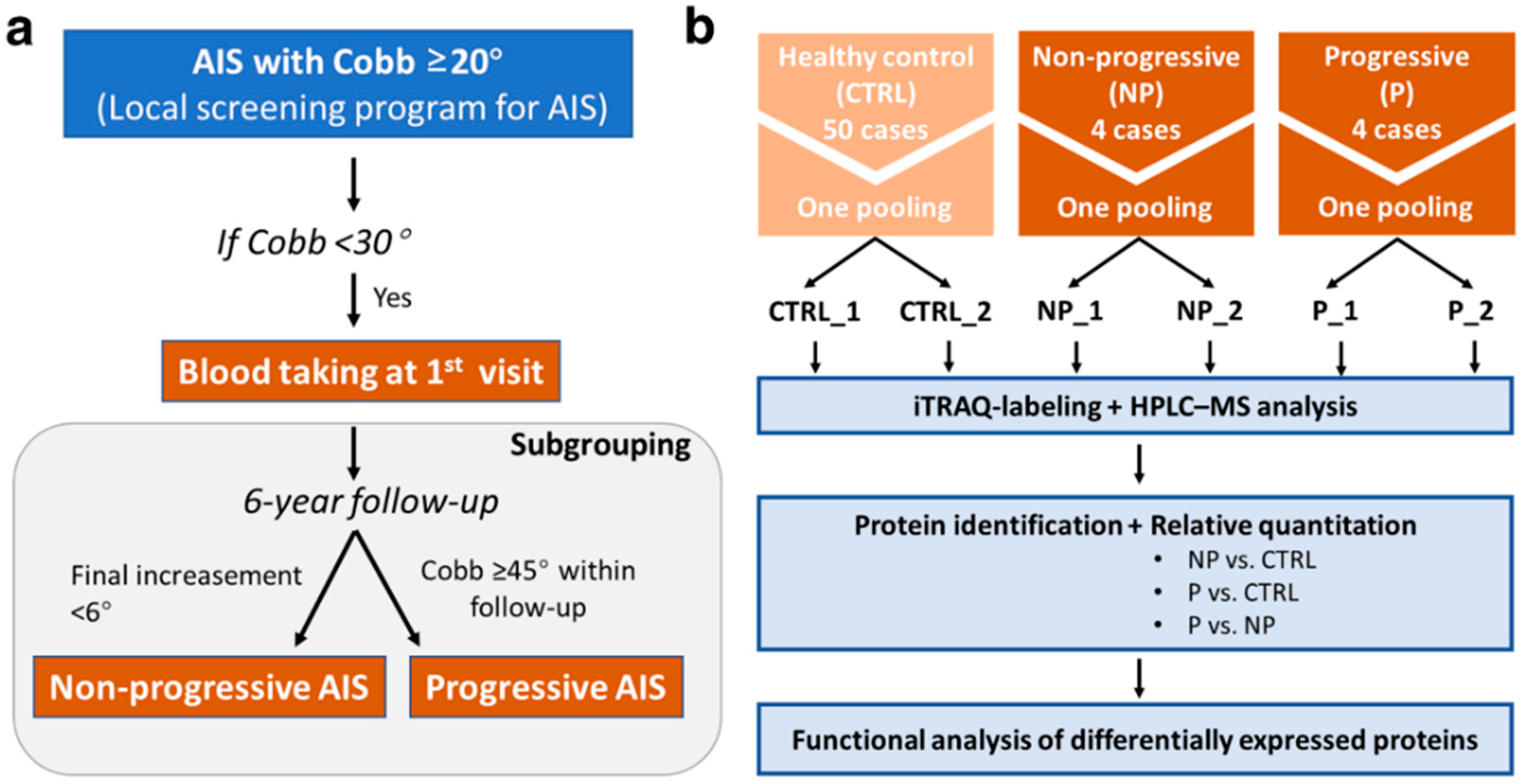
2.1. Subjects Recruitment and Blood Taking
2.2. Sample Preparation for iTRAQ-Based Proteomic Analysis
2.3. HPLC–MS and Bioinformatic Analysis
2.4. Statistical Analysis
3. Results
3.1. Subject Characteristics
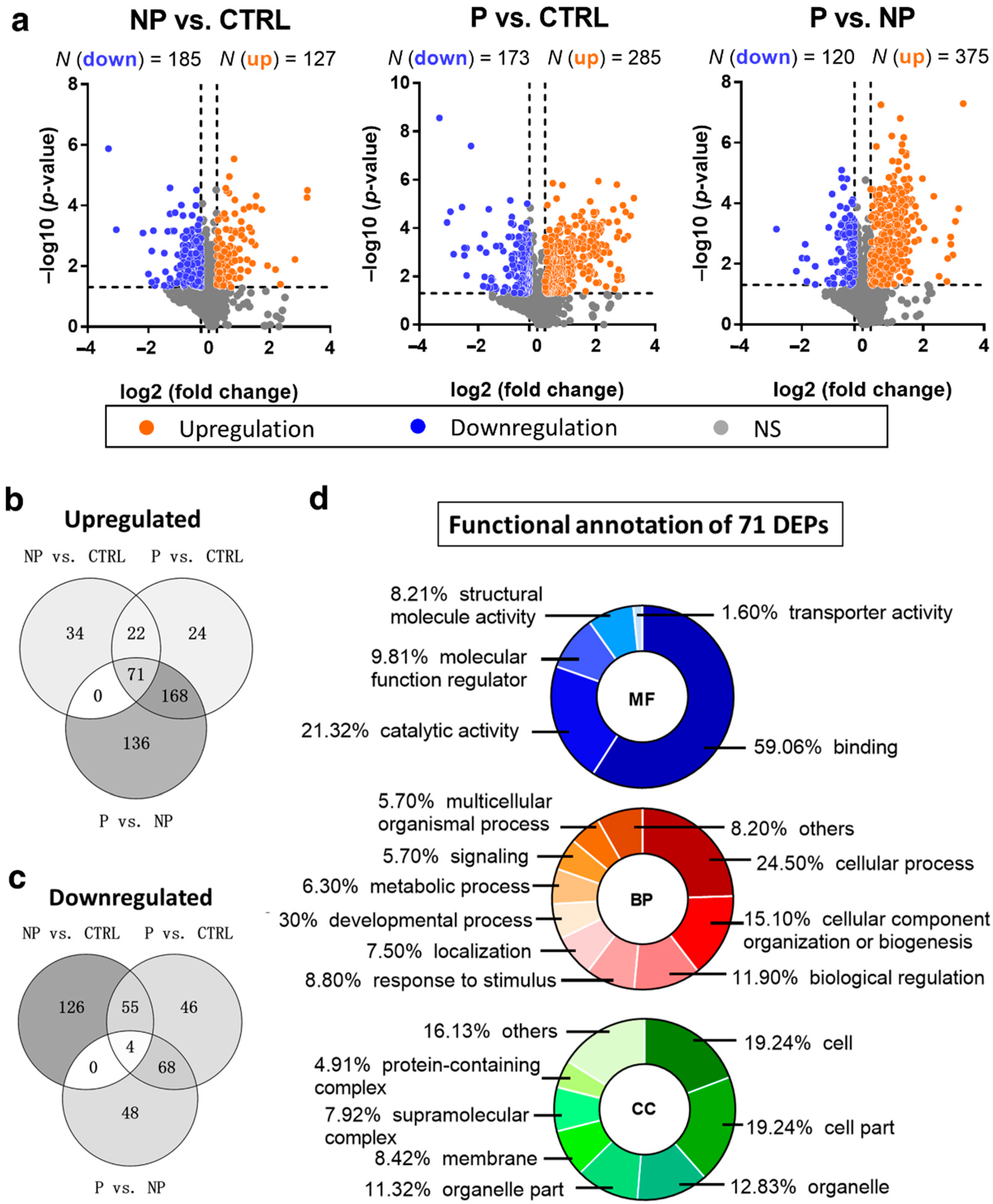
3.2. Up- and Downregulated DEPs in Pairwise Comparison Analysis
3.3. Gradually Changing DEPs Accompanying a Higher Risk of Scoliosis Progression
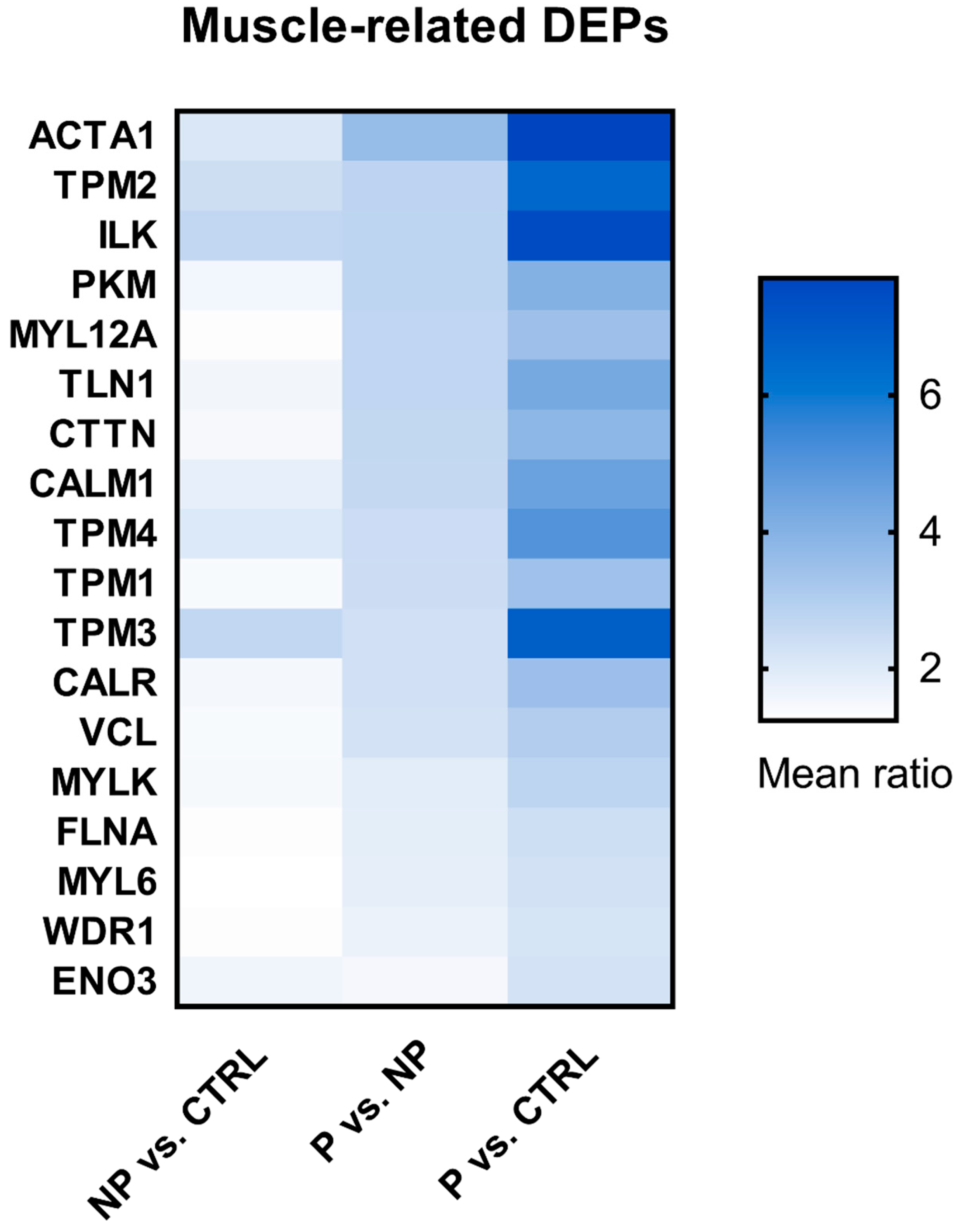
3.4. Shortlist of Muscle-Related DEPs
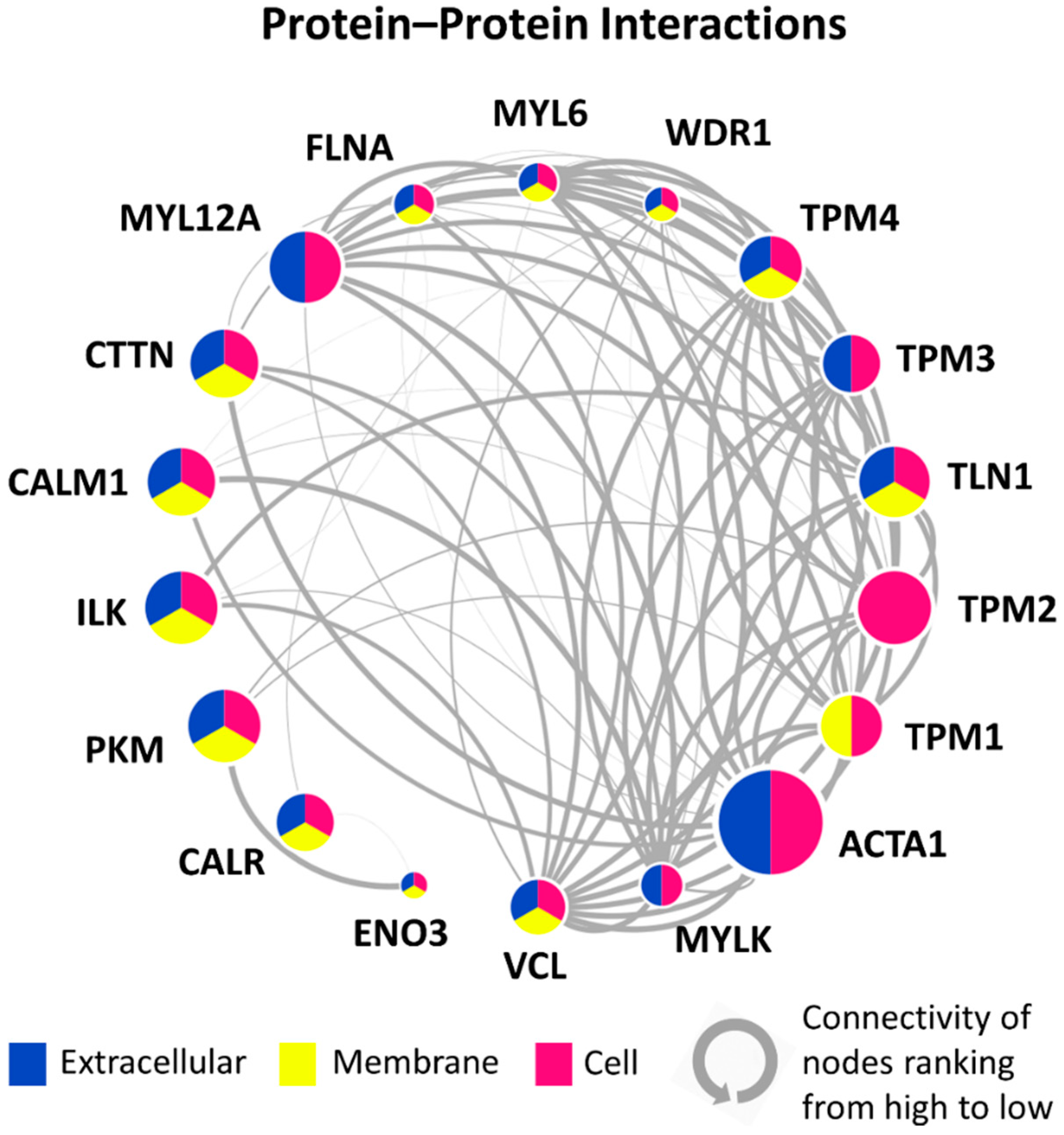
3.5. PPI Network of 18 Candidate DEPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weinstein, S.L.; Dolan, L.A.; Cheng, J.C.; Danielsson, A.; Morcuende, J.A. Adolescent idiopathic scoliosis. Nat. Rev. Dis. Primers 2015, 1, 15030. [Google Scholar] [CrossRef]
- Ueno, M.; Takaso, M.; Nakazawa, T.; Imura, T.; Saito, W.; Shintani, R.; Uchida, K.; Fukuda, M.; Takahashi, K.; Ohtori, S.; et al. A 5-year epidemiological study on the prevalence rate of idiopathic scoliosis in Tokyo: School screening of more than 250,000 children. J. Orthop. Sci. 2011, 16, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Luk, K.D.; Lee, C.F.; Cheung, K.M.; Cheng, J.C.; Ng, B.K.; Lam, T.P.; Mak, K.H.; Yip, P.S.; Fong, D.Y. Clinical Effectiveness of School Screening for Adolescent Idiopathic Scoliosis: A Large Population-Based Retrospective Cohort Study. Spine 2010, 35, 1607–1614. [Google Scholar] [CrossRef]
- Weinstein, S.L.; Dolan, L.A.; Wright, J.G.; Dobbs, M.B. Effects of Bracing in Adolescents with Idiopathic Scoliosis. N. Engl. J. Med. 2013, 369, 1512–1521. [Google Scholar] [CrossRef]
- Schiller, J.R.; Thakur, N.A.; Eberson, C.P. Brace Management in Adolescent Idiopathic Scoliosis. Clin. Orthop. Relat. Res. 2010, 468, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Llorens, J.; Ramirez, M.; Colomina, M.J.; Bago, J.; Molina, A.; Caceres, E.; Gea, J. Muscle dysfunction and exercise limitation in adolescent idiopathic scoliosis. Eur. Respir. J. 2010, 36, 393–400. [Google Scholar] [CrossRef]
- Yu, W.S.; Chan, K.Y.; Yu, F.W.P.; Yeung, H.Y.; Lee, K.M.; Ng, K.W.; Qiu, Y.; Lam, T.P.; Cheng, J.C.Y. Lower handgrip strength in girls with adolescent idiopathic scoliosis (AIS) a case-control study. Stud. Health Technol. Inf. 2012, 176, 175. [Google Scholar]
