Single-Nucleotide Polymorphisms in MICA and MICB Genes Could Play a Role in the Outcome in AML Patients after HSCT
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cohort Description
2.2. Sample Preparation and Sequencing
2.3. Sequence Evaluation
2.4. Statistical Analysis
3. Results
3.1. Clinical Data Evaluation
3.2. Distribution of Exon Groups and Polymorphisms within the Cohort
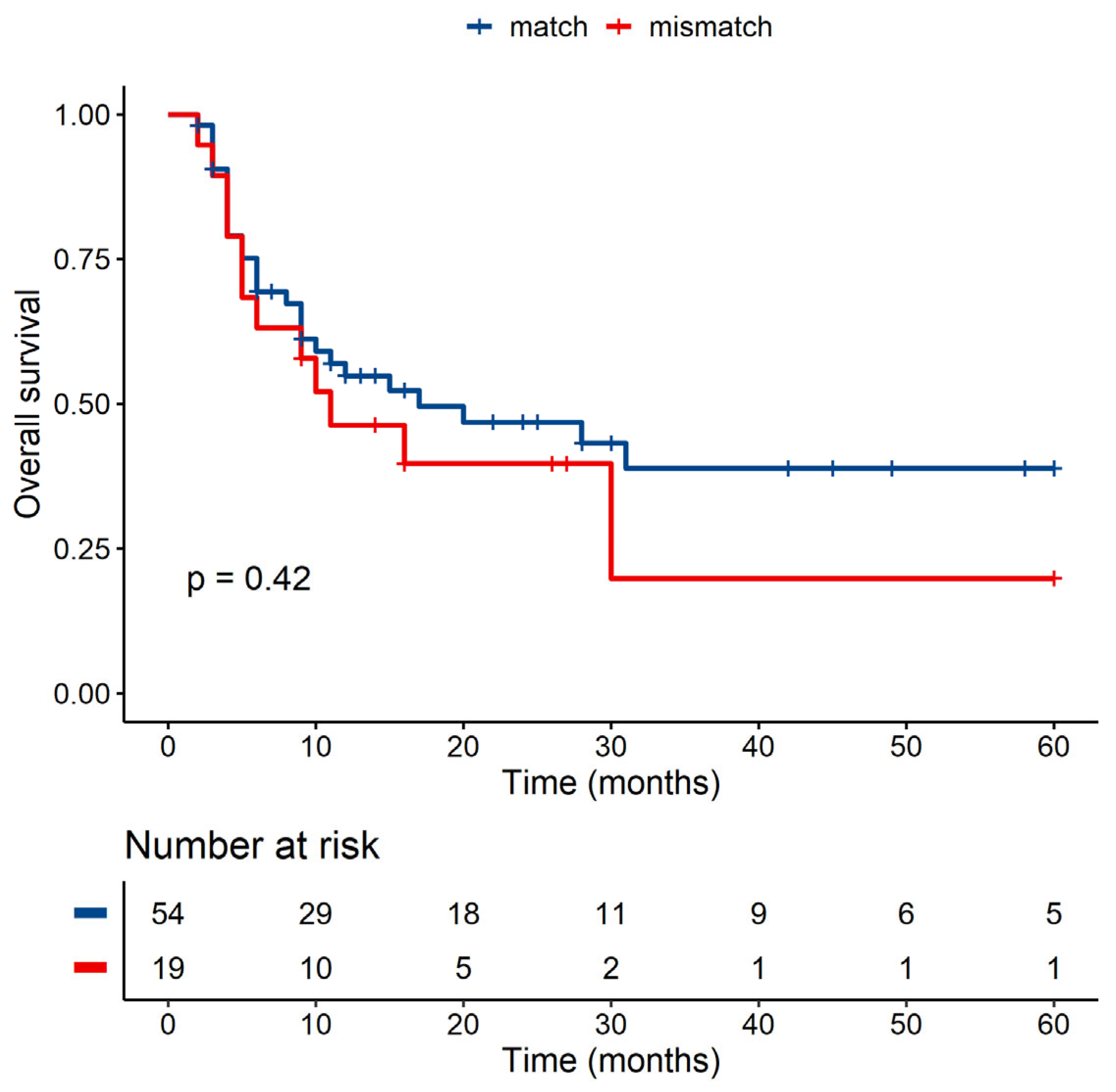
3.3. Association of Polymorphism with Clinical Data
3.3.1. Donor MICA Exon 2 Plays a Role in Overall Survival
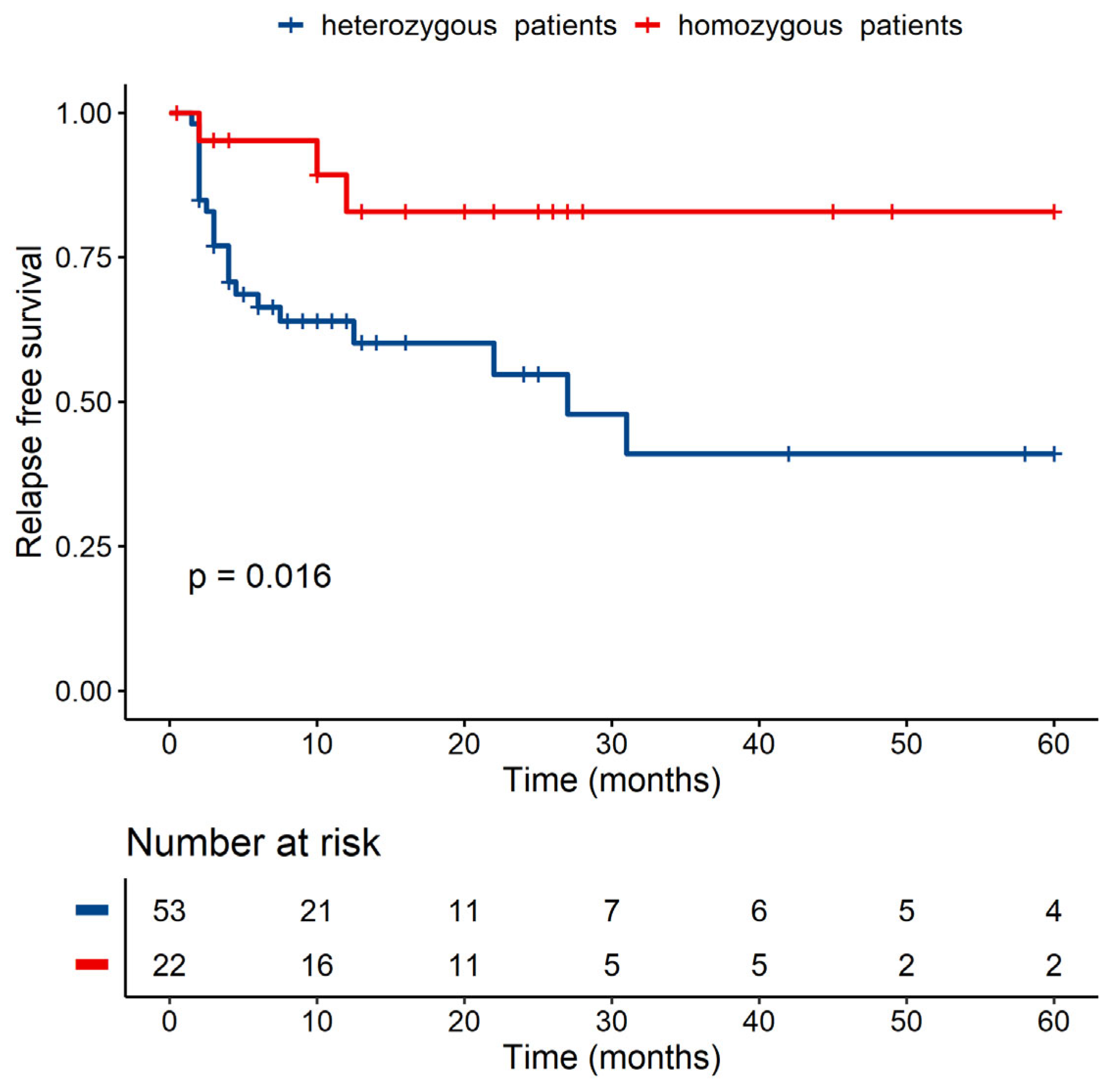
3.3.2. Patients’ Homozygosity within MICB Seems to Be Linked to a Lower Risk of Relapse in Univariate but Not in Multivariate Analysis
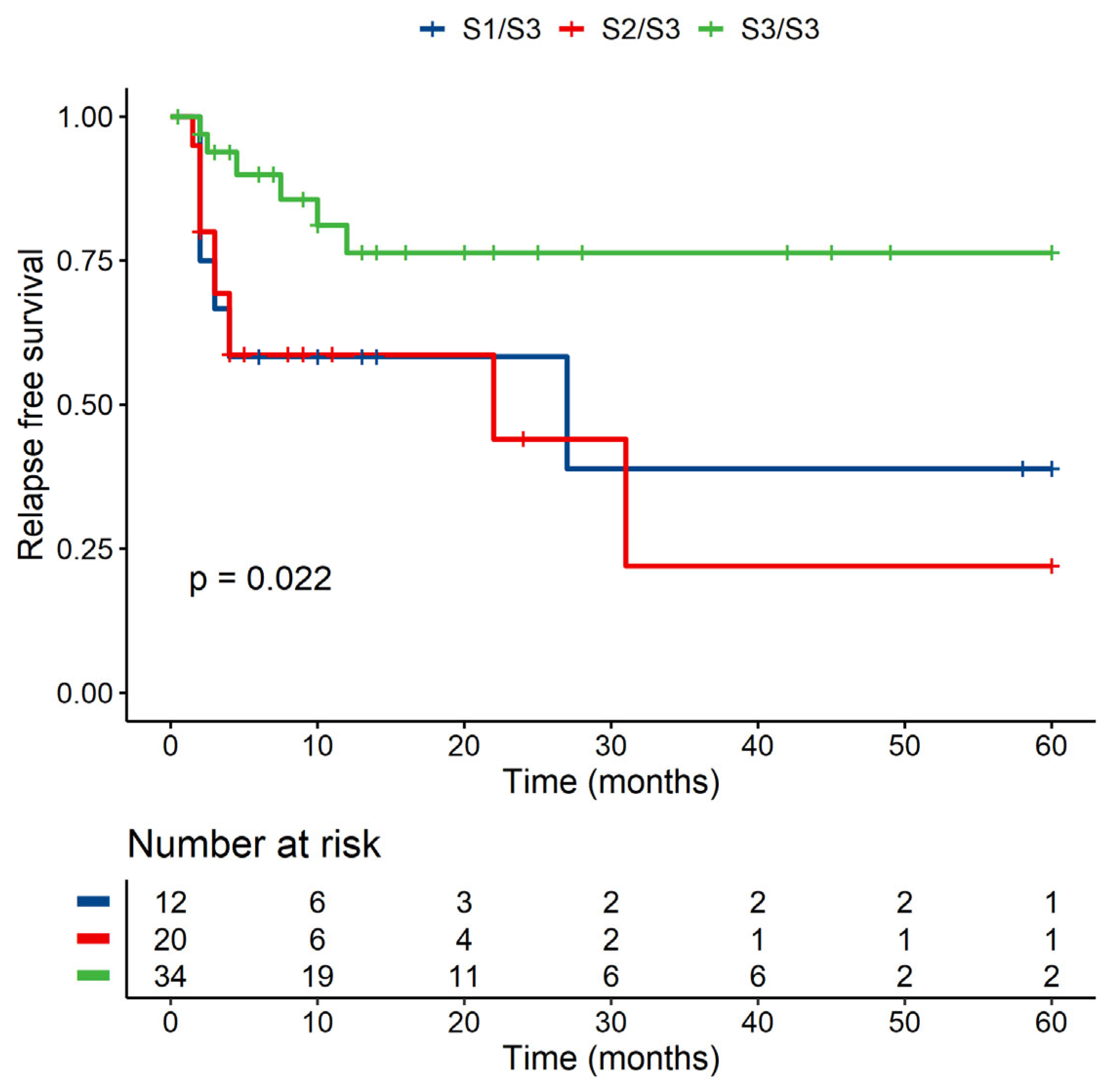
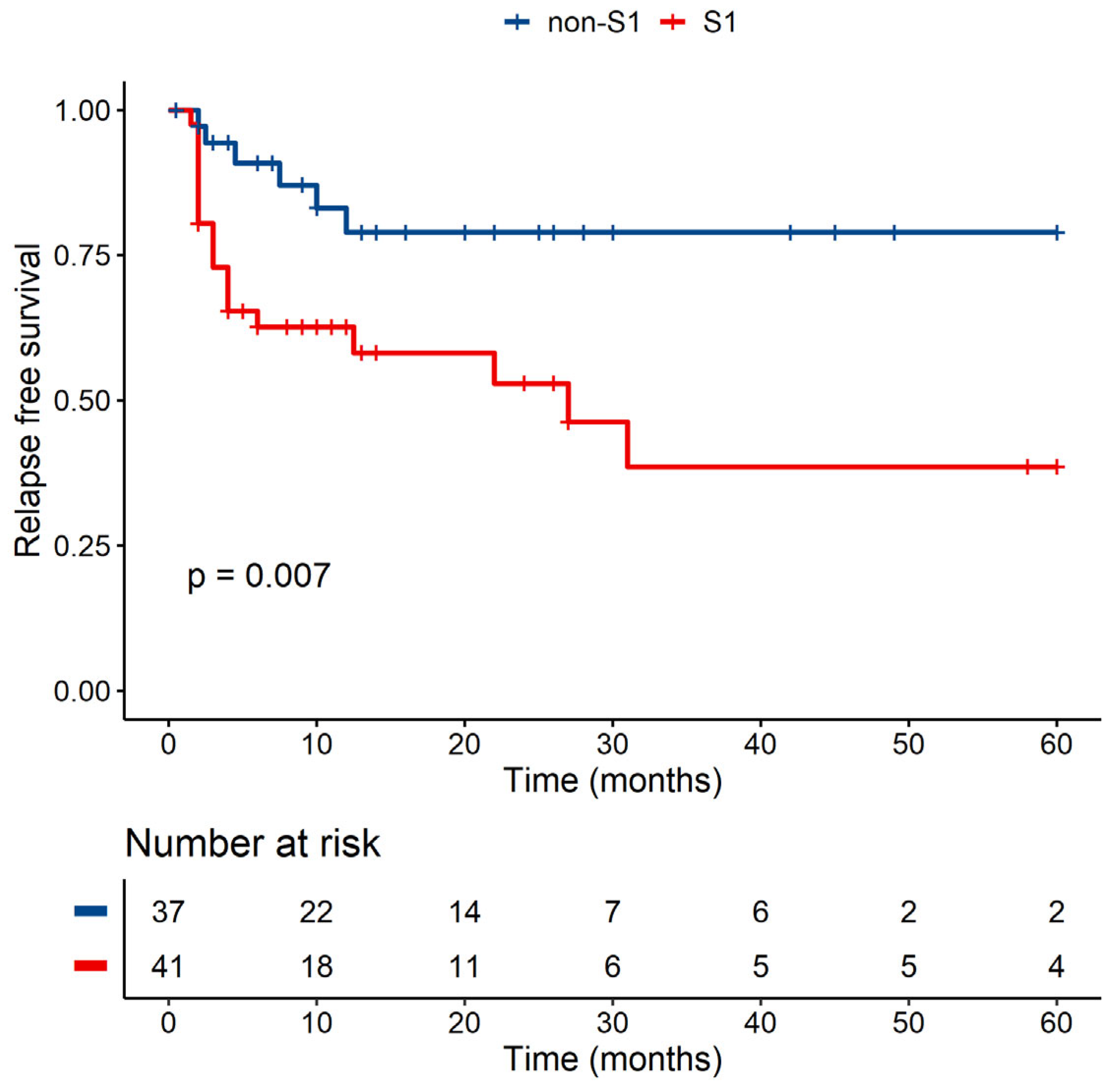
3.3.3. MICB-58Lys Can Be Linked to a Lower Risk of Relapse
3.3.4. Role of Known Polymorphisms Was Not Evident in Our Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gratwohl, A.; Brand, R.; Frassoni, F.; Rocha, V.; Niederwieser, D.; Reusser, P.; Einsele, H.; Cordonnier, C.; Parties, A.a.C.L.W.; Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation. Cause of death after allogeneic haematopoietic stem cell transplantation (HSCT) in early leukaemias: An EBMT analysis of lethal infectious complications and changes over calendar time. Bone Marrow Transpl. 2005, 36, 757–769. [Google Scholar] [CrossRef] [Green Version]
