Thermal Pain Thresholds Are Significantly Associated with Plasma Proteins of the Immune System in Chronic Widespread Pain—An Exploratory Pilot Study Using Multivariate and Network Analyses
Abstract
:1. Introduction
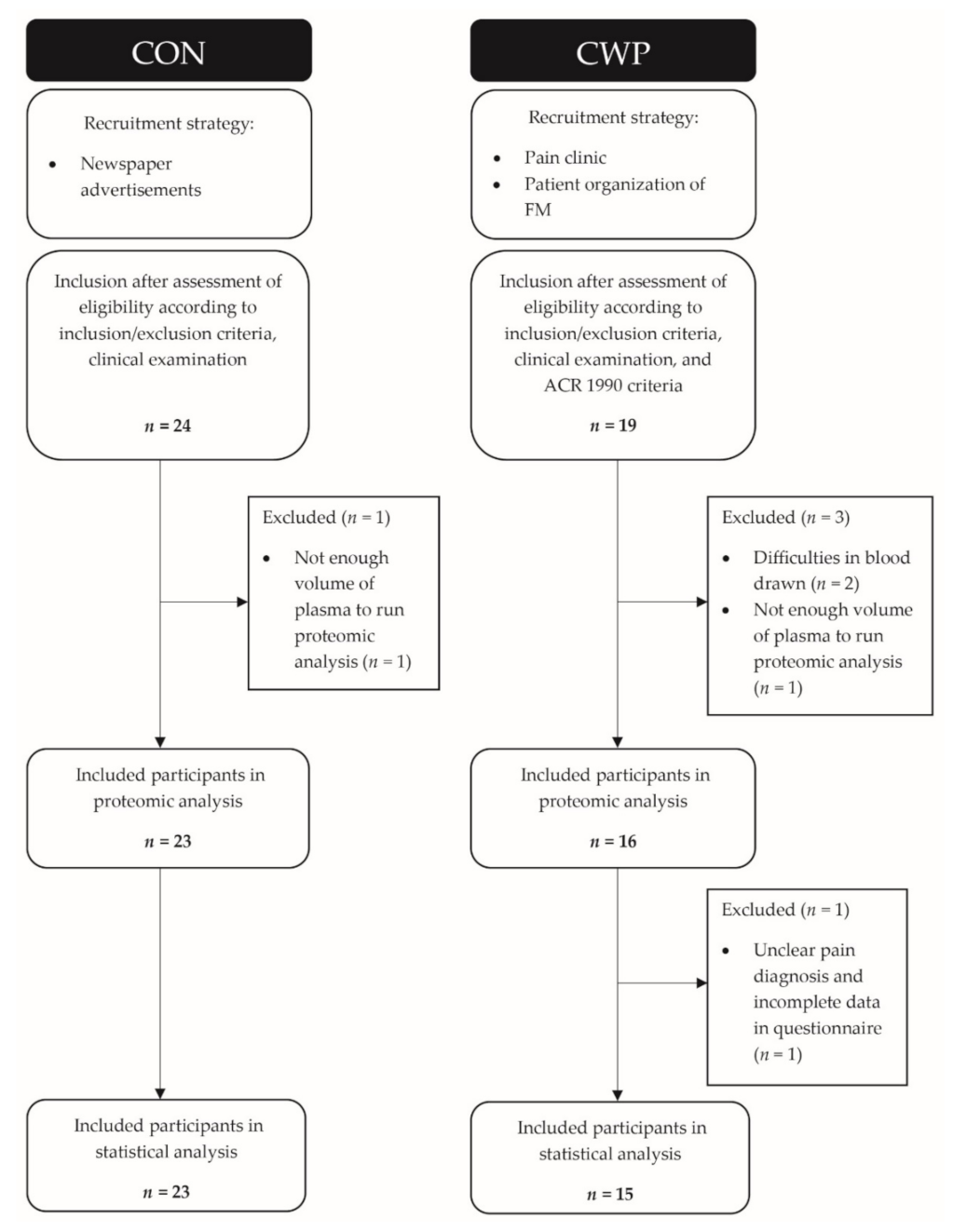
2. Materials and Methods
2.1. Subjects
2.2. Clinical Variables
2.3. Pain Thresholds for Cold and Heat
2.4. Proteins and Other Biochemical Substances
2.4.1. Sample Collection
Blood Sampling
Muscle Biopsy Sampling
2.4.2. Biochemical Analyses
Proteomics of Muscle Biopsy and Plasma—Two-Dimensional Gel Electrophoresis (2-DE)
Protein Identification
Database Search
Proximal Extension Assay for Identifying Cytokines, Chemokines, and Growth Factors
2.5. Statistics
2.6. Network Analysis
3. Results
3.1. Clinical Variables
3.2. Thermal Pain Thresholds