- Lin, M.C.; Liaw, M.Y.; Chen, W.J.; Cheng, P.T.; Wong, A.M.; Chiou, W.K. Pulmonary function and spinal characteristics: Their relationships in persons with idiopathic and postpoliomyelitic scoliosis. Arch. Phys. Med. Rehabil. 2001, 82, 335–341. [Google Scholar] [CrossRef]
- Bogduk NaM, J.E.; Pearcy, M.J. A universal model of the lumbar back muscles in the upright position. Spine 1992, 17, 897–913. [Google Scholar] [CrossRef]
- Wong, C. Mechanism of right thoracic adolescent idiopathic scoliosis at risk for progression; a unifying pathway of development by normal growth and imbalance. Scoliosis 2015, 10, 2. [Google Scholar] [CrossRef]
- Modi, H.N.; Suh, S.-W.; Yang, J.-H.; Hong, J.-Y.; Venkatesh, K.P.; Muzaffar, N. Spontaneous regression of curve in immature idiopathic scoliosis—Does spinal column play a role to balance? An observation with literature review. J. Orthop. Surg. Res. 2010, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Zapata, K.A.; Wang-Price, S.S.; Sucato, D.J.; Dempsey-Robertson, M. Ultrasonographic Measurements of Paraspinal Muscle Thickness in Adolescent Idiopathic Scoliosis: A Comparison and Reliability Study. Pediatr. Phys. Ther. 2015, 27, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Mannion, A.F.; Meier, M.; Grob, D.; Müntener, M. Paraspinal