- Bosch, M.; Khan, F.M.; Storek, J. Immune reconstitution after hematopoietic cell transplantation. Curr. Opin. Hematol. 2012, 19, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Simonetta, F.; Alvarez, M.; Negrin, R.S. Natural Killer Cells in Graft-versus-Host-Disease after Allogeneic Hematopoietic Cell Transplantation. Front. Immunol. 2017, 8, 465. [Google Scholar] [CrossRef]
- Olson, J.A.; Leveson-Gower, D.B.; Gill, S.; Baker, J.; Beilhack, A.; Negrin, R.S. NK cells mediate reduction of GVHD by inhibiting activated, alloreactive T cells while retaining GVT effects. Blood 2010, 115, 4293–4301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Yang, S.; Zou, Y.; Yan, X.; Wu, H.; Zhou, M.; Sun, Y.C.; Zhang, Y.; Zhu, H.; Xu, K.; et al. NK cell predicts the severity of acute graft-versus-host disease in patients after allogeneic stem cell transplantation using antithymocyte globulin (ATG) in pretreatment scheme. BMC Immunol. 2019, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef]
- Gao, F.; Ye, Y.; Gao, Y.; Huang, H.; Zhao, Y. Influence of KIR and NK Cell Reconstitution in the Outcomes of Hematopoietic Stem Cell Transplantation. Front. Immunol. 2020, 11, 2022. [Google Scholar] [CrossRef]
- Zingoni, A.; Molfetta, R.; Fionda, C.; Soriani, A.; Paolini, R.; Cippitelli, M.; Cerboni, C.; Santoni, A. NKG2D and Its Ligands: “One for All, All for One”. Front. Immunol. 2018, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Wensveen, F.M.; Jelenčić, V.; Polić, B. NKG2D: A Master Regulator of Immune Cell Responsiveness. Front. Immunol. 2018, 9, 441. [Google Scholar] [CrossRef]
- Hayashi, T.; Imai, K.; Morishita, Y.; Hayashi, I.; Kusunoki, Y.; Nakachi, K. Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance. Cancer Res. 2006, 66, 563–570. [Google Scholar] [CrossRef] [Green Version]
