Evaluation of a Targeted Next-Generation Sequencing Panel for the Non-Invasive Detection of Variants in Circulating DNA of Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Blood and Tissue Samples
2.3. Isolation of cfDNA from Plasma
2.4. Analysis of KRAS Variants in Tumor Tissues
2.5. Detection of Gene Variants in cfDNA by Digital PCR
2.6. Analysis of cfDNA by Targeted NGS with TST170
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
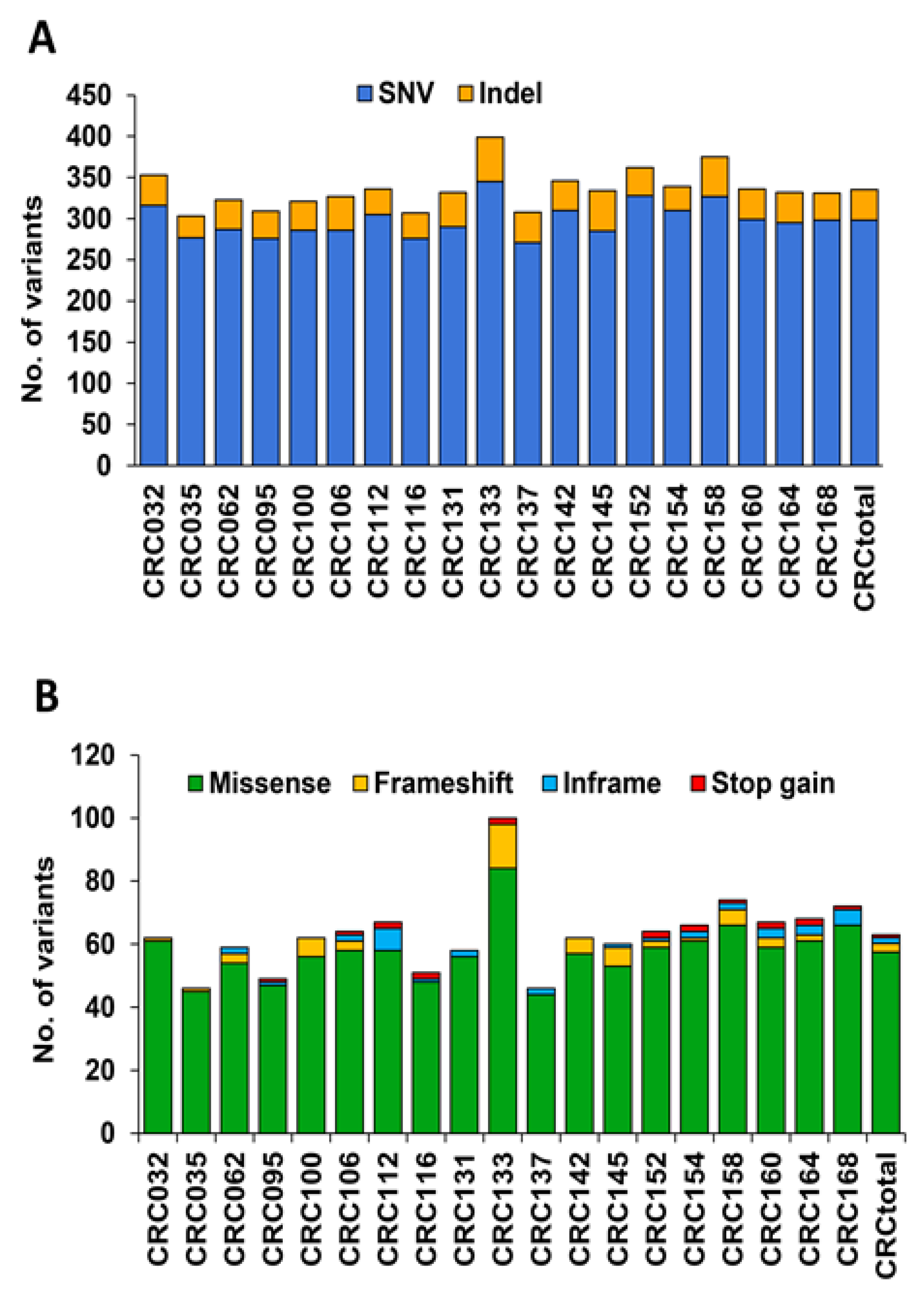
3.2. Analysis of Variants in cfDNA of mCRC Patients Using the TST170 Targeted Panel
3.3. Identification of Cancer-Associated Variants with Clinical Significance in cfDNA of mCRC Patients
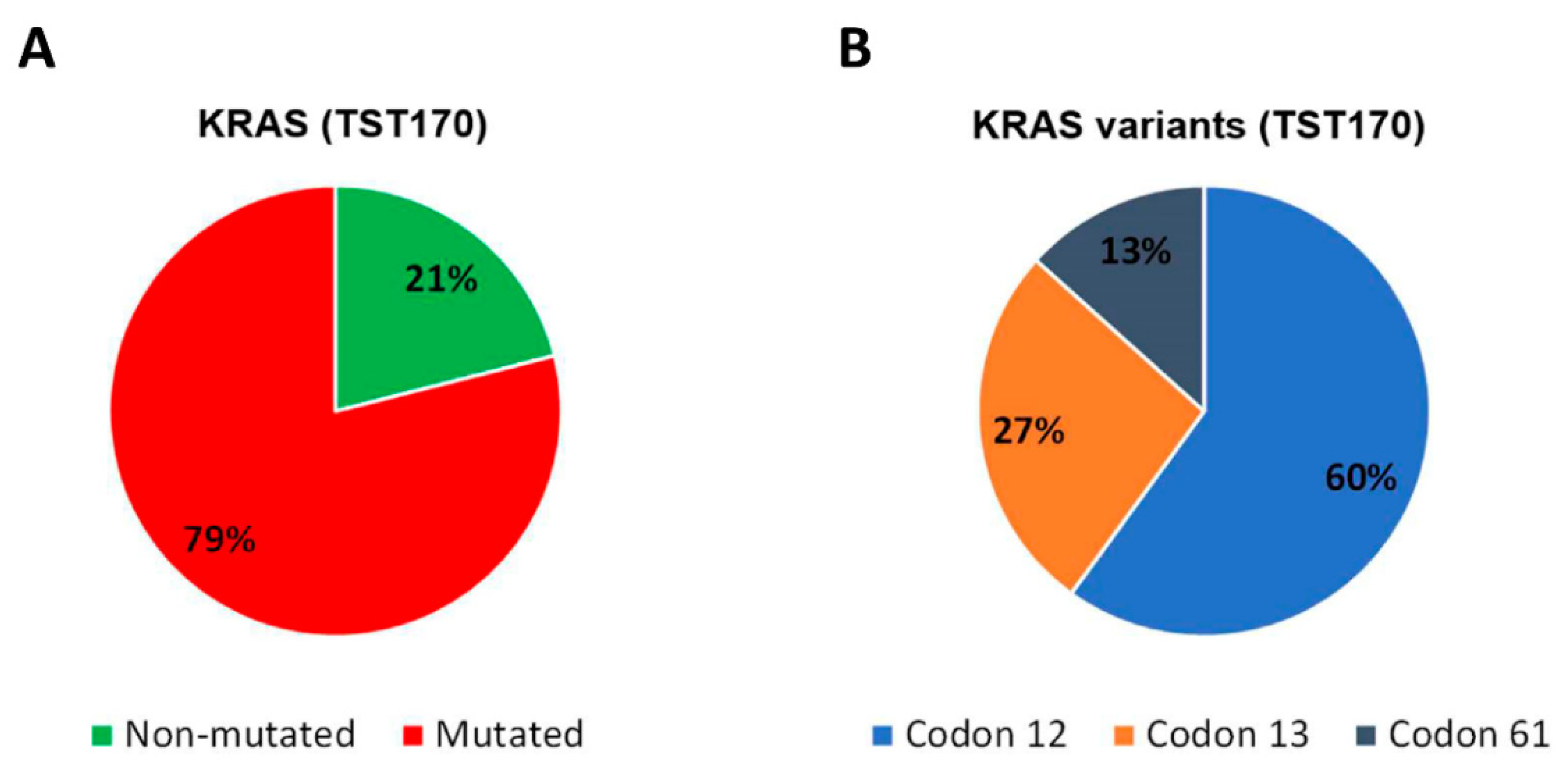
3.4. Analysis of KRAS Variants in cfDNA of mCRC Patients and Concordance with Tissue Analysis
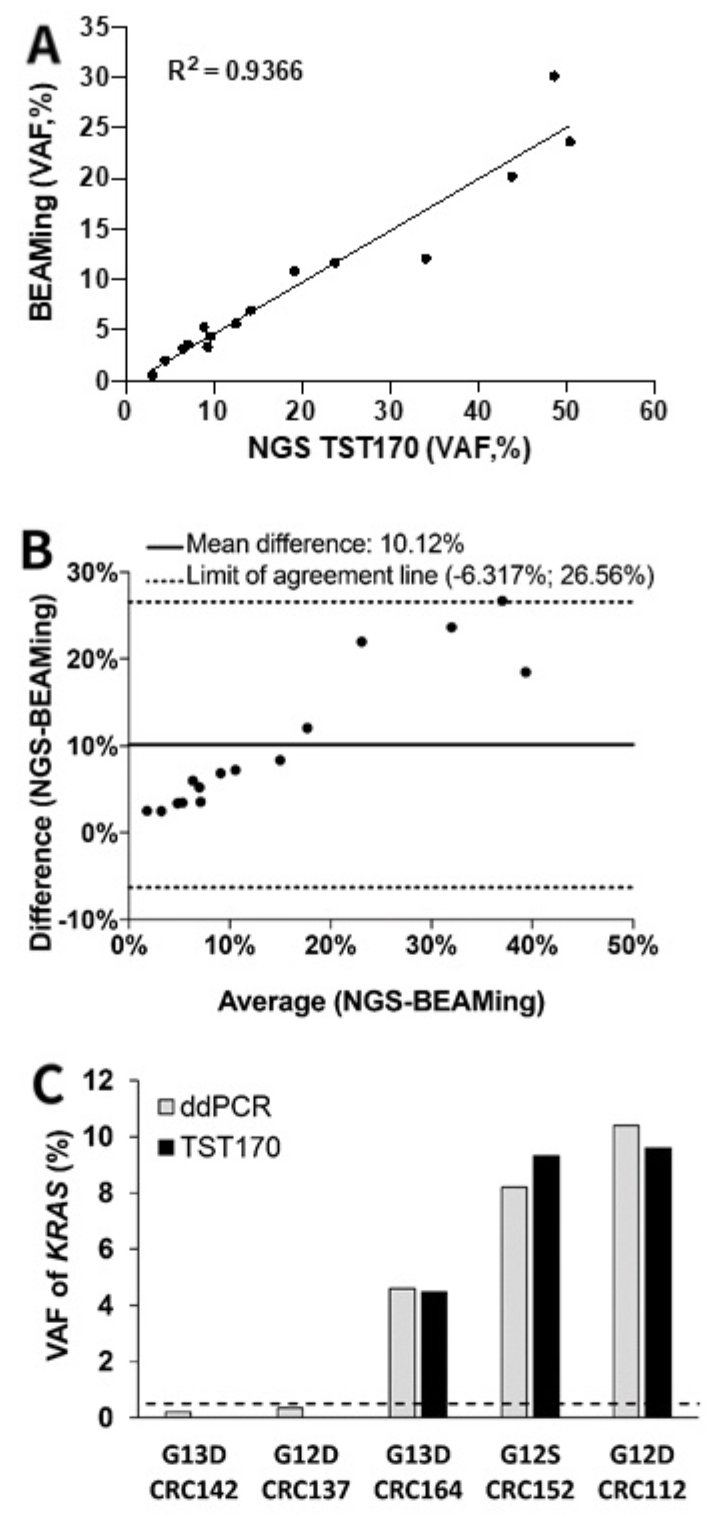
3.5. Concordance of KRAS Variants in cfDNA of mCRC Patients by BEAMing and TST170
3.6. Description of KRAS Variants in Discordant Samples Detected in cfDNA by TST170
3.7. KRAS Analysis in cfDNA According to Clinical–Pathological Characteristics of Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douillard, J.Y.; Oliner, K.S.; Siena, S.; Tabernero, J.; Burkes, R.; Barugel, M.; Humblet, Y.; Bodoky, G.; Cunningham, D.; Jassem, J.; et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 2013, 369, 1023–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lech, G.; Slotwinski, R.; Slodkowski, M.; Krasnodebski, I.W. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J. Gastroenterol. 2016, 22, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, L.; Zavridou, M.; Kasimir-Bauer, S.; Lianidou, E.S. Liquid biopsy in ovarian cancer: The potential of circulating miRNAs and exosomes. Transl. Res. J. Lab. Clin. Med. 2019, 205, 77–91. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Howell, J.A.; Khan, S.A.; Knapp, S.; Thursz, M.R.; Sharma, R. The clinical role of circulating free tumor DNA in gastrointestinal malignancy. Transl. Res. J. Lab. Clin. Med. 2017, 183, 137–154. [Google Scholar] [CrossRef]