muscle fibre type alterations associated with scoliosis: An old problem revisited with new evidence. Eur. Spine J. 1998, 7, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Stetkarova, I.; Zamecnik, J.; Bocek, V.; Vasko, P.; Brabec, K.; Krbec, M. Electrophysiological and histological changes of paraspinal muscles in adolescent idiopathic scoliosis. Eur. Spine J. 2016, 25, 3146–3153. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Meng, Y.; Jin, X.; Zhang, C.; Zhao, J.; Wang, C.; Gao, R.; Zhou, X. Volumetric and Fatty Infiltration Imbalance of Deep Paravertebral Muscles in Adolescent Idiopathic Scoliosis. Med. Sci. Monit. 2017, 23, 2089–2095. [Google Scholar] [CrossRef]
- Chan, Y.L.; Cheng, J.C.Y.; Guo, X.; King, A.D.; Griffith, J.F.; Metreweli, C. MRI evaluation of multifidus muscles in adolescent idiopathic scoliosis. Pediatr. Radiol. 1999, 29, 360–363. [Google Scholar] [CrossRef]
- Jiang, H.; Meng, Y.; Jin, X.; Zhang, C.; Zhao, J.; Wang, C.; Gao, R.; Zhou, X. Fiber Type-Specific Morphological and Cellular Changes of Paraspinal Muscles in Patients with Severe Adolescent Idiopathic Scoliosis. Med. Sci. Monit. 2020, 26, e924415. [Google Scholar]