3.3. Clinical Variables as Regressors of CPT and HPT
3.4. Proteins as Regressors of CPT and HPT
3.4.1. CPT
3.4.2. HPT
3.5. Both Proteins and Clinical Variables as Regressors of CPT and HPT
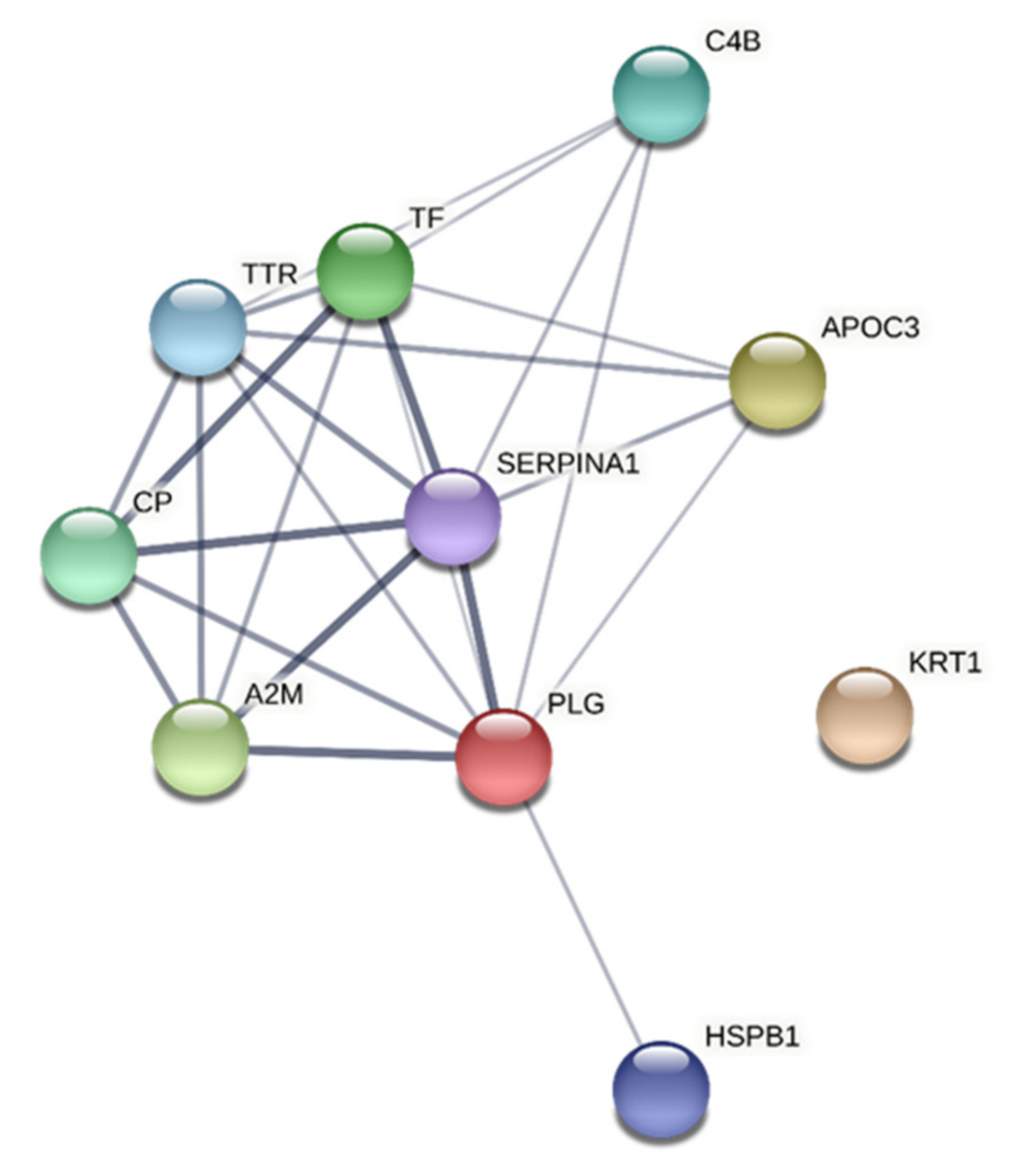
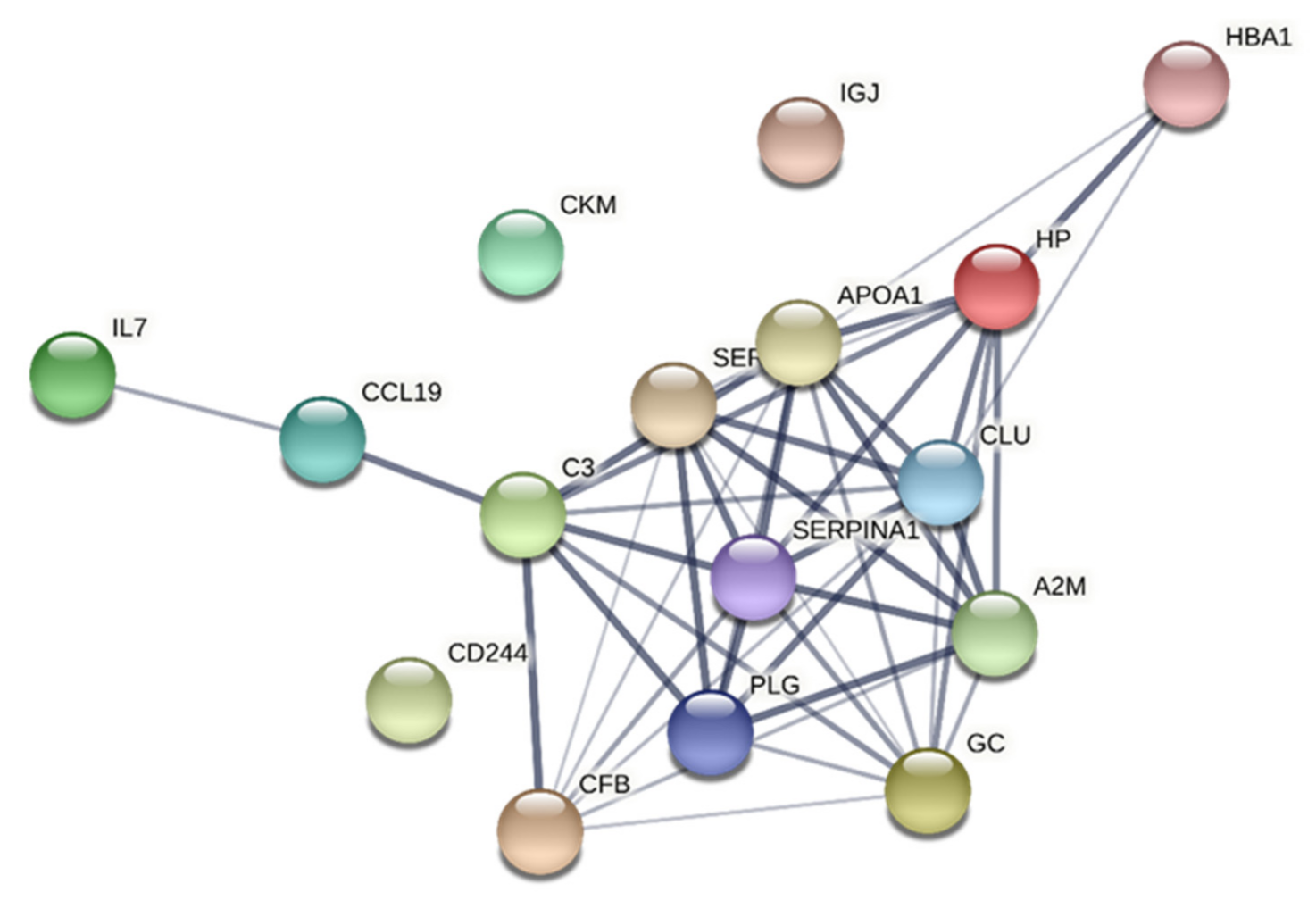
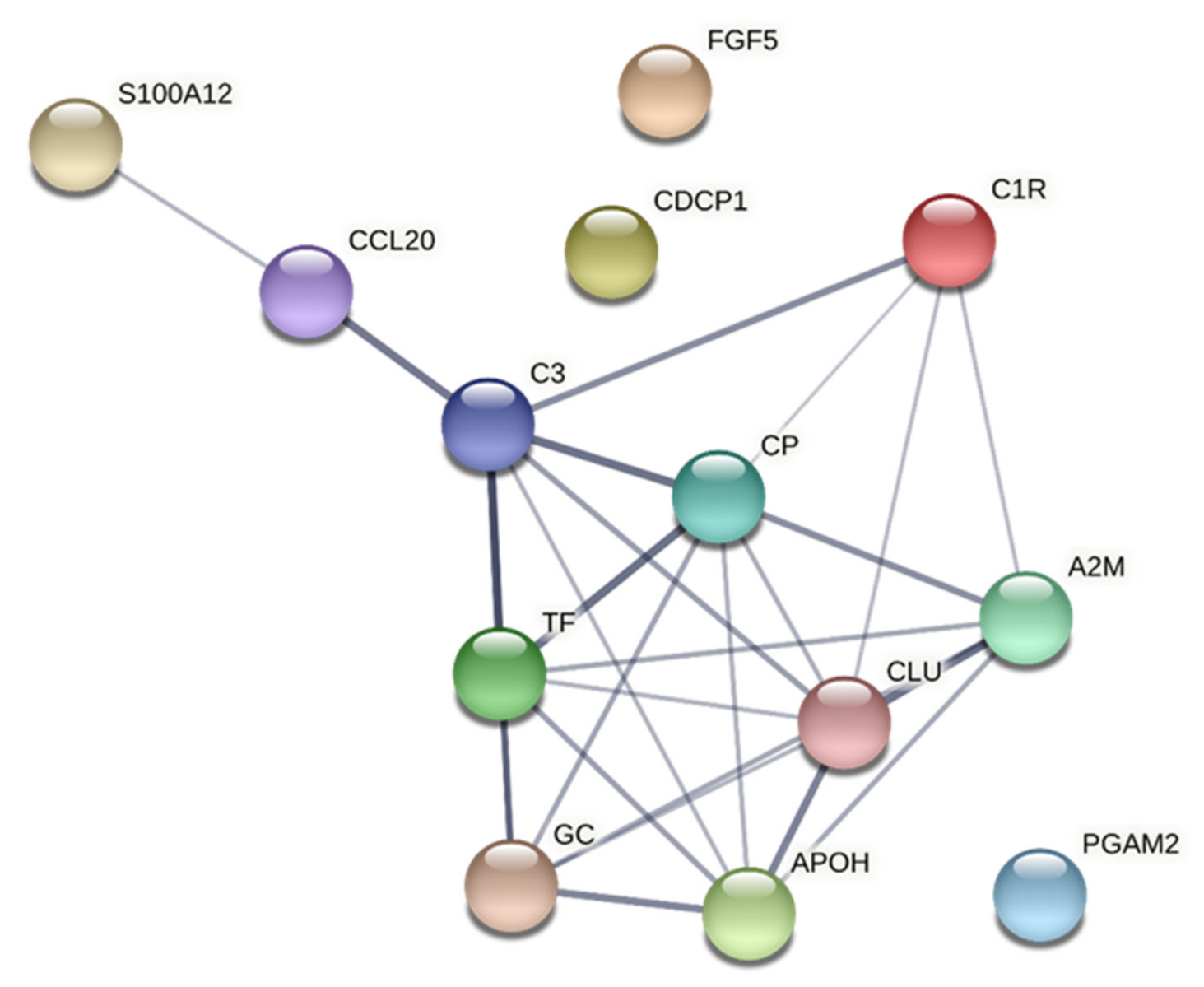
3.6. Network Analyses
3.6.1. CPT
3.6.2. HPT
4. Discussion
- Patients with CWP had lowered pain thresholds for thermal stimulus; these levels were generally not related to the included clinical variables except for HPT in CWP.
- Patterns of highly interacting proteins mainly from plasma showed strong associations with CPT and HPT both in CWP and in CON.
- Differences in the important proteins for the two thermal pain thresholds were noted between CWP and CON; more complex patterns emerged in CWP.