- Samandari, M.; Julia, M.G.; Rice, A.; Chronopoulos, A.; Del Rio Hernandez, A.E. Liquid biopsies for management of pancreatic cancer. Transl. Res. J. Lab. Clin. Med. 2018, 201, 98–127. [Google Scholar] [CrossRef]
- Leung, F.; Kulasingam, V.; Diamandis, E.P.; Hoon, D.S.; Kinzler, K.; Pantel, K.; Alix-Panabieres, C. Circulating Tumor DNA as a Cancer Biomarker: Fact or Fiction? Clin. Chem. 2016, 62, 1054–1060. [Google Scholar] [CrossRef] [Green Version]
- Dressman, D.; Yan, H.; Traverso, G.; Kinzler, K.W.; Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. USA 2003, 100, 8817–8822. [Google Scholar] [CrossRef] [Green Version]
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; Rodriguez, C.; Brozos, E.; et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef]
- Garcia-Foncillas, J.; Alba, E.; Aranda, E.; Diaz-Rubio, E.; Lopez-Lopez, R.; Tabernero, J.; Vivancos, A. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: An expert taskforce review. Ann. Oncol. 2017, 28, 2943–2949. [Google Scholar] [CrossRef] [PubMed]
- Casas-Arozamena, C.; Diaz, E.; Moiola, C.P.; Alonso-Alconada, L.; Ferreiros, A.; Abalo, A.; Gil, C.L.; Oltra, S.S.; de Santiago, J.; Cabrera, S.; et al. Genomic Profiling of Uterine Aspirates and cfDNA as an Integrative Liquid Biopsy Strategy in Endometrial Cancer. J. Clin. Med. 2020, 9, 585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, K.; Guttery, D.S.; Fernandez-Garcia, D.; Hills, A.; Hastings, R.K.; Luo, J.; Goddard, K.; Shahin, V.; Woodley-Barker, L.; Rosales, B.M.; et al. Next Generation Sequencing of Circulating Cell-Free DNA for Evaluating Mutations and Gene Amplification in Metastatic Breast Cancer. Clin. Chem. 2017, 63, 532–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, J.; Forestier, J.; Dusserre, E.; Wozny, A.S.; Geiguer, F.; Merle, P.; Tissot, C.; Ferraro-Peyret, C.; Jones, F.S.; Edelstein, D.L.; et al. Cross-platform comparison for the detection of RAS mutations in cfDNA (ddPCR Biorad detection assay, BEAMing assay, and NGS strategy). Oncotarget 2018, 9, 21122–21131. [Google Scholar] [CrossRef]