- Espinoza, J.L.; Takami, A.; Onizuka, M.; Sao, H.; Akiyama, H.; Miyamura, K.; Okamoto, S.; Inoue, M.; Kanda, Y.; Ohtake, S.; et al. NKG2D gene polymorphism has a significant impact on transplant outcomes after HLA-fully-matched unrelated bone marrow transplantation for standard risk hematologic malignancies. Haematologica 2009, 94, 1427–1434. [Google Scholar] [CrossRef]
- Bahram, S. MIC genes: From genetics to biology. Adv. Immunol. 2000, 76, 1–60. [Google Scholar] [CrossRef]
- Machuldova, A.; Holubova, M.; Caputo, V.S.; Cedikova, M.; Jindra, P.; Houdova, L.; Pitule, P. Role of Polymorphisms of NKG2D Receptor and Its Ligands in Acute Myeloid Leukemia and Human Stem Cell Transplantation. Front. Immunol. 2021, 12, 1010. [Google Scholar] [CrossRef]
- Isernhagen, A.; Malzahn, D.; Viktorova, E.; Elsner, L.; Monecke, S.; von Bonin, F.; Kilisch, M.; Wermuth, J.M.; Walther, N.; Balavarca, Y.; et al. The MICA-129 dimorphism affects NKG2D signaling and outcome of hematopoietic stem cell transplantation. EMBO Mol. Med. 2015, 7, 1480–1502. [Google Scholar] [CrossRef]
- Boukouaci, W.; Busson, M.; de Latour, R.P.; Rocha, V.; Suberbielle, C.; Bengoufa, D.; Dulphy, N.; Haas, P.; Scieux, C.; Amroun, H.; et al. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood 2009, 114, 5216–5224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuerst, D.; Neuchel, C.; Niederwieser, D.; Bunjes, D.; Gramatzki, M.; Wagner, E.; Wulf, G.; Glass, B.; Pfreundschuh, M.; Einsele, H.; et al. Matching for the MICA-129 polymorphism is beneficial in unrelated hematopoietic stem cell transplantation. Blood 2016, 128, 3169–3176. [Google Scholar] [CrossRef] [Green Version]