Strengths and Limitations
5. Conclusions and Clinical Implications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mansfield, K.E.; Sim, J.; Jordan, J.L.; Jordan, K.P. A systematic review and meta-analysis of the prevalence of chronic widespread pain in the general population. Pain 2016, 157, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, M.A.; Ferrone, C.; Cutolo, M. Epidemiology of chronic musculoskeletal pain. Best Pract. Res. Clin. Rheumatol. 2011, 25, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Bergman, S.; Herrstrom, P.; Hogstrom, K.; Petersson, I.F.; Svensson, B.; Jacobsson, L.T. Chronic musculoskeletal pain, prevalence rates, and sociodemographic associations in a Swedish population study. J. Rheumatol. 2001, 28, 1369–1377. [Google Scholar] [PubMed]
- Perez de Heredia-Torres, M.; Huertas-Hoyas, E.; Maximo-Bocanegra, N.; Palacios-Cena, D.; Fernandez-De-Las-Penas, C. Cognitive performance in women with fibromyalgia: A case-control study. Aust. Occup. Ther. J. 2016, 63, 329–337. [Google Scholar] [CrossRef]
- Aparicio, V.A.; Ortega, F.B.; Carbonell-Baeza, A.; Gatto-Cardia, C.; Sjostrom, M.; Ruiz, J.R.; Delgado-Fernandez, M. Fibromyalgia’s key symptoms in normal-weight, overweight, and obese female patients. Pain Manag. Nurs. 2013, 14, 268–276. [Google Scholar] [CrossRef]
- Wolfe, F.; Smythe, H.A.; Yunus, M.B.; Bennett, R.M.; Bombardier, C.; Goldenberg, D.L.; Tugwell, P.; Campbell, S.M.; Abeles, M.; Clark, P.; et al. The American College of Rheumatology 1990 Criteria for the Classification of Fibromyalgia. Report of the Multicenter Criteria Committee. Arthritis Rheum. 1990, 33, 160–172. [Google Scholar] [CrossRef]
- Breivik, H.; Collett, B.; Ventafridda, V.; Cohen, R.; Gallacher, D. Survey of chronic pain in Europe: Prevalence, impact on daily life, and treatment. Eur. J. Pain 2006, 10, 287–333. [Google Scholar] [CrossRef]
- Tracey, I.; Woolf, C.J.; Andrews, N.A. Composite Pain Biomarker Signatures for Objective Assessment and Effective Treatment. Neuron 2019, 101, 783–800. [Google Scholar] [CrossRef] [Green Version]
- Galvez-Sanchez, C.M.; Reyes Del Paso, G.A. Diagnostic Criteria for Fibromyalgia: Critical Review and Future Perspectives. J. Clin. Med. 2020, 9, 1219. [Google Scholar] [CrossRef]
- Wåhlén, K.; Olausson, P.; Carlsson, A.; Ghafouri, N.; Gerdle, B.; Ghafouri, B. Systemic alterations in plasma proteins from women with chronic widespread pain compared to healthy controls: A proteomic study. J. Pain Res. 2017, 10, 797–809. [Google Scholar] [CrossRef] [Green Version]
- Olausson, P.; Gerdle, B.; Ghafouri, N.; Larsson, B.; Ghafouri, B. Identification of proteins from interstitium of trapezius muscle in women with chronic myalgia using microdialysis in combination with proteomics. PLoS ONE 2012, 7, e52560. [Google Scholar] [CrossRef] [Green Version]
- Hadrevi, J.; Bjorklund, M.; Kosek, E.; Hallgren, S.; Antti, H.; Fahlstrom, M.; Hellstrom, F. Systemic differences in serum metabolome: A cross sectional comparison of women with localised and widespread pain and controls. Sci. Rep. 2015, 5, 15925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culic, O.; Cordero, M.D.; Zanic-Grubisic, T.; Somborac-Bacura, A.; Pucar, L.B.; Detel, D.; Varljen, J.; Barisic, K. Serum activities of adenosine deaminase, dipeptidyl peptidase IV and prolyl endopeptidase in patients with fibromyalgia: Diagnostic implications. Clin. Rheumatol. 2016, 35, 2565–2571. [Google Scholar] [CrossRef] [PubMed]
- Olausson, P.; Ghafouri, B.; Backryd, E.; Gerdle, B. Clear differences in cerebrospinal fluid proteome between women with chronic widespread pain and healthy women—A multivariate explorative cross-sectional study. J. Pain Res. 2017, 10, 575–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanette, S.A.; Dussan-Sarria, J.A.; Souza, A.; Deitos, A.; Torres, I.L.S.; Caumo, W. Higher serum S100B and BDNF levels are correlated with a lower pressure-pain threshold in fibromyalgia. Mol. Pain 2014, 10, 46. [Google Scholar] [CrossRef] [Green Version]
- Bazzichi, L.; Ciregia, F.; Giusti, L.; Baldini, C.; Giannaccini, G.; Giacomelli, C.; Sernissi, F.; Bombardieri, S.; Lucacchini, A. Detection of potential markers of primary fibromyalgia syndrome in human saliva. Proteom. Clin. Appl. 2009, 3, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Gerdle, B.; Ghafouri, B.; Ghafouri, N.; Backryd, E.; Gordh, T. Signs of ongoing inflammation in female patients with chronic widespread pain: A multivariate, explorative, cross-sectional study of blood samples. Medicine 2017, 96, e6130. [Google Scholar] [CrossRef]
- Jablochkova, A.; Backryd, E.; Kosek, E.; Mannerkorpi, K.; Ernberg, M.; Gerdle, B.; Ghafouri, B. Unaltered low nerve growth factor and high brain-derived neurotrophic factor levels in plasma from patients with fibromyalgia after a 15-week progressive resistance exercise. J. Rehabil. Med. 2019, 51, 779–787. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Varela, D.; Barry, A.M.; Schmidt, M. Proteome-based systems biology in chronic pain. J. Proteom. 2019, 190, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Niederberger, E.; Geisslinger, G. Proteomics in neuropathic pain research. J. Am. Soc. Anesthesiol. 2008, 108, 314–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emilsson, V.; Ilkov, M.; Lamb, J.R.; Finkel, N.; Gudmundsson, E.F.; Pitts, R.; Hoover, H.; Gudmundsdottir, V.; Horman, S.R.; Aspelund, T.; et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018, 361, 769–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Olausson, P.; Gerdle, B.; Ghafouri, N.; Sjostrom, D.; Blixt, E.; Ghafouri, B. Protein alterations in women with chronic widespread pain--An explorative proteomic study of the trapezius muscle. Sci. Rep. 2015, 5, 11894. [Google Scholar] [CrossRef] [Green Version]
- Han, C.L.; Sheng, Y.C.; Wang, S.Y.; Chen, Y.H.; Kang, J.H. Serum proteome profiles revealed dysregulated proteins and mechanisms associated with fibromyalgia syndrome in women. Sci. Rep. 2020, 10, 12347. [Google Scholar] [CrossRef]
- Wåhlén, K.; Ernberg, M.; Kosek, E.; Mannerkorpi, K.; Gerdle, B.; Ghafouri, B. Significant correlation between plasma proteome profile and pain intensity, sensitivity, and psychological distress in women with fibromyalgia. Sci. Rep. 2020, 10, 12508. [Google Scholar] [CrossRef]
- Ramirez-Tejero, J.A.; Martinez-Lara, E.; Rus, A.; Camacho, M.V.; Del Moral, M.L.; Siles, E. Insight into the biological pathways underlying fibromyalgia by a proteomic approach. J. Proteom. 2018, 186, 47–55. [Google Scholar] [CrossRef]
- Olausson, P.; Ghafouri, B.; Ghafouri, N.; Gerdle, B. Specific proteins of the trapezius muscle correlate with pain intensity and sensitivity—An explorative multivariate proteomic study of the trapezius muscle in women with chronic widespread pain. J. Pain Res. 2016, 9, 345–356. [Google Scholar] [CrossRef] [Green Version]
- Wåhlén, K.; Ghafouri, B.; Ghafouri, N.; Gerdle, B. Plasma Protein Pattern Correlates With Pain Intensity and Psychological Distress in Women With Chronic Widespread Pain. Front. Psychol. 2018, 9, 2400. [Google Scholar] [CrossRef]