- Kastrisiou, M.; Zarkavelis, G.; Pentheroudakis, G.; Magklara, A. Clinical Application of Next-Generation Sequencing as A Liquid Biopsy Technique in Advanced Colorectal Cancer: A Trick or A Treat? Cancers 2019, 11, 1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Lee, S.; Sung, J.S.; Chung, H.J.; Lim, A.R.; Kim, J.W.; Choi, Y.J.; Park, K.H.; Kim, Y.H. Clinical Application of Targeted Deep Sequencing in Metastatic Colorectal Cancer Patients: Actionable Genomic Alteration in K-MASTER Project. Cancer Res. Treat. 2021, 53, 123–130. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, D.; Kim, H.S.; Na, K.; Lee, J.Y.; Nam, E.J.; Kim, S.W.; Kim, S.; Kim, Y.T. Integrating a Next Generation Sequencing Panel into Clinical Practice in Ovarian Cancer. Yonsei Med. J. 2019, 60, 914–923. [Google Scholar] [CrossRef]
- Na, K.; Kim, H.S.; Shim, H.S.; Chang, J.H.; Kang, S.G.; Kim, S.H. Targeted next-generation sequencing panel (TruSight Tumor 170) in diffuse glioma: A single institutional experience of 135 cases. J. Neurooncol. 2019, 142, 445–454. [Google Scholar] [CrossRef]
- Tabernero, J.; Lenz, H.J.; Siena, S.; Sobrero, A.; Falcone, A.; Ychou, M.; Humblet, Y.; Bouche, O.; Mineur, L.; Barone, C.; et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015, 16, 937–948. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [Green Version]
- Choi, I.S.; Kato, S.; Fanta, P.T.; Leichman, L.; Okamura, R.; Raymond, V.M.; Lanman, R.B.; Lippman, S.M.; Kurzrock, R. Genomic Profiling of Blood-Derived Circulating Tumor DNA from Patients with Colorectal Cancer: Implications for Response and Resistance to Targeted Therapeutics. Mol. Cancer Ther. 2019, 18, 1852–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spindler, K.L.; Pallisgaard, N.; Andersen, R.F.; Brandslund, I.; Jakobsen, A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS ONE 2015, 10, e0108247. [Google Scholar] [CrossRef]