- Horgan, R.P.; Kenny, L.C. ‘Omic’ technologies: Genomics, transcriptomics, proteomics and metabolomics. Obstet. Gynaecol. 2011, 13, 189–195. [Google Scholar] [CrossRef]
- Sun, Z.-J.; Jia, H.-M.; Qiu, G.-X.; Zhou, C.; Guo, S.; Zhang, J.-G.; Shen, J.-X.; Zhao, Y.; Zou, Z.-M. Identification of candidate diagnostic biomarkers for adolescent idiopathic scoliosis using UPLC/QTOF-MS analysis: A first report of lipid metabolism profiles. Sci. Rep. 2016, 6, 22274. [Google Scholar] [CrossRef]
- Shen, N.; Chen, N.; Zhou, X.; Zhao, B.; Huang, R.; Liang, J.; Yang, X.; Chen, M.; Song, Y.; Du, Q. Alterations of the gut microbiome and plasma proteome in Chinese patients with adolescent idiopathic scoliosis. Bone 2019, 120, 364–370. [Google Scholar] [CrossRef]
- Makino, H.; Seki, S.; Kitajima, I.; Motomura, H.; Nogami, M.; Yahara, Y.; Ejiri, N.; Kimura, T. Differential proteome analysis in adolescent idiopathic scoliosis patients with thoracolumbar/lumbar curvatures. BMC Musculoskelet Disord. 2019, 20, 247. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, H.; Leung, R.K.; Choy, K.W.; Lam, T.P.; Ng, B.K.; Qiu, Y.; Feng, J.Q.; Cheng, J.C.; Lee, W.Y. Aberrant miR-145-5p/β-catenin signal impairs osteocyte function in adolescent idiopathic scoliosis. FASEB J. 2018, 32, 6537–6549. [Google Scholar] [CrossRef] [PubMed]
- Richards, B.S.; Bernstein, R.M.; D’Amato, C.R.; Thompson, G.H. Standardization of Criteria for Adolescent Idiopathic Scoliosis Brace Studies: SRS Committee on Bracing and Nonoperative Management. Spine 2005, 30, 2068–2075. [Google Scholar] [CrossRef] [PubMed]
- Takagi, Y.; Yasuhara, T.; Gomi, K. Creatine kinase and its isozymes. Rinsho Byori. 2001, 116, 52–61. [Google Scholar] [PubMed]
- iTRAQ Reagents—8plex Protocol. Available online: https://www.sciex.com/content/dam/SCIEX/pdf/brochures/mass-spectrometry-4375249C.pdf (accessed on 21 May 2021).
- Savitski, M.M.; Wilhelm, M.; Hahne, H.; Kuster, B.; Bantscheff, M. A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Mol. Cell Proteom. 2015, 14, 2394–2404. [Google Scholar] [CrossRef]
- Wen, B.; Zhou, R.; Feng, Q.; Wang, Q.; Wang, J.; Liu, S. IQuant: An automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 2014, 14, 2280–2285. [Google Scholar] [CrossRef]
- Oliveros, J.C. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 17 September 2021).