- Kosek, E.; Cohen, M.; Baron, R.; Gebhart, G.F.; Mico, J.A.; Rice, A.S.; Rief, W.; Sluka, A.K. Do we need a third mechanistic descriptor for chronic pain states? Pain 2016, 157, 1382–1386. [Google Scholar] [CrossRef]
- Gerdle, B.; Bäckryd, E.; Novo, M.; Roeck Hansen, E.; Rothman, M.; Stålnacke, B.-M.; Westergren, H.; Rivano Fischer, M. Smärtanalys—Diagnos, Smärtmekanismer, Psykologisk Och Social Bedömning; Studentlitteratur AB: Lund, Sweden, 2020. [Google Scholar]
- Nijs, J.; George, S.; Clauw, D.; Fernández-de-las-Peñas, C.; Kosek, E.; Ickmans, K.; Fernández-Carnero, J.; Polli, A.; Kapreli, E.; Huysmans, E.; et al. Central sensitisation in chronic pain conditions: Latest discoveries and their potential for precision medicine. Lancet Rheumatol. 2021, 3, e383–e392. [Google Scholar] [CrossRef]
- Kosek, E.; Clauw, D.; Nijs, J.; Baron, R.; Gilron, I.; Harris, R.E.; Mico, J.A.; Rice, A.S.; Sterling, M. Chronic nociplastic pain affecting the musculoskeletal system: Clinical criteria and grading system. Pain 2021, in press. [Google Scholar] [CrossRef]
- Potvin, S.; Marchand, S. Pain facilitation and pain inhibition during conditioned pain modulation in fibromyalgia and in healthy controls. Pain 2016, 157, 1704–1710. [Google Scholar] [CrossRef]
- Gerdle, B.; Wåhlén, K.; Ghafouri, B. Plasma protein patterns are strongly correlated with pressure pain thresholds in women with chronic widespread pain and in healthy controls-an exploratory case-control study. Medicine 2020, 99, e20497. [Google Scholar] [CrossRef]
- Muller, M.; Butikofer, L.; Andersen, O.K.; Heini, P.; Arendt-Nielsen, L.; Juni, P.; Curatolo, M. Cold pain hypersensitivity predicts trajectories of pain and disability after low back surgery: A prospective cohort study. Pain 2021, 162, 184–194. [Google Scholar] [CrossRef]
- Gerdle, B.; Ghafouri, B. Proteomic studies of common chronic pain conditions—A systematic review and associated network analyses. Expert Rev. Proteom. 2020, 17, 483–505. [Google Scholar] [CrossRef]
- Wallin, M.; Liedberg, G.; Börsbo, B.; Gerdle, B. Thermal detection and pain thresholds but not pressure pain thresholds are correlated with psychological factors in women with chronic whiplash-associated pain. Clin. J. Pain 2012, 28, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Grundstrom, H.; Larsson, B.; Arendt-Nielsen, L.; Gerdle, B.; Kjolhede, P. Pain catastrophizing is associated with pain thresholds for heat, cold and pressure in women with chronic pelvic pain. Scand. J. Pain 2020, 20, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Gerdle, B.; Larsson, B.; Forsberg, F.; Ghafouri, N.; Karlsson, L.; Stensson, N.; Ghafouri, B. Chronic widespread pain: Increased glutamate and lactate concentrations in the trapezius muscle and plasma. Clin. J. Pain 2014, 30, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Gerdle, B.; Soderberg, K.; Salvador Puigvert, L.; Rosendal, L.; Larsson, B. Increased interstitial concentrations of pyruvate and lactate in the trapezius muscle of patients with fibromyalgia: A microdialysis study. J. Rehabil. Med. 2010, 42, 679–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghafouri, N.; Ghafouri, B.; Larsson, B.; Stensson, N.; Fowler, C.J.; Gerdle, B. Palmitoylethanolamide and stearoylethanolamide levels in the interstitium of the trapezius muscle of women with chronic widespread pain and chronic neck-shoulder pain correlate with pain intensity and sensitivity. Pain 2013, 154, 1649–1658. [Google Scholar] [CrossRef]
- Ferreira-Valente, M.A.; Pais-Ribeiro, J.L.; Jensen, M.P. Validity of four pain intensity rating scales. Pain 2011, 152, 2399–2404. [Google Scholar] [CrossRef] [PubMed]
- Zigmond, A.S.; Snaith, R.P. The hospital anxiety and depression scale. Acta Psychiatr. Scand. 1983, 67, 361–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LoMartire, R.; Äng, B.; Gerdle, B.; Vixner, L. Psychometric properties of SF-36, EQ-5D and HADS in patients with chronic pain. Pain 2020, 161, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.L.; Bishop, S.R.; Pivik, J. The Pain Catastrophizing Scale: Development and validation. Psychol. Assess. 1995, 7, 524–532. [Google Scholar] [CrossRef]
- Burckhardt, C.S.; Anderson, K.L. The Quality of Life Scale (QOLS): Reliability, validity, and utilization. Health Qual. Life Outcomes 2003, 1, 60. [Google Scholar] [CrossRef] [Green Version]
- Raak, R.; Wallin, M. Thermal Thresholds and Catastrophizing in Individuals with Chronic Pain after Whiplash Injury. Biol. Res. Nurs. 2006, 8, 138–146. [Google Scholar] [CrossRef]
- Wallin, M.K.; Raak, R.I. Quality of life in subgroups of individuals with whiplash associated disorders. Eur. J. Pain 2008, 12, 842–849. [Google Scholar] [CrossRef]
- Fruhstorfer, H.; Lindblom, U.; Schmidt, W. Method for quantitative estimation of thermal thresholds in patients. J. Neurol. Neurosurg. Psychiatry 1976, 39, 1071–1075. [Google Scholar] [CrossRef] [Green Version]
- Gorg, A.; Drews, O.; Luck, C.; Weiland, F.; Weiss, W. 2-DE with IPGs. Electrophoresis 2009, 30 (Suppl. S1), S122–S132. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, A.M.; Wheelock, C.E. Trials and tribulations of omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. Biosyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikström, C. Multi-and Megavariate Data Analysis: Basic Principles and Applications; Third Revised Edition; MKS Umetrics AB: Malmö, Sweden, 2013. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Blumenstiel, K.; Gerhardt, A.; Rolke, R.; Bieber, C.; Tesarz, J.; Friederich, H.C.; Eich, W.; Treede, R.D. Quantitative sensory testing profiles in chronic back pain are distinct from those in fibromyalgia. Clin. J. Pain 2011, 27, 682–690. [Google Scholar] [CrossRef]
- Hurtig, I.M.; Raak, R.I.; Kendall, S.A.; Gerdle, B.; Wahren, L.K. Quantitative sensory testing in fibromyalgia patients and in healthy subjects: Identification of subgroups. Clin. J. Pain 2001, 17, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.W.; Tooley, E.M.; Montague, E.Q.; Robinson, A.E.; Cosper, C.J.; Mullins, P.G. Habituation and sensitization to heat and cold pain in women with fibromyalgia and healthy controls. Pain 2008, 140, 420–428. [Google Scholar] [CrossRef]
- Brietzke, A.P.; Antunes, L.C.; Carvalho, F.; Elkifury, J.; Gasparin, A.; Sanches, P.R.S.; da Silva Junior, D.P.; Dussan-Sarria, J.A.; Souza, A.; da Silva Torres, I.L.; et al. Potency of descending pain modulatory system is linked with peripheral sensory dysfunction in fibromyalgia: An exploratory study. Medicine 2019, 98, e13477. [Google Scholar] [CrossRef] [PubMed]
- Rehm, S.; Sachau, J.; Hellriegel, J.; Forstenpointner, J.; Borsting Jacobsen, H.; Harten, P.; Gierthmuhlen, J.; Baron, R. Pain matters for central sensitization: Sensory and psychological parameters in patients with fibromyalgia syndrome. Pain Rep. 2021, 6, e901. [Google Scholar] [CrossRef] [PubMed]