- Garcia-Foncillas, J.; Tabernero, J.; Elez, E.; Aranda, E.; Benavides, M.; Camps, C.; Jantus-Lewintre, E.; Lopez, R.; Muinelo-Romay, L.; Montagut, C.; et al. Prospective multicenter real-world RAS mutation comparison between OncoBEAM-based liquid biopsy and tissue analysis in metastatic colorectal cancer. Br. J. Cancer 2018, 119, 1464–1470. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Attard, G. Plasma DNA Analysis in Prostate Cancer: Opportunities for Improving Clinical Management. Clin. Chem. 2019, 65, 100–107. [Google Scholar] [CrossRef] [Green Version]
- Froyen, G.; Le Mercier, M.; Lierman, E.; Vandepoele, K.; Nollet, F.; Boone, E.; Van der Meulen, J.; Jacobs, K.; Lambin, S.; Vander Borght, S.; et al. Standardization of Somatic Variant Classifications in Solid and Haematological Tumours by a Two-Level Approach of Biological and Clinical Classes: An Initiative of the Belgian ComPerMed Expert Panel. Cancers 2019, 11, 2030. [Google Scholar] [CrossRef] [Green Version]
- Kondelin, J.; Salokas, K.; Saarinen, L.; Ovaska, K.; Rauanheimo, H.; Plaketti, R.M.; Hamberg, J.; Liu, X.; Yadav, L.; Gylfe, A.E.; et al. Comprehensive evaluation of coding region point mutations in microsatellite-unstable colorectal cancer. EMBO Mol. Med. 2018, 10, e8552. [Google Scholar] [CrossRef]
- Mamlouk, S.; Childs, L.H.; Aust, D.; Heim, D.; Melching, F.; Oliveira, C.; Wolf, T.; Durek, P.; Schumacher, D.; Blaker, H.; et al. DNA copy number changes define spatial patterns of heterogeneity in colorectal cancer. Nat. Commun. 2017, 8, 14093. [Google Scholar] [CrossRef]