- Parmar, S.; Del Lima, M.; Zou, Y.; Patah, P.A.; Liu, P.; Cano, P.; Rondon, G.; Pesoa, S.; de Padua Silva, L.; Qazilbash, M.H.; et al. Donor-recipient mismatches in MHC class I chain-related gene A in unrelated donor transplantation lead to increased incidence of acute graft-versus-host disease. Blood 2009, 114, 2884–2887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carapito, R.; Aouadi, I.; Pichot, A.; Spinnhirny, P.; Morlon, A.; Kotova, I.; Macquin, C.; Rolli, V.; Cesbron, A.; Gagne, K.; et al. Compatibility at amino acid position 98 of MICB reduces the incidence of graft-versus-host disease in conjunction with the CMV status. Bone Marrow Transpl. 2020, 55, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Carapito, R.; Jung, N.; Kwemou, M.; Untrau, M.; Michel, S.; Pichot, A.; Giacometti, G.; Macquin, C.; Ilias, W.; Morlon, A.; et al. Matching for the nonconventional MHC-I MICA gene significantly reduces the incidence of acute and chronic GVHD. Blood 2016, 128, 1979–1986. [Google Scholar] [CrossRef]
- Armand, P.; Kim, H.T.; Logan, B.R.; Wang, Z.; Alyea, E.P.; Kalaycio, M.E.; Maziarz, R.T.; Antin, J.H.; Soiffer, R.J.; Weisdorf, D.J.; et al. Validation and refinement of the Disease Risk Index for allogeneic stem cell transplantation. Blood 2014, 123, 3664–3671. [Google Scholar] [CrossRef]
- Sorror, M.L.; Maris, M.B.; Storb, R.; Baron, F.; Sandmaier, B.M.; Maloney, D.G.; Storer, B. Hematopoietic cell transplantation (HCT)-specific comorbidity index: A new tool for risk assessment before allogeneic HCT. Blood 2005, 106, 2912–2919. [Google Scholar] [CrossRef] [Green Version]