- Grundstrom, H.; Gerdle, B.; Alehagen, S.; Bertero, C.; Arendt-Nielsen, L.; Kjolhede, P. Reduced pain thresholds and signs of sensitization in women with persistent pelvic pain and suspected endometriosis. Acta Obstet. Gynecol. Scand. 2019, 98, 327–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, Y.; Zhang, B.; Cao, J.; Wilson, G.; Zhang, Z.; Kong, J. Identifying inter-individual differences in pain threshold using brain connectome: A test-retest reproducible study. Neuroimage 2019, 202, 116049. [Google Scholar] [CrossRef]
- Jin, P.; Lan, J.; Wang, K.; Baker, M.S.; Huang, C.; Nice, E.C. Pathology, proteomics and the pathway to personalised medicine. Expert Rev. Proteom. 2018, 15, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Khoonsari, P.E.; Musunri, S.; Herman, S.; Svensson, C.I.; Tanum, L.; Gordh, T.; Kultima, K. Systematic analysis of the cerebrospinal fluid proteome of fibromyalgia patients. J. Proteom. 2019, 190, 35–43. [Google Scholar] [CrossRef]
- Bäckryd, E.; Tanum, L.; Lind, A.L.; Larsson, A.; Gordh, T. Evidence of both systemic inflammation and neuroinflammation in fibromyalgia patients, as assessed by a multiplex protein panel applied to the cerebrospinal fluid and to plasma. J. Pain Res. 2017, 10, 515–525. [Google Scholar] [CrossRef] [Green Version]
- Obreja, O.; Rukwied, R.; Nagler, L.; Schmidt, M.; Schmelz, M.; Namer, B. Nerve growth factor locally sensitizes nociceptors in human skin. Pain 2018, 159, 416–426. [Google Scholar] [CrossRef]
- Yurkovich, J.T.; Hood, L. Blood Is a Window into Health and Disease. Clin. Chem. 2019, 65, 1204–1206. [Google Scholar] [CrossRef] [PubMed]
- Bruderer, R.; Muntel, J.; Muller, S.; Bernhardt, O.M.; Gandhi, T.; Cominetti, O.; Macron, C.; Carayol, J.; Rinner, O.; Astrup, A.; et al. Analysis of 1508 Plasma Samples by Capillary-Flow Data-Independent Acquisition Profiles Proteomics of Weight Loss and Maintenance. Mol. Cell. Proteom. 2019, 18, 1242–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baral, P.; Udit, S.; Chiu, I.M. Pain and immunity: Implications for host defence. Nat. Rev. Immunol. 2019, 19, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.V.; Soulika, A.M. The Dynamics of the Skin’s Immune System. Int. J. Mol. Sci. 2019, 20, 1811. [Google Scholar] [CrossRef] [Green Version]
- Kabashima, K.; Honda, T.; Ginhoux, F.; Egawa, G. The immunological anatomy of the skin. Nat. Rev. Immunol. 2019, 19, 19–30. [Google Scholar] [CrossRef]
- Krausgruber, T.; Fortelny, N.; Fife-Gernedl, V.; Senekowitsch, M.; Schuster, L.C.; Lercher, A.; Nemc, A.; Schmidl, C.; Rendeiro, A.F.; Bergthaler, A.; et al. Structural cells are key regulators of organ-specific immune responses. Nature 2020, 583, 296–302. [Google Scholar] [CrossRef]
- Rice, F.L.; Kruger, L.; Albrecht, P.J. Histology of Nociceptors. In Encyclopedia of Pain; Gebhart, G.F., Schmidt, R.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1482–1490. [Google Scholar]
- Lacagnina, M.; Heijnen, C.; Watkins, L.; Grace, P. Autoimmune regulation of chronic pain. Pain Rep. 2021, 6, e905. [Google Scholar] [CrossRef]
- Abdo, H.; Calvo-Enrique, L.; Lopez, J.M.; Song, J.; Zhang, M.D.; Usoskin, D.; El Manira, A.; Adameyko, I.; Hjerling-Leffler, J.; Ernfors, P. Specialized cutaneous Schwann cells initiate pain sensation. Science 2019, 365, 695–699. [Google Scholar] [CrossRef]
- Bray, E.R.; Cheret, J.; Yosipovitch, G.; Paus, R. Schwann cells as underestimated, major players in human skin physiology and pathology. Exp. Dermatol. 2020, 29, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Karshikoff, B.; Tadros, M.; Mackey, S.; Zouikr, I. Neuroimmune modulation of pain across the developmental spectrum. Curr. Opin. Behav. Sci. 2019, 28, 85–92. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coskun Benlidayi, I. Role of inflammation in the pathogenesis and treatment of fibromyalgia. Rheumatol. Int. 2019, 39, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Goncalves Dos Santos, G.; Delay, L.; Yaksh, T.L.; Corr, M. Neuraxial Cytokines in Pain States. Front. Immunol. 2019, 10, 3061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerdle, B.; Forsgren, M.; Bengtsson, A.; Dahlqvist Leinhard, O.; Sören, B.; Karlsson, A.; Brandejsky, V.; Lund, E.; Lundberg, P. Decreased muscle concentrations of ATP and PCR in the quadriceps muscle of fibromyalgia patients—A 31P MRS study. Eur. J. Pain 2013, 17, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Phothimat, P.; Oates, C.T.; Hernanz-Schulman, M.; Olsen, N.J. Use of P-31 magnetic resonance spectroscopy to detect metabolic abnormalities in muscles of patients with fibromyalgia. Arthritis Rheum. 1998, 41, 406–413. [Google Scholar] [CrossRef]
- Gerdle, B.; Ghafouri, B.; Lund, E.; Bengtsson, A.; Lundberg, P.; Ettinger-Veenstra, H.V.; Leinhard, O.D.; Forsgren, M.F. Evidence of Mitochondrial Dysfunction in Fibromyalgia: Deviating Muscle Energy Metabolism Detected Using Microdialysis and Magnetic Resonance. J. Clin. Med. 2020, 9, 3527. [Google Scholar] [CrossRef]
- Sluka, K.A.; Clauw, D.J. Neurobiology of fibromyalgia and chronic widespread pain. Neuroscience 2016, 338, 114–129. [Google Scholar] [CrossRef] [Green Version]
- Schrepf, A.; Harper, D.E.; Harte, S.E.; Wang, H.; Ichesco, E.; Hampson, J.P.; Zubieta, J.K.; Clauw, D.J.; Harris, R.E. Endogenous opioidergic dysregulation of pain in fibromyalgia: A PET and fMRI study. Pain 2016, 157, 2217–2225. [Google Scholar] [CrossRef] [Green Version]
- Üçeyler, N.; Sommer, C. Small Nerve Fiber Pathology. In Fibromylagia Syndrome and Widespread Pain—From Construction to Relevant Recognition; Häuser, W., Perrot, S., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2018; pp. 204–214. [Google Scholar]
- Gerdle, B.; Larsson, B. Muscle. In Fibromyalgia Syndrome and Widespread Pain—From Construction to Relevant Recognition; Häuser, W., Perrot, S., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2018; pp. 215–231. [Google Scholar]
- Albrecht, D.S.; Forsberg, A.; Sandstrom, A.; Bergan, C.; Kadetoff, D.; Protsenko, E.; Lampa, J.; Lee, Y.C.; Hoglund, C.O.; Catana, C.; et al. Brain glial activation in fibromyalgia—A multi-site positron emission tomography investigation. Brain Behav. Immun. 2019, 75, 72–83. [Google Scholar] [CrossRef]
- Jensen, K.B.; Kosek, E.; Petzke, F.; Carville, S.; Fransson, P.; Marcus, H.; Williams, S.C.R.; Choy, E.; Giesecke, T.; Mainguy, Y.; et al. Evidence of dysfunctional pain inhibition in Fibromyalgia reflected in rACC during provoked pain. Pain 2009, 144, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Littlejohn, G.; Guymer, E. Neurogenic inflammation in fibromyalgia. Semin. Immunopathol. 2018, 40, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Van Ettinger-Veenstra, H.; Lundberg, P.; Alfoldi, P.; Sodermark, M.; Graven-Nielsen, T.; Sjors, A.; Engstrom, M.; Gerdle, B. Chronic widespread pain patients show disrupted cortical connectivity in default mode and salience networks, modulated by pain sensitivity. J. Pain Res. 2019, 12, 1743–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goubert, D.; Danneels, L.; Graven-Nielsen, T.; Descheemaeker, F.; Meeus, M. Differences in Pain Processing Between Patients with Chronic Low Back Pain, Recurrent Low Back Pain, and Fibromyalgia. Pain Physician 2017, 20, 307–318. [Google Scholar]