- Sun, X.; Huang, T.; Cheng, F.; Huang, K.; Liu, M.; He, W.; Li, M.; Zhang, X.; Xu, M.; Chen, S.; et al. Monitoring colorectal cancer following surgery using plasma circulating tumor DNA. Oncol. Lett. 2018, 15, 4365–4375. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Lu, L.; Zhou, Z.; Liu, J.; Zhang, D.; Nan, K.; Zhao, X.; Li, F.; Tian, L.; Dong, H.; et al. CNV Detection from Circulating Tumor DNA in Late Stage Non-Small Cell Lung Cancer Patients. Genes 2019, 10, 926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigemizu, D.; Fujimoto, A.; Akiyama, S.; Abe, T.; Nakano, K.; Boroevich, K.A.; Yamamoto, Y.; Furuta, M.; Kubo, M.; Nakagawa, H.; et al. A practical method to detect SNVs and indels from whole genome and exome sequencing data. Sci. Rep. 2013, 3, 2161. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Mol. Diagn. 2017, 19, 187–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janku, F.; Lee, J.J.; Tsimberidou, A.M.; Hong, D.S.; Naing, A.; Falchook, G.S.; Fu, S.; Luthra, R.; Garrido-Laguna, I.; Kurzrock, R. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS ONE 2011, 6, e22769. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, H.; Bardelli, A.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B.; Velculescu, V.E. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002, 418, 934. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra168. [Google Scholar] [CrossRef] [PubMed]
- Bando, H.; Nakamura, Y.; Taniguchi, H.; Shiozawa, M.; Yasui, H.; Esaki, T.; Ohta, T.; Denda, T.; Satoh, T.; Yamazaki, K.; et al. Impact of a metastatic site on circulating tumor DNA (ctDNA) analysis in patients (pts) with metastatic colorectal cancer (mCRC). J. Clin. Oncol. 2021, 39, 3554. [Google Scholar] [CrossRef]
- Demuth, C.; Spindler, K.G.; Johansen, J.S.; Pallisgaard, N.; Nielsen, D.; Hogdall, E.; Vittrup, B.; Sorensen, B.S. Measuring KRAS Mutations in Circulating Tumor DNA by Droplet Digital PCR and Next-Generation Sequencing. Transl. Oncol. 2018, 11, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]

| Characteristics | Patients (N = 19) | |
|---|---|---|
| No. | % | |
| Age (years) | ||
| <60 | 5 | 26 |
| 60–69 | 5 | 26 |