- Kabalak, G.; Thomas, R.M.; Martin, J.; Ortego-Centeno, N.; Jimenez-Alonso, J.; de Ramón, E.; Buyny, S.; Hamsen, S.; Gross, W.L.; Schnarr, S.; et al. Association of an NKG2D gene variant with systemic lupus erythematosus in two populations. Hum. Immunol. 2010, 71, 74–78. [Google Scholar] [CrossRef]
- Asl, A.A.; Nezamdoust, F.V.; Fesahat, F.; Astani, A.; Barati, M.; Raee, P.; Asadi-Saghandi, A. Association between rs1049174 NKG2D gene polymorphism and idiopathic recurrent spontaneous abortion in Iranian women: A case-control study. J. Obstet. Gynaecol. 2020, 41, 774–778. [Google Scholar] [CrossRef]
- Field, S.F.; Nejentsev, S.; Walker, N.M.; Howson, J.M.M.; Godfrey, L.M.; Jolley, J.D.; Hardy, M.P.A.; Todd, J.A. Sequencing-Based Genotyping and Association Analysis of the MICA and MICB Genes in Type 1 Diabetes. Diabetes 2008, 57, 1753–1756. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Gonzalez, W.G.; Martinez-Alvarez, J.C.; Arrazola-Garcia, A.; Perez-Rodriguez, M. Haplotype block 1 variant (HB-1v) of the NKG2 family of receptors. Hum. Immunol. 2019, 80, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.; Barker, D.J.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Marsh, S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2019, 48, D948–D955. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 2000, 28, 1102, 1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. 2018. Available online: https://www.R-project.org/ (accessed on 6 October 2021).