- Evdokimov, D.; Frank, J.; Klitsch, A.; Unterecker, S.; Warrings, B.; Serra, J.; Papagianni, A.; Saffer, N.; Meyer Zu Altenschildesche, C.; Kampik, D.; et al. Reduction of skin innervation is associated with a severe fibromyalgia phenotype. Ann. Neurol. 2019, 86, 504–516. [Google Scholar] [CrossRef] [Green Version]
- Fasolino, A.; Di Stefano, G.; Leone, C.; Galosi, E.; Gioia, C.; Lucchino, B.; Terracciano, A.; Di Franco, M.; Cruccu, G.; Truini, A. Small-fibre pathology has no impact on somatosensory system function in patients with fibromyalgia. Pain 2020, 161, 2385–2393. [Google Scholar] [CrossRef]
- Sawaddiruk, P.; Paiboonworachat, S.; Chattipakorn, N.; Chattipakorn, S.C. Alterations of brain activity in fibromyalgia patients. J. Clin. Neurosci. 2017, 38, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Iordanova Schistad, E.; Kong, X.Y.; Furberg, A.S.; Backryd, E.; Grimnes, G.; Emaus, N.; Rosseland, L.A.; Gordh, T.; Stubhaug, A.; Engdahl, B.; et al. A population-based study of inflammatory mechanisms and pain sensitivity. Pain 2020, 161, 338–350. [Google Scholar] [CrossRef] [PubMed]

| Group | CON | n = 23 | CWP | n = 15 | Statistics |
|---|---|---|---|---|---|
| Variables | Mean | SD | Mean | SD | p-Value |
| Age (y) | 41.0 | 10.2 | 49.2 | 8.9 | 0.014 |
| BMI (kg/m2) | 24.0 | 2.8 | 26.0 | 5.0 | 0.185 |
| NRS-neck | 0.0 | 0.0 | 5.7 | 2.4 | <0.001 |
| NRS-shoulders | 0.0 | 0.0 | 5.7 | 1.9 | <0.001 |
| NRS-whole body | 0.0 | 0.0 | 4.9 | 2.0 | <0.001 |
| HADS | 3.3 | 2.8 | 14.0 | 5.3 | <0.001 |
| PCS | 6.4 | 6.4 | 13.0 | 7.5 | 0.010 |
| QOLS | 93.1 | 9.7 | 82.5 | 13.1 | 0.013 |
| CPT (°C) | 11.6 | 4.0 | 15.8 | 5.7 | 0.020 |
| HPT (°C) | 48.0 | 1.4 | 46.6 | 2.0 | 0.027 |
| Spot No. | Accession No. | Protein | VIPpred | p(corr) | Characterization |
|---|---|---|---|---|---|
| 3420 | P01009 | Alpha-1-antitrypsin | 2.16 | −0.45 | Acute phase protein |
| 3712 | P01009 | Alpha-1-antitrypsin | 2.14 | 0.44 | Acute phase protein |
| 5811 | P01023 | Alpha-2-macroglobulin | 3.12 | 0.64 | Acute phase protein |
| 4902 | P01023 | Alpha-2-macroglobulin | 2.19 | 0.45 | Acute phase protein |
| 1055 | P02656 | Apolipoprotein C-III | 3.42 | 0.73 | Antiinflammation and enhance immunity |
| 1054 | P02656 | Apolipoprotein C-III | 2.15 | 0.46 | Antiinflammation and enhance immunity |
| 4814 | P00450 | Ceruloplasmin | 3.20 | 0.67 | Acute phase protein |
| 3817 | P00450 | Ceruloplasmin | 2.78 | 0.56 | Acute phase protein |
| 3904 | P00450 | Ceruloplasmin | 2.32 | 0.47 | Acute phase protein |
| 4832 | P00450 | Ceruloplasmin | 2.20 | 0.43 | Acute phase protein |
| 4816 | P00450 | Ceruloplasmin | 2.19 | 0.44 | Acute phase protein |
| 7216 | P0C0L5 | Complement C4-B | 2.95 | 0.60 | Complement factors |
| 8101 | P0C0L5 | Complement C4-B | 2.57 | 0.53 | Complement factors |
| B3540 | P04792 | Heat shock protein beta-1 | 2.59 | 0.63 | Protective |
| B1834 | P04264 | Keratin, type II cytoskeletal I | 2.99 | 0.82 | Mitochondrial function |
| 8901 | P00747 | Plasminogen | 2.53 | 0.52 | Acute phase protein |
| 7820 | P00747 | Plasminogen | 2.51 | 0.52 | Acute phase protein |
| 7819 | P00747 | Plasminogen | 2.42 | 0.50 | Acute phase protein |
| 7702 | P02787 | Serotransferrin | 2.29 | 0.50 | Acute phase protein |
| 1003 | P02766 | Transthyretin | 2.38 | 0.49 | Acute phase protein |
| R2 = 0.94 | |||||
| Q2 = 0.76 | |||||
| CV-ANOVA p = 0.0047 |
| Spot No. | Accession No. | Protein | VIPpred | p(corr) | Characterization |
|---|---|---|---|---|---|
| 3712 | P01009 | Alpha-1-antitrypsin | 2.17 | 0.58 | Acute phase protein + |
| 5906 | P01023 | Alpha-2-macroglobulin | 2.62 | 0.70 | Acute phase protein + |
| 4608 | P08697 | Alpha-2-antiplasmin | 2.10 | 0.56 | Acute phase protein + |
| 3606 | P08697 | Alpha-2-antiplasmin | 1.98 | 0.53 | Acute phase protein + |
| 1102 | P02647 | Apolipoprotein A-I | 2.81 | −0.75 | Antiinflammation |
| NA | Q99731 | CCL19 | 2.32 | 0.66 | Chemokine |
| NA | Q9BZW8 | CD244 | 2.23 | 0.63 | Immunity, Ig superfamily |
| 3214 | P10909 | Clusterin | 2.73 | −0.73 | Neuroprotective, etc. |
| 7901 | P00751 | Complement factor B | 2.48 | 0.66 | Complement factors |
| 6841 | P01024 | Complement C3c alpha chain | 1.98 | 0.53 | Complement factors |
| 145 | P01024 | Complement C3c alpha chain fragment 2 | 1.92 | −0.52 | Complement factors |
| B5736 | P06732 | Creatine kinase M-type | 2.46 | 0.66 | ATP production |
| 3105 | P00738 | Haptoglobin | 2.06 | −0.55 | Acute phase protein + |
| 9009 | P69905 | Haemoglobin subunit alpha | 1.98 | −0.53 | Blood—haemoglobin + |
| 9001 | P01834 | Ig kappa chain C region | 2.63 | −0.71 | Immunoglobulin |
| 166 | P01591 | Ig J chain | 2.59 | −0.69 | Immunoglobulin |
| NA | P13232 | IL-7 | 1.97 | 0.56 | Cytokine |
| 8901 | P00747 | Plasminogen | 1.99 | 0.53 | Acute phase protein + |
| 3408 | P02774 | Vitamin D-binding protein | 2.43 | −0.65 | Immunity, etc. |
| 2502 | P02774 | Vitamin D-binding protein | 2.05 | 0.55 | Immunity, etc. |
| R2 = 0.88 | |||||
| Q2 = 0.81 | |||||
| CV-ANOVA p < 0.001 |
| Spot No. | Accession No. | Protein | VIPpred | p(corr) | Characterization |
|---|---|---|---|---|---|
| 5811 | P01023 | Alpha-2-macroglobulin | 2.77 | −0.59 | Acute phase protein |
| 4902 | P01023 | Alpha-2-macroglobulin | 2.65 | −0.58 | Acute phase protein |
| 5821 | P01023 | Alpha-2-macroglobulin | 2.01 | −0.43 | Acute phase protein |
| 6528 | P02749 | Beta-2-glycoprotein 1 | 2.40 | 0.51 | Immunity, regulation of complement/coagulation |
| NA | P78556 | CCL20 | 2.05 | −0.48 | Chemokine |
| NA | Q9H5V8 | CDCP1 | 2.11 | 0.50 | Cytokine |
| 4830 | P00450 | Ceruloplasmin | 2.59 | −0.53 | Acute phase protein |
| 3817 | P00450 | Ceruloplasmin | 2.45 | −0.51 | Acute phase protein |
| 4832 | P00450 | Ceruloplasmin | 2.13 | −0.44 | Acute phase protein |
| 1105 | P10909 | Clusterin | 2.21 | −0.46 | Neuroprotective, etc. |
| 4914 | P00736 | Complement C1r subcomponent | 2.15 | −0.44 | Complement factors |
| 6842 | P01024 | Complement C3 alpha chain | 2.62 | 0.55 | Complement factors |
| NA | P80511 | EN-RAGE | 2.04 | 0.48 | Immunity, etc. |
| NA | P12034 | FGF-5 | 2.09 | 0.48 | Tissue repair and regulator Schwann cells |
| 5511 | P01877 | Ig alpha-2 chain C region | 2.28 | 0.46 | Immunoglobulin |
| 8001 | P01834 | Ig kappa chain C region | 2.05 | −0.43 | Immunoglobulin |
| B7525 | P15259 | Phosphoglycerate mutase 2 | 2.02 | −0.51 | Glycolytic pathway |
| 7744 | P02787 | Serotransferrin | 2.51 | 0.54 | Acute phase protein |
| 7731 | P02787 | Serotransferrin | 2.84 | 0.61 | Acute phase protein |
| 3408 | P02774 | Vitamin D-binding protein | 2.19 | −0.47 | Immunity, etc. |
| R2 = 0.77 | |||||
| Q2 = 0.63 | |||||
| CV-ANOVA p < 0.001 |
| Spot No. | Accession No. | Protein | VIPpred | p(corr) | Characterization |
|---|---|---|---|---|---|
| 3619 | P08697 | Alpha-2-antiplasmin | 2.02 | 0.51 | Acute phase protein |
| 6903 | P01023 | Alpha-2-macroglobulin | 2.02 | 0.51 | Acute phase protein |
| 1102 | P02647 | Apolipoprotein A-I | 2.00 | 0.51 | Antiinflammation |
| 3214 | P10909 | Clusterin | 2.71 | 0.69 | Neuroprotective, etc. |