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Sadat-Ali, M.; Al-Habdan, I.; Al-Othman, A. Adolescent idiopathic scoliosis. Is low melatonin a cause? Jt. Bone Spine 2000, 67, 62–64. [Google Scholar]
- Fagan, A.B.; Kennaway, D.J.; Sutherland, A.D. Total 24-Hour Melatonin Secretion in Adolescent Idiopathic Scoliosis: A Case-Control Study. Spine 1998, 23, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Kindsfater, K.; Lowe, T.; Lawellin, D.; Weinstein, D.; Akmakjian, J. Levels of platelet calmodulin for the prediction of progression and severity of adolescent idiopathic scoliosis. JBJS 1994, 76, 1186–1192. [Google Scholar] [CrossRef]
- Lowe, T.G.; Burwell, R.G.; Dangerfield, P.H. Platelet calmodulin levels in adolescent idiopathic scoliosis (AIS): Can they predict curve progression and severity? Summary of an electronic focus group debate of the IBSE. Eur. Spine J. 2004, 13, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Noshchenko, A.; Hoffecker, L.; Lindley, E.M.; Burger, E.L.; Cain, C.M.; Patel, V.V.; Bradford, A.P. Predictors of spine deformity progression in adolescent idiopathic scoliosis: A systematic review with meta-analysis. World J. Orthop. 2015, 6, 537–558. [Google Scholar] [CrossRef]
- Zhang, J.; Cheuk, K.Y.; Xu, L.; Wang, Y.; Feng, Z.; Sit, T.; Cheng, K.-L.; Nepotchatykh, E.; Lam, T.-P.; Liu, Z.; et al. A validated composite model to predict risk of curve progression in adolescent idiopathic scoliosis. Eclinicalmedicine 2020, 18, 100236. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Amat, A.; Boulaiz, H.; Prados, J.; Marchal, J.A.; Puche, P.P.; Caba, O.; Rodríguez-Serrano, F.; Aránega, A. Release of alpha-actin into serum after skeletal muscle damage. Br. J. Sports Med. 2005, 39, 830–834. [Google Scholar] [CrossRef]
- Amat, A.M.; Corrales, J.A.M.; Serrano, F.R.; Boulaiz, H.; Salazar, J.C.P.; Contreras, F.H.; Perez, O.C.; Delgado, E.C.; Martín, I.; Jimenez, A.A. Role of alpha-actin in muscle damage of injured athletes in comparison with traditional markers. Br. J. Sports Med. 2007, 41, 442–446. [Google Scholar] [CrossRef]
- Tekus, E.; Vaczi, M.; Horvath-Szalai, Z.; Ludany, A.; Koszegi, T.; Wilhelm, M. Plasma Actin, Gelsolin and Orosomucoid Levels after Eccentric Exercise. J. Hum. Kinet. 2017, 56, 99–108. [Google Scholar] [CrossRef]
- Martínez-Amat, A.; Marchal, J.; Prados, J.; Hita, F.; Rodriguez-Serrano, F.; Boulaiz, H.; Martin, I.; Melguizo, C.; Caba, O.; Velez, C.; et al. Release of muscle α-actin into serum after intensive exercise. Biol. Sport 2010, 27, 263–268. [Google Scholar] [CrossRef]
- Kanungo, S.; Wells, K.; Tribett, T.; El-Gharbawy, A. Glycogen metabolism and glycogen storage disorders. Ann. Transl. Med. 2018, 6, 474. [Google Scholar] [CrossRef] [PubMed]
- Marttila, M.; Lehtokari, V.-L.; Marston, S.; Nyman, T.; Barnerias, C.; Beggs, A.; Bertini, E.; Ceyhan-Birsoy, Ö.; Cintas, P.; Gerard, M.; et al. Mutation update and genotype-phenotype correlations of novel and previously described mutations in TPM2 and TPM3 causing congenital myopathies. Hum. Mutat. 2014, 35, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.W.; Wang, G.; Baek, S.J.; Shen, R.-F. Comparative Study of Three Proteomic Quantitative Methods, DIGE, cICAT, and iTRAQ, Using 2D Gel- or LC−MALDI TOF/TOF. J. Proteome Res. 2006, 5, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Truebano, M.; Skibinski, D.O.F. The consequences of sample pooling in proteomics: An empirical study. Electrophoresis 2009, 30, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
| Groups | CTRL (N = 50) | NP (N = 4) | P (N = 4) | p-Value 1 |
|---|---|---|---|---|