- Gratwohl, A. The EBMT risk score. Bone Marrow Transpl. 2012, 47, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Weisdorf, D.; Hakke, R.; Blazar, B.; Miller, W.; McGlave, P.; Ramsay, N.; Kersey, J.; Filipovich, A. Risk factors for acute graft-versus-host disease in histocompatible donor bone marrow transplantation. Transplantation 1991, 51, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.J.; Levine, D.M.; Storer, B.E.; Nelson, S.C.; Dong, X.; Hansen, J.A. Recipient and donor genetic variants associated with mortality after allogeneic hematopoietic cell transplantation. Blood Adv. 2020, 4, 3224–3233. [Google Scholar] [CrossRef]
- Chen, D.; Gyllensten, U. MICA polymorphism: Biology and importance in cancer. Carcinogenesis 2014, 35, 2633–2642. [Google Scholar] [CrossRef]
- Ashiru, O.; López-Cobo, S.; Fernández-Messina, L.; Pontes-Quero, S.; Pandolfi, R.; Reyburn, H.T.; Valés-Gómez, M. A GPI anchor explains the unique biological features of the common NKG2D-ligand allele MICA*008. Biochem. J. 2013, 454, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Onyeaghala, G.; Lane, J.; Pankratz, N.; Nelson, H.H.; Thyagarajan, B.; Walcheck, B.; Anderson, K.E.; Prizment, A.E. Association between MICA polymorphisms, s-MICA levels, and pancreatic cancer risk in a population-based case-control study. PLoS ONE 2019, 14, e0217868. [Google Scholar] [CrossRef] [Green Version]
- Isernhagen, A.; Schilling, D.; Monecke, S.; Shah, P.; Elsner, L.; Walter, L.; Multhoff, G.; Dressel, R. The MICA-129Met/Val dimorphism affects plasma membrane expression and shedding of the NKG2D ligand MICA. Immunogenetics 2016, 68, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nice, T.J.; Coscoy, L.; Raulet, D.H. Posttranslational regulation of the NKG2D ligand Mult1 in response to cell stress. J. Exp. Med. 2009, 206, 287–298. [Google Scholar] [CrossRef] [Green Version]
- Apithy, M.-J.; Charbonnier, A.; Desoutter, J.; Diouf, M.; Morel, P.; Garçon, L.; Marolleau, J.-P.; Guillaume, N. Impact of MICA and NKG2D polymorphisms in HLA-fully matched related and unrelated hematopoietic stem cell transplantation. Bone Marrow Transplant. 2018, 53, 918–922. [Google Scholar] [CrossRef] [Green Version]


| Patient Age Group | Type of Donor | ||
|---|---|---|---|
| <50 | 32 | Related donor (full match) | 25 |
| 50–65 | 65 | Unrelated donor | 64 |
| >65 | 27 | Haploidentical donor | 35 |
| Median (years) | 58 | Conditioning therapy | |
| Range (years) | 23–74 | Myeloablative | 23 |
| Patient’s sex | Reduced | 101 | |
| Male | 72 | GVHD prophylaxis | |
| Female | 52 | With PTCY **** | 34 |
| Diagnosis | Without PTCY | 90 | |
| AML | 118 | aGVHD | |
| MDS | 6 | Yes | 96 |
| Disease Risk Index (DRI) * | No | 28 | |
| Low | 5 | cGVHD | |
| Intermediate | 67 | Yes | 38 |
| High | 33 | No | 64 |
| Very high | 7 | Unknown | 22 |
| Unknown | 12 | Relapse | |
| AML as secondary malignancy ** | Yes | 41 | |
| Yes | 37 | No | 82 |
| No | 87 | Unknown | 1 |
| Karyotype | Outcome | ||
| Normal karyotype | 58 | Dead | 62 |
| Complex karyotype *** | 16 | Alive | 62 |
| Other karyotype changes | 11 | Cause of death | |
| Unknown | 39 | Relapse | 29 |
| Disease status during HSCT | Infection | 16 | |
| Active disease | 43 | Organ failure | 7 |
| Complete remission | 81 | GVHD | 6 |
| Graft source | Graft rejection | 1 | |
| Bone marrow | 24 | Unknown | 3 |
| Peripheral blood stem cells | 100 | HLA mismatch | |
| CMV match/mismatch | None | 73 | |
| Match | 75 | Haploidentical | 35 |
| Mismatch | 49 | ABC/DP-DR | 16 |