| 5819 | P00736 | Complement C1r subcomponent | 1.96 | −0.50 | Complement factors |
| 6844 | P10643 | Complement component C7 | 2.33 | 0.59 | Complement factors |
| 6845 | P10643 | Complement component C7 | 1.97 | 0.50 | Complement factors |
| B5736 | P06732 | Creatine kinase M-type | 2.09 | −0.53 | ATP production |
| NA | P09603 | CSF-1 | 2.48 | −0.66 | Cytokine |
| NA | Q9NSA1 | FGF-21 | 2.02 | −0.54 | Cytokine |
| 8621 | P02671 | Fibrinogen alpha chain | 2.35 | −0.60 | Acute phase protein |
| NA | P49771 | FIt3L | 2.35 | −0.62 | Cytokine |
| 9006 | Q0KKI6 | Ig light chain | 2.15 | 0.55 | Immunoglobulin |
| 9008 | Q0KKI6 | Ig light chain | 2.18 | 0.56 | Immunoglobulin |
| NA | Q08334 | IL-10RB | 2.56 | −0.68 | Cytokine |
| NA | P20783 | Neurotrophin 3 | 1.97 | −0.52 | Neurons- repair and growth |
| B7523 | P15259 | Phosphoglycerate mutase 2 | 1.97 | 0.50 | Glycolytic pathway |
| NA | Q07011 | TNFRSF9 | 2.02 | −0.53 | Inflammation development |
| NA | P15692 | VEGF-A | 1.99 | −0.53 | Neuroprotective and pro-nociceptive |
| 2502 | P02774 | Vitamin D-binding protein | 1.97 | −0.50 | Immunity, etc. |
| R2 = 0.77 | |||||
| Q2 = 0.61 | |||||
| CV-ANOVA p = 0.0033 |
| Category. | Term ID | Term Description | Gene Count | Strength | FDR | Matching Proteins in Network |
|---|---|---|---|---|---|---|
| CC | GO:0005576 | extracellular region | 9 | 0.85 | 1.08 × 10−5 | APOC3, TTR, KRT1, CP, PLG, A2M, TF, C4B, SERPINA1 |
| CC | GO:0034774 | secretory granule lumen | 5 | 1.48 | 1.96 × 10−5 | TTR, PLG, A2M, TF, SERPINA1 |
| CC | GO:0030141 | secretory granule | 6 | 1.15 | 2.77 × 10−5 | TTR, KRT1, PLG, A2M, TF, SERPINA1 |
| CC | GO:0031093 | platelet alpha granule lumen | 3 | 1.94 | 0.00010 | PLG, A2M, SERPINA1 |
| CC | GO:0031232 | extrinsic component of external side of plasma membrane | 2 | 2.75 | 0.00014 | PLG, TF |
| CC | GO:0031410 | cytoplasmic vesicle | 7 | 0.79 | 0.00028 | APOC3, TTR, KRT1, PLG, A2M, TF, SERPINA1 |
| MF | GO:0004857 | enzyme inhibitor activity | 5 | 1.4 | 7.89 × 10−5 | APOC3, HSPB1, A2M, C4B, SERPINA1 |
| MF | GO:0005515 | protein binding | 10 | 0.47 | 0.0010 | APOC3, TTR, HSPB1, KRT1, CP, PLG, A2M, TF, C4B, SERPINA1 |
| BP | GO:0002576 | platelet degranulation | 4 | 1.78 | 0.00011 | PLG, A2M, TF, SERPINA1 |
| BP | GO:0006810 | transport | 10 | 0.68 | 0.00011 | APOC3, TTR, HSPB1, KRT1, CP, PLG, A2M, TF, C4B, SERPINA1 |
| BP | GO:0045055 | regulated exocytosis | 6 | 1.23 | 0.00011 | TTR, KRT1, PLG, A2M, TF, SERPINA1 |
| BP | GO:0065008 | regulation of biological quality | 9 | 0.69 | 0.00018 | APOC3, TTR, HSPB1, KRT1, CP, PLG, A2M, TF, SERPINA1 |
| BP | GO:0016192 | vesicle-mediated transport | 7 | 0.91 | 0.00024 | TTR, KRT1, PLG, A2M, TF, C4B, SERPINA1 |
| BP | GO:0072376 | protein activation cascade | 3 | 1.9 | 0.00037 | KRT1, A2M, C4B |
| BP | GO:0019538 | protein metabolic process | 9 | 0.62 | 0.00038 | APOC3, TTR, KRT1, CP, PLG, A2M, TF, C4B, SERPINA1 |
| BP | GO:0043062 | extracellular structure organization | 4 | 1.36 | 0.00082 | APOC3, TTR, PLG, A2M |
| Category | Term ID | Term Description | Gene Count | Strength | FDR | Matching Proteins in Network |
|---|---|---|---|---|---|---|
| CC | GO:0060205 | cytoplasmic vesicle lumen | 9 | 1.51 | 1.88 × 10−10 | APOA1, C3, PLG, CLU, SERPINF2, HBA1, A2M, HP, SERPINA1 |
| CC | GO:0005576 | extracellular region | 14 | 0.83 | 1.14 × 10−9 | APOA1, C3, IGJ, IL7, CCL19, PLG, CLU, SERPINF2, HBA1, A2M, HP, SERPINA1, CFB, GC |
| CC | GO:0005615 | extracellular space | 11 | 1.07 | 1.96 × 10−9 | APOA1, C3, IGJ, IL7, CCL19, PLG, CLU, SERPINF2, HBA1, HP, SERPINA1 |
| CC | GO:0034774 | secretory granule lumen | 8 | 1.48 | 1.96 × 10−9 | APOA1, C3, PLG, CLU, SERPINF2, A2M, HP, SERPINA1 |
| CC | GO:0031093 | platelet alpha granule lumen | 5 | 1.95 | 4.95 × 10−8 | PLG, CLU, SERPINF2, A2M, SERPINA1 |
| CC | GO:0034366 | spherical high-density lipoprotein particle | 3 | 2.61 | 1.37 × 10−6 | APOA1, CLU, HP |
| CC | GO:0071682 | endocytic vesicle lumen | 3 | 2.29 | 6.95 × 10−6 | APOA1, HBA1, HP |
| CC | GO:0031838 | haptoglobin-haemoglobin complex | 2 | 2.79 | 6.56 × 10−5 | HBA1, HP |
| CC | GO:0009986 | cell surface | 5 | 0.95 | 0.00098 | APOA1, PLG, CLU, SERPINF2, CD244 |
| MF | GO:0005102 | signalling receptor binding | 9 | 0.86 | 9.94 × 10−5 | APOA1, C3, IGJ, IL7, CCL19, PLG, CLU, A2M, CD244 |
| MF | GO:0004857 | enzyme inhibitor activity | 5 | 1.2 | 0.00070 | APOA1, C3, SERPINF2, A2M, SERPINA1 |
| MF | GO:0004866 | endopeptidase inhibitor activity | 4 | 1.46 | 0.00070 | C3, SERPINF2, A2M, SERPINA1 |
| MF | GO:0005515 | protein binding | 14 | 0.41 | 0.00070 | APOA1, C3, IGJ, IL7, CCL19, PLG, CLU, SERPINF2, A2M, HP, CD244, SERPINA1, CFB, GC |
| BP | GO:0002576 | platelet degranulation | 6 | 1.75 | 8.75 × 10−7 | APOA1, PLG, CLU, SERPINF2, A2M, SERPINA1 |
| BP | GO:0002697 | regulation of immune effector process | 7 | 1.37 | 4.74 × 10−6 | APOA1, C3, CCL19, CLU, A2M, CD244, CFB |
| BP | GO:0032940 | secretion by cell | 9 | 1.06 | 5.70 × 10−6 | APOA1, C3, CCL19, PLG, CLU, SERPINF2, A2M, HP, SERPINA1 |
| BP | GO:0006950 | response to stress | 13 | 0.69 | 7.61 × 10−6 | APOA1, C3, IGJ, CCL19, PLG, CLU, SERPINF2, HBA1, A2M, HP, CD244, SERPINA1, CFB |
| BP | GO:0006959 | humoral immune response | 6 | 1.46 | 7.61 × 10−6 | C3, IGJ, IL7, CCL19, CLU, CFB |
| BP | GO:0045055 | regulated exocytosis | 8 | 1.15 | 7.61 × 10−6 | APOA1, C3, PLG, CLU, SERPINF2, A2M, HP, SERPINA1 |
| BP | GO:0070613 | regulation of protein processing | 5 | 1.72 | 7.61 × 10−6 | C3, CLU, SERPINF2, A2M, CFB |
| BP | GO:0030449 | regulation of complement activation | 4 | 1.97 | 1.16 × 10−5 | C3, CLU, A2M, CFB |
| BP | GO:0006952 | defence response | 9 | 0.95 | 1.23 × 10−5 | C3, IGJ, CCL19, CLU, SERPINF2, HP, CD244, SERPINA1, CFB |
| BP | GO:0016192 | vesicle-mediated transport | 10 | 0.86 | 1.23 × 10−5 | APOA1, C3, IGJ, PLG, CLU, SERPINF2, HBA1, A2M, HP, SERPINA1 |
| BP | GO:2000257 | regulation of protein activation cascade | 4 | 1.96 | 1.23 × 10−5 | C3, CLU, A2M, CFB |
| BP | GO:0001817 | regulation of cytokine production | 7 | 1.14 | 2.26 × 10−5 | APOA1, C3, IL7, CCL19, CLU, SERPINF2, CD244 |
| BP | GO:0032101 | regulation of response to external stimulus | 8 | 1.01 | 2.26 × 10−5 | APOA1, C3, CCL19, PLG, CLU, SERPINF2, A2M, CFB |
| BP | GO:0072376 | protein activation cascade | 4 | 1.82 | 2.73 × 10−5 | C3, CLU, A2M, CFB |
| BP | GO:0001819 | positive regulation of cytokine production | 6 | 1.27 | 2.82 × 10−5 | C3, IL7, CCL19, CLU, SERPINF2, CD244 |
| BP | GO:0002673 | regulation of acute inflammatory response | 4 | 1.73 | 5.46 × 10−5 | C3, CLU, A2M, CFB |
| BP | GO:0006955 | immune response | 9 | 0.85 | 5.46 × 10−5 | C3, IGJ, IL7, CCL19, CLU, HP, CD244, SERPINA1, CFB |
| BP | GO:0006954 | inflammatory response | 6 | 1.18 | 8.23 × 10−5 | C3, CCL19, CLU, SERPINF2, HP, SERPINA1 |
| BP | GO:0043086 | negative regulation of catalytic activity | 7 | 1.02 | 9.16 × 10−5 | APOA1, C3, IL7, SERPINF2, A2M, HP, SERPINA1 |
| BP | GO:0098542 | defence response to other organisms | 7 | 1.0 | 0.00013 | C3, IGJ, CCL19, CLU, HP, CD244, CFB |