| 70–79 | 7 | 37 |
| >80 | 2 | 11 |
| Gender | ||
| Female | 6 | 32 |
| Male | 13 | 68 |
| Histology | ||
| Adenocarcinoma | 18 | 95 |
| Mucinous adenocarcinoma | 1 | 5 |
| Primary tumor location | ||
| Right colon | 8 | 42 |
| Left colon/rectum | 11 | 58 |
| Number of metastatic locations | ||
| 1 | 9 | 47 |
| ≥2 | 10 | 53 |
| Metastatic location | ||
| Liver | 10 | 53 |
| Lung | 10 | 53 |
| Peritoneum | 5 | 26 |
| Previously resected primary tumor | ||
| Yes | 10 | 53 |
| No | 9 | 47 |
| Previous systemic treatment 1 | ||
| Yes | 6 | 32 |
| No | 13 | 68 |
| MSI status 2 | ||
| Negative | 16 | 84 |
| Unknown | 3 | 16 |
| KRAS status in tissue | ||
| Wild type | 1 | 5 |
| Mutated | 16 | 84 |
| Unknown | 2 | 11 |
| KRAS status in cfDNA | ||
| Wild type | 0 | 0 |
| Mutated | 19 | 100 |
| NRAS status in cfDNA | ||
| Wild type | 15 | 79 |
| Mutated | 1 | 5 |
| Unknown | 3 | 16 |
| Tissue biopsy location | ||
| Primary Tumor | 14 | 74 |
| Metastasis | 2 | 10 |
| Unknown | 3 | 16 |
| Sample ID | KRAS Variants | ||
|---|---|---|---|
| Tumor Tissue | cfDNA | ||
| BEAMing (VAF, %) | TST170 (VAF, %) | ||
| CRC032 | p.Q61L | KR3Cdn61 (0.55) | p.Q61L (3.08) |
| CRC035 | p.G12S | KR2Cdn12 (11.68) | p.G12S (23.72) |
| CRC062 | p.G12V | KR2Cdn12 (23.68) | p.G12V (50.37) |
| CRC095 | p.G12V | KR2Cdn12 (5.33) | p.G12V (8.87) |
| CRC100 | p.G13D | KR2Cdn13 (6.96) | p.G13D (14.18) |
| CRC106 | p.G12D | KR2Cdn12 (0.32) | ND |
| CRC112 | NA | KR2Cdn12 (4.38) | p.G12D (9.6) |
| CRC116 | p.G13D | KR2Cdn13 (30.12) | p.G13D (48.61) |
| CRC131 | p.G12V | KR2Cdn12 (12.10) | p.G12V (34.09) |
| CRC133 | NA | KR2Cdn13 (10.84) | p.G13D (19.19) |
| CRC137 | p.G12D | KR2Cdn12 (0.21) | ND |
| CRC142 | p.G13D | KR2Cdn13 (0.11) | ND |
| CRC145 | NA | KR2Cdn12 (0.70) | ND |
| CRC152 | p.G12D | KR2Cdn12 (3.35) | p.G12S (9.32) * |
| CRC154 | p.Q61L | KR3Cdn61 (20.17) | p.Q61L (43.83) |
| CRC158 | p.G12V | KR2Cdn12 (5.69) | p.G12V (12.53) |