| Pretransplant T-cell depletion (patient) | Sex match/mismatch | ||
| Yes | 64 | Match | 65 |
| No | 60 | Mismatch | 59 |
| EBMT risk score | HCT-CI ***** | ||
| 1–2 | 24 | ≥3 | 34 |
| 3 | 39 | <3 | 81 |
| 4 | 30 | unknown | 9 |
| 5–6 | 29 | ||
| NA | 2 | ||
| Sequence of | Length of the Sequence | PCR Primers | Published in |
|---|---|---|---|
| NKG2D-Hb1 | 253 bp | F: TGCGAGGTATTTATGTTCTG R: ACAGTTTAGGAATACAGCAC | [22] |
| NKG2D-Hb2 | 230 bp | F: TTAAGGCTGGAGAATAATGC | [23] |
| R: TCAGTGAAGGAAGAGAAGG | |||
| MICA | 1.9 kbp (exons 2–4) | F: CCCCCTTCTTCTGTTCATCA R: TGACTCTGAAGCACCAGCAC | [24] |
| MICB | 2.1 kbp (exons 2–5) | F: GGACAGCAGACCTGTGTGTTA R: AAAGGAGCTTTCCCATCTCC | [24] |
| OS and RFS Compared within Parameters |
|---|
| MICA/B exons 2–4 |
| MICA-129 |
| MICB-98 |
| MICA/B hetero vs. homozygosity |
| NKG2D haploblocks 1–2 |
| Match and mismatch within exons 2–4 (MICA/B) |
| Match and mismatch within MICA-129 |
| Match and mismatch within MICB-98 |
| Match and mismatch NKG2D haploblocks 1–2 |
| Each specific group of each exon compared to the other groups |
| OS | RFS | |||||||
|---|---|---|---|---|---|---|---|---|
| HR * | Lower CI ** | Upper CI | p-Value | HR | Lower CI | Upper CI | p-Value | |
| Patient age group | ||||||||
| <50 | 1 | 1 | ||||||
| 50–65 | 1.706 | 0.922 | 3.155 | 0.089 | 1.274 | 0.602 | 2.699 | 0.527 |
| >65 | 1.105 | 0.490 | 2.494 | 0.809 | 0.552 | 0.173 | 1.763 | 0.316 |
| Patient’s sex | ||||||||
| Female | 1 | 1 | ||||||
| Male | 0.939 | 0.567 | 1.557 | 0.809 | 0.956 | 0.493 | 1.856 | 0.895 |
| Diagnosis | ||||||||
| AML | 1 | 1 | ||||||
| MDS | 0.332 | 0.046 | 2.398 | 0.275 | 0.563 | 0.077 | 4.109 | 0.571 |
| DRI | ||||||||
| Low | 1 | 1 | ||||||
| Intermediate | 0.920 | 0.278 | 3.050 | 0.892 | 1.181 | 0.152 | 9.149 | 0.874 |
| High | 3.821 | 1.151 | 12.686 | 0.029 | 7.450 | 0.989 | 56.129 | 0.051 |
| Very high | 8.256 | 2.095 | 32.545 | 0.003 | 23.261 | 2.747 | 196.974 | 0.004 |
| AML as secondary malignancy | ||||||||
| No | 1 | 1 | ||||||
| Yes | 2.106 | 1.263 | 3.513 | 0.004 | 2.188 | 1.123 | 4.261 | 0.021 |
| Karyotype | ||||||||
| Normal karyotype | 1 | 1 | ||||||
| Other changes | 1.542 | 0.866 | 2.745 | 0.141 | 1.703 | 0.746 | 3.887 | 0.206 |
| Complex karyotype | 4.162 | 2.100 | 8.251 | <0.001 | 8.404 | 3.658 | 19.306 | <0.001 |
| Disease status during HSCT | ||||||||
| Active disease | 1 | 1 | ||||||
| Complete remission | 0.527 | 0.320 | 0.869 | 0.012 | 0.510 | 0.265 | 0.983 | 0.044 |
| Graft source | ||||||||
| Bone marrow | 1 | 1 | ||||||
| PBSC | 0.829 | 0.457 | 1.505 | 0.538 | 0.646 | 0.312 | 1.341 | 0.241 |
| Type of donor | ||||||||
| Haploidentical | 1 | 1 | ||||||
| Related | 0.648 | 0.323 | 1.298 | 0.221 | 1.346 | 0.529 | 3.424 | 0.533 |
| Unrelated | 0.595 | 0.333 | 1.063 | 0.080 | 0.974 | 0.423 | 2.243 | 0.950 |
| Conditioning | ||||||||
| Myeloablative | 1 | 1 | ||||||
| Reduced | 1.564 | 0.743 | 3.291 | 0.239 | 0.969 | 0.424 | 2.215 | 0.940 |
| GVHD prophylaxis | ||||||||
| No | 1 | 1 | ||||||
| Yes | 1.578 | 0.910 | 2.736 | 0.104 | 0.796 | 0.348 | 1.821 | 0.589 |
| aGVHD | ||||||||
| No | 1 | 1 | ||||||
| Yes | 1.139 | 0.617 | 2.101 | 0.678 | 0.904 | 0.425 | 1.923 | 0.793 |
| cGVHD | ||||||||
| No | 1 | 1 | ||||||
| Yes | 0.476 | 0.243 | 0.933 | 0.031 | 0.232 | 0.080 | 0.673 | 0.007 |
| HCT-CI | ||||||||
| ≥3 | 1 | 1 | ||||||
| <3 | 0.753 | 0.436 | 1.302 | 0.311 | 0.850 | 0.414 | 1.745 | 0.659 |
| S4+ Grafts | Non-S4 Grafts | |
|---|---|---|
| Alive patients | 41% | 56% |
| Dead patients | 59% | 44% |
| Causes of death | ||