| Category | Term ID | Term Description | Gene Count | Strength | FDR | Matching Proteins in Network |
|---|---|---|---|---|---|---|
| CC | GO:0005576 | extracellular region | 12 | 0.86 | 2.37 × 10−8 | APOH, C3, CP, CDCP1, FGF5, CLU, A2M, CCL20, S100A12, TF, GC, C1R |
| CC | GO:0034774 | secretory granule lumen | 6 | 1.45 | 1.74 × 10−6 | APOH, C3, CLU, A2M, S100A12, TF |
| CC | GO:0005615 | extracellular space | 7 | 0.97 | 5.86 × 10−5 | APOH, C3, CP, FGF5, CLU, CCL20, C1R |
| MF | GO:0005102 | signalling receptor binding | 7 | 0.84 | 0.0025 | C3, FGF5, CLU, A2M, CCL20, S100A12, TF |
| MF | GO:0005507 | copper ion binding | 2 | 1.73 | 0.0435 | CP, S100A12 |
| BP | GO:0072376 | protein activation cascade | 5 | 2.01 | 1.05 × 10−6 | APOH, C3, CLU, A2M, C1R |
| BP | GO:0030449 | regulation of complement activation | 4 | 2.06 | 1.88 × 10−5 | C3, CLU, A2M, C1R |
| BP | GO:2000257 | regulation of protein activation cascade | 4 | 2.05 | 1.88 × 10−5 | C3, CLU, A2M, C1R |
| BP | GO:0002673 | regulation of acute inflammatory response | 4 | 1.82 | 6.69 × 10−5 | C3, CLU, A2M, C1R |
| BP | GO:0006959 | humoral immune response | 5 | 1.48 | 6.69 × 10−5 | C3, CLU, CCL20, S100A12, C1R |
| BP | GO:0070613 | regulation of protein processing | 4 | 1.72 | 0.00012 | C3, CLU, A2M, C1R |
| BP | GO:0002576 | platelet degranulation | 4 | 1.67 | 0.00014 | APOH, CLU, A2M, TF |
| BP | GO:0006958 | complement activation, classical pathway | 3 | 2.12 | 0.00016 | C3, CLU, C1R |
| BP | GO:0050727 | regulation of inflammatory response | 5 | 1.35 | 0.00016 | C3, CLU, A2M, S100A12, C1R |
| BP | GO:0045055 | regulated exocytosis | 6 | 1.12 | 0.00019 | APOH, C3, CLU, A2M, S100A12, TF |
| BP | GO:0032101 | regulation of response to external stimulus | 6 | 0.98 | 0.00087 | APOH, C3, CLU, A2M, S100A12, C1R |
| Category | Term ID | Term Description | Gene Count | Strength | FDR | Matching Proteins in Network |
|---|---|---|---|---|---|---|
| CC | GO:0005576 | extracellular region | 14 | 0.81 | 1.36 × 10−8 | APOA1, FGA, CLU, SERPINF2, C7, A2M, CSF1, NTF3, GC, C1R, FLT3LG, FGF21, VEGFA, TNFRSF9 |
| CC | GO:0031093 | platelet alpha granule lumen | 5 | 1.93 | 1.67 × 10−7 | FGA, CLU, SERPINF2, A2M, VEGFA |
| CC | GO:0005615 | extracellular space | 10 | 1.01 | 1.76 × 10−7 | APOA1, FGA, CLU, SERPINF2, CSF1, C1R, FLT3LG, FGF21, VEGFA, TNFRSF9 |
| CC | GO:0034774 | secretory granule lumen | 6 | 1.33 | 4.05 × 10−6 | APOA1, FGA, CLU, SERPINF2, A2M, VEGFA |
| CC | GO:0009986 | cell surface | 7 | 1.07 | 1.11 × 10−5 | APOA1, FGA, CLU, SERPINF2, FLT3LG, VEGFA, TNFRSF9 |
| CC | GO:0005577 | fibrinogen complex | 2 | 2.46 | 0.00031 | FGA, SERPINF2 |
| CC | GO:0034366 | spherical high-density lipoprotein particle | 2 | 2.41 | 0.00035 | APOA1, CLU |
| MF | GO:0005102 | signalling receptor binding | 9 | 0.84 | 0.00020 | APOA1, FGA, CLU, A2M, CSF1, NTF3, FLT3LG, FGF21, VEGFA |
| MF | GO:0048018 | receptor ligand activity | 6 | 1.18 | 0.00020 | APOA1, CSF1, NTF3, FLT3LG, FGF21, VEGFA |
| MF | GO:0008083 | growth factor activity | 4 | 1.46 | 0.00037 | CSF1, NTF3, FGF21, VEGFA |
| BP | GO:0002576 | platelet degranulation | 6 | 1.75 | 7.32 × 10−7 | APOA1, FGA, CLU, SERPINF2, A2M, VEGFA |
| BP | GO:0032101 | regulation of response to external stimulus | 10 | 1.11 | 7.32 × 10−7 | APOA1, FGA, CLU, SERPINF2, C7, A2M, CSF1, NTF3, C1R, VEGFA |
| BP | GO:2000257 | regulation of protein activation cascade | 5 | 2.05 | 7.32 × 10−7 | FGA, CLU, C7, A2M, C1R |
| BP | GO:0072376 | protein activation cascade | 5 | 1.92 | 1.47 × 10−6 | FGA, CLU, C7, A2M, C1R |
| BP | GO:0051246 | regulation of protein metabolic process | 12 | 0.74 | 1.02 × 10−5 | APOA1, FGA, CLU, SERPINF2, C7, A2M, CSF1, NTF3, C1R, FLT3LG, FGF21, VEGFA |
| BP | GO:0070613 | regulation of protein processing | 5 | 1.72 | 1.02 × 10−5 | CLU, SERPINF2, C7, A2M, C1R |
| BP | GO:0019220 | regulation of phosphate metabolic process | 10 | 0.87 | 1.80 × 10−5 | APOA1, PGAM2, FGA, CLU, SERPINF2, CSF1, NTF3, FLT3LG, FGF21, VEGFA |
| BP | GO:0030449 | regulation of complement activation | 4 | 1.97 | 1.80 × 10−5 | CLU, C7, A2M, C1R |
| BP | GO:0001934 | positive regulation of protein phosphorylation | 8 | 1.02 | 3.63 × 10−5 | FGA, CLU, SERPINF2, CSF1, NTF3, FLT3LG, FGF21, VEGFA |
| BP | GO:0001932 | regulation of protein phosphorylation | 9 | 0.9 | 3.74 × 10−5 | APOA1, FGA, CLU, SERPINF2, CSF1, NTF3, FLT3LG, FGF21, VEGFA |
| BP | GO:0002682 | regulation of immune system process | 9 | 0.9 | 3.74 × 10−5 | APOA1, FGA, CLU, C7, A2M, CSF1, C1R, FLT3LG, VEGFA |
| BP | GO:1902533 | positive regulation of intracellular signal transduction | 8 | 1.01 | 3.74 × 10−5 | APOA1, FGA, CLU, SERPINF2, CSF1, NTF3, FGF21, VEGFA |
| BP | GO:0048584 | positive regulation of response to stimulus | 10 | 0.77 | 5.42 × 10−5 | APOA1, FGA, CLU, SERPINF2, C7, CSF1, NTF3, C1R, FGF21, VEGFA |
| BP | GO:0002673 | regulation of acute inflammatory response | 4 | 1.73 | 6.68 × 10−5 | CLU, C7, A2M, C1R |
| BP | GO:0009605 | response to external stimulus | 10 | 0.75 | 7.61 × 10−5 | APOA1, IL10RB, FGA, CLU, C7, CSF1, NTF3, C1R, FGF21, VEGFA |
| BP | GO:0010811 | positive regulation of cell-substrate adhesion | 4 | 1.65 | 0.00011 | APOA1, FGA, CSF1, VEGFA |
| BP | GO:1902531 | regulation of intracellular signal transduction | 9 | 0.8 | 0.00015 | APOA1, FGA, CLU, SERPINF2, A2M, CSF1, NTF3, FGF21, VEGFA |
| BP | GO:0002684 | positive regulation of immune system process | 7 | 0.99 | 0.00017 | FGA, CLU, C7, CSF1, C1R, FLT3LG, VEGFA |
| BP | GO:0006958 | complement activation, classical pathway | 3 | 2.03 | 0.00017 | CLU, C7, C1R |
| BP | GO:0048583 | regulation of response to stimulus | 12 | 0.58 | 0.00017 | APOA1, FGA, CLU, SERPINF2, C7, A2M, CSF1, NTF3, C1R, FLT3LG, FGF21, VEGFA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerdle, B.; Wåhlén, K.; Gordh, T.; Ghafouri, B. Thermal Pain Thresholds Are Significantly Associated with Plasma Proteins of the Immune System in Chronic Widespread Pain—An Exploratory Pilot Study Using Multivariate and Network Analyses. J. Clin. Med. 2021, 10, 3652. https://doi.org/10.3390/jcm10163652
Gerdle B, Wåhlén K, Gordh T, Ghafouri B. Thermal Pain Thresholds Are Significantly Associated with Plasma Proteins of the Immune System in Chronic Widespread Pain—An Exploratory Pilot Study Using Multivariate and Network Analyses. Journal of Clinical Medicine. 2021; 10(16):3652. https://doi.org/10.3390/jcm10163652
Chicago/Turabian StyleGerdle, Björn, Karin Wåhlén, Torsten Gordh, and Bijar Ghafouri. 2021. "Thermal Pain Thresholds Are Significantly Associated with Plasma Proteins of the Immune System in Chronic Widespread Pain—An Exploratory Pilot Study Using Multivariate and Network Analyses" Journal of Clinical Medicine 10, no. 16: 3652. https://doi.org/10.3390/jcm10163652
APA StyleGerdle, B., Wåhlén, K., Gordh, T., & Ghafouri, B. (2021). Thermal Pain Thresholds Are Significantly Associated with Plasma Proteins of the Immune System in Chronic Widespread Pain—An Exploratory Pilot Study Using Multivariate and Network Analyses. Journal of Clinical Medicine, 10(16), 3652. https://doi.org/10.3390/jcm10163652