| CRC160 | p.G12D | KR2Cdn12 (3.59) | p.G12A (7.04) * |
| CRC164 | WT | KR2Cdn13 (1.98) | p.G13D (4.48) * |
| CRC168 | p.G12A | KR2Cdn12 (3.16) | p.G12A (6.53) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Casanova, A.; Bao-Caamano, A.; Lago-Lestón, R.M.; Brozos-Vázquez, E.; Costa-Fraga, N.; Ferreirós-Vidal, I.; Abdulkader, I.; Vidal-Insua, Y.; Rivera, F.V.; Candamio Folgar, S.; et al. Evaluation of a Targeted Next-Generation Sequencing Panel for the Non-Invasive Detection of Variants in Circulating DNA of Colorectal Cancer. J. Clin. Med. 2021, 10, 4487. https://doi.org/10.3390/jcm10194487
Rodríguez-Casanova A, Bao-Caamano A, Lago-Lestón RM, Brozos-Vázquez E, Costa-Fraga N, Ferreirós-Vidal I, Abdulkader I, Vidal-Insua Y, Rivera FV, Candamio Folgar S, et al. Evaluation of a Targeted Next-Generation Sequencing Panel for the Non-Invasive Detection of Variants in Circulating DNA of Colorectal Cancer. Journal of Clinical Medicine. 2021; 10(19):4487. https://doi.org/10.3390/jcm10194487
Chicago/Turabian StyleRodríguez-Casanova, Aitor, Aida Bao-Caamano, Ramón M. Lago-Lestón, Elena Brozos-Vázquez, Nicolás Costa-Fraga, Isabel Ferreirós-Vidal, Ihab Abdulkader, Yolanda Vidal-Insua, Francisca Vázquez Rivera, Sonia Candamio Folgar, and et al. 2021. "Evaluation of a Targeted Next-Generation Sequencing Panel for the Non-Invasive Detection of Variants in Circulating DNA of Colorectal Cancer" Journal of Clinical Medicine 10, no. 19: 4487. https://doi.org/10.3390/jcm10194487
APA StyleRodríguez-Casanova, A., Bao-Caamano, A., Lago-Lestón, R. M., Brozos-Vázquez, E., Costa-Fraga, N., Ferreirós-Vidal, I., Abdulkader, I., Vidal-Insua, Y., Rivera, F. V., Candamio Folgar, S., López-López, R., Muinelo-Romay, L., & Diaz-Lagares, A. (2021). Evaluation of a Targeted Next-Generation Sequencing Panel for the Non-Invasive Detection of Variants in Circulating DNA of Colorectal Cancer. Journal of Clinical Medicine, 10(19), 4487. https://doi.org/10.3390/jcm10194487