| Relapse | 41% | 54% |
| Infection | 36% | 22% |
| Organ failure | 14% | 11% |
| GVHD | 9% | 11% |
| Graft rejection | 0% | 3% |
| MICA Exon 2 Amino Acid Sequence from Position 3 to 37 of MICA Protein | |
|---|---|
| Group | Sequence |
| Group S1 | HSLRYNLTVLSWDGSVQSGFLAEVHLDGQPFLRYD |
| Group S2 | HSLRYNLTVLSWDGSVQSGFLAEVHLDGQPFLRCD |
| Group S4 | HSLRYNLTVLSGDGSVQSGFLAEVHLDGQPFLRCD |
| Group S6 | HSLRYNLTVLSWDGSVQSGFLTEVHLDGQPFLRCD |
| Group S7 | HSLPYNLTVLSWDGSVQSGFLAEVHLDGQPFLRYD |
| MICB Exon 2 Amino Acid Sequence from Position 49 to 63 of MICB Protein | |
|---|---|
| Group | Sequence |
| Group S1 | QWAEDVLGAETWDTE |
| Group S2 | QWAEDVLGAKTWDTE |
| Group S3 | QWAENVLGAKTWDTE |
| MICA Exon 2—Comparison of Group Combinations | ||||
|---|---|---|---|---|
| p-Value | Hazard Ratio | Lower CI* | Upper CI | |
| S1/S1 | 1 | |||
| S1/S2 | 2.062 | 0.427 | 0.088 | 2.062 |
| S1/S4 | 0.028 | 2.745 | 1.113 | 6.771 |
| S1/S6 | 0.120 | 0.193 | 0.024 | 1.538 |
| MICA exon 2 (MICA-14Gly)—MICA-14Gly (S4) presence versus absence (non-S4) | ||||
| p-Value | Hazard ratio | Lower CI | Upper CI | |
| Non-S4 | 1 | |||
| S4+ | 0.035 | 2.254 | 1.058 | 4.801 |
| MICA-129 match/mismatch | ||||
| p-Value | Hazard ratio | Lower CI | Upper CI | |
| Match | 1 | |||
| Mismatch | 0.155 | 2.407 | 0.718 | 8.069 |
| Donor—MICB Homozygote Versus Heterozygote | ||||
|---|---|---|---|---|
| p-Value | Hazard Ratio | Lower CI* | Upper CI | |
| Homozygote | 1 | |||
| Heterozygote | 0.433 | 1.693 | 0.454 | 6.309 |
| Patient—MICB Homozygote Versus Heterozygote | ||||
| p-Value | Hazard ratio | Lower CI | Upper CI | |
| Homozygote | 1 | |||
| Heterozygote | 0.322 | 2.101 | 0.483 | 9.134 |
| MICB Exon 3—Heterozygotes (S1/S3 + S2/S3) Versus Homozygote (S3/S3) | ||||
| p-Value | Hazard ratio | Lower CI | Upper CI | |
| S1/S3 + S2/S3 | 1 | |||
| S3/S3 | 0.376 | 0.506 | 0.112 | 2.286 |
| MICB Exon 2 (MICB-58Glu)—MICB-58Glu (S1) Presence Versus Absence (non-S1) | ||||
| p-Value | Hazard ratio | Lower CI | Upper CI | |
| Non-S1 | 1 | |||
| S1+ | 0.069 | 3.764 | 0.902 | 15.707 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machuldova, A.; Houdova, L.; Kratochvilova, K.; Leba, M.; Jindra, P.; Ostasov, P.; Maceckova, D.; Klieber, R.; Gmucova, H.; Sramek, J.; et al. Single-Nucleotide Polymorphisms in MICA and MICB Genes Could Play a Role in the Outcome in AML Patients after HSCT. J. Clin. Med. 2021, 10, 4636. https://doi.org/10.3390/jcm10204636
Machuldova A, Houdova L, Kratochvilova K, Leba M, Jindra P, Ostasov P, Maceckova D, Klieber R, Gmucova H, Sramek J, et al. Single-Nucleotide Polymorphisms in MICA and MICB Genes Could Play a Role in the Outcome in AML Patients after HSCT. Journal of Clinical Medicine. 2021; 10(20):4636. https://doi.org/10.3390/jcm10204636
Chicago/Turabian StyleMachuldova, Alena, Lucie Houdova, Katerina Kratochvilova, Martin Leba, Pavel Jindra, Pavel Ostasov, Diana Maceckova, Robin Klieber, Hana Gmucova, Jiri Sramek, and et al. 2021. "Single-Nucleotide Polymorphisms in MICA and MICB Genes Could Play a Role in the Outcome in AML Patients after HSCT" Journal of Clinical Medicine 10, no. 20: 4636. https://doi.org/10.3390/jcm10204636
APA StyleMachuldova, A., Houdova, L., Kratochvilova, K., Leba, M., Jindra, P., Ostasov, P., Maceckova, D., Klieber, R., Gmucova, H., Sramek, J., & Holubova, M. (2021). Single-Nucleotide Polymorphisms in MICA and MICB Genes Could Play a Role in the Outcome in AML Patients after HSCT. Journal of Clinical Medicine, 10(20), 4636. https://doi.org/10.3390/jcm10204636