| Age (years) | 13.1 ± 0.5 | 13.4 ± 0.5 | 12.6 ± 0.9 | 0.155 |
| Body Weight (kg) | 49.6 ± 9.8 | 42.2 ± 3.2 | 34.3 ± 3.8 * | 0.002 |
| Body Height (cm) | 157.4 ± 6.1 | 154.0 ± 10.6 | 145.8 ± 3.4 * | 0.014 |
| Arm Span (cm) | 156.1 ± 6.9 | 152.4 ± 12.2 | 146.9 ± 8.1 | 0.090 |
| BMI (kg/m2) | 20.23 ± 3.0 | 18.3 ± 2.3 | 15.9 ± 0.5 * | 0.005 |
| AIS Subgroups | Non-Progressive (NP) | Progressive (P) | |||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | p 1 | |
| Selection criteria of the subgroups | |||||
| Cobb at first visit (°) | 23.5 | 2.4 | 26.5 | 3 | 0.189 |
| (Individual: 21, 22, 25, 26) | (Individual: 23, 25, 29, 29) | ||||
| Change of Cobb angle at latest follow-up | Less than 6° | Final Cobb larger than 45° | / | ||
| (Individual: 24, 21, 20, 28) | (Individual: 57, 59, 49, 50) | ||||
| With bracing | No (0/4) | Yes (4/4) | / | ||
| Sexual and skeletal maturity | |||||
| Time since onset of menarche (years) | 1.6 | 0.8 | −0.9 | 0.4 | 0.021 |
| (range: 0.62~2.57) | (range: −1.38~−0.34) | ||||
| Breast stage | 3.3 | 0.5 | 2.0 | 0.8 | 0.044 |
| (range: 3~4) | (range: 1~3) | ||||
| Pubic hair | 3.0 | 0.0 | 1.8 | 0.5 | 0.011 |
| (range: 3~3) | (range: 1~2) | ||||
| Risser sign | 3.5 | 0.6 | 0.8 | 1.0 | 0.019 |
| (range: 3~4) | (range: 0~2) | ||||
| Body composition | |||||
| Skeletal muscle mass (kg) | 16.8 | 2.7 | 14.1 | 1.2 | 0.149 |
| Body fat mass (kg) | 9.9 | 2.1 | 7.1 | 2.0 | 0.149 |
| Fat-free mass (kg) | 32.0 | 4.7 | 27.5 | 2.2 | 0.149 |
| Right arm lean mass (kg) | 1.2 | 0.2 | 0.9 | 0.1 | 0.029 |
| Left arm lean mass (kg) | 1.2 | 0.2 | 0.9 | 0.1 | 0.043 |
| Trunk lean mass (kg) | 13.4 | 1.4 | 10.9 | 0.8 | 0.043 |
| Right leg lean mass (kg) | 4.9 | 1.2 | 3.8 | 0.3 | 0.083 |
| Left leg lean mass (kg) | 4.8 | 1.1 | 3.7 | 0.3 | 0.083 |
| Handgrip strength | |||||
| Dominant hand (kg) | 17.8 | 3.9 | 14.4 | 3.4 | 0.146 |
| Non-dominant hand (kg) | 18.4 | 5.3 | 13.8 | 3.6 | 0.149 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Chen, H.; Zhang, J.; Lam, T.-p.; Hung, A.L.H.; Cheng, J.C.Y.; Lee, W.Y.W. Potential Muscle-Related Biomarkers in Predicting Curve Progression to the Surgical Threshold in Adolescent Idiopathic Scoliosis—A Pilot Proteomic Study Comparing Four Non-Progressive vs. Four Progressive Patients vs. A Control Cohort. J. Clin. Med. 2021, 10, 4927. https://doi.org/10.3390/jcm10214927
Wang Y, Chen H, Zhang J, Lam T-p, Hung ALH, Cheng JCY, Lee WYW. Potential Muscle-Related Biomarkers in Predicting Curve Progression to the Surgical Threshold in Adolescent Idiopathic Scoliosis—A Pilot Proteomic Study Comparing Four Non-Progressive vs. Four Progressive Patients vs. A Control Cohort. Journal of Clinical Medicine. 2021; 10(21):4927. https://doi.org/10.3390/jcm10214927
Chicago/Turabian StyleWang, Yujia, Huanxiong Chen, Jiajun Zhang, Tsz-ping Lam, A.L.H. Hung, J.C.Y. Cheng, and W.Y.W. Lee. 2021. "Potential Muscle-Related Biomarkers in Predicting Curve Progression to the Surgical Threshold in Adolescent Idiopathic Scoliosis—A Pilot Proteomic Study Comparing Four Non-Progressive vs. Four Progressive Patients vs. A Control Cohort" Journal of Clinical Medicine 10, no. 21: 4927. https://doi.org/10.3390/jcm10214927
APA StyleWang, Y., Chen, H., Zhang, J., Lam, T.-p., Hung, A. L. H., Cheng, J. C. Y., & Lee, W. Y. W. (2021). Potential Muscle-Related Biomarkers in Predicting Curve Progression to the Surgical Threshold in Adolescent Idiopathic Scoliosis—A Pilot Proteomic Study Comparing Four Non-Progressive vs. Four Progressive Patients vs. A Control Cohort. Journal of Clinical Medicine, 10(21), 4927. https://doi.org/10.3390/jcm10214927






