Impartially Validated Multiple Deep-Chain Models to Detect COVID-19 in Chest X-ray Using Latent Space Radiomics
Abstract
:1. Introduction
- The proposed approach trains and independently uses a 2D U-Net model for segmenting the lung lobes in CXR images;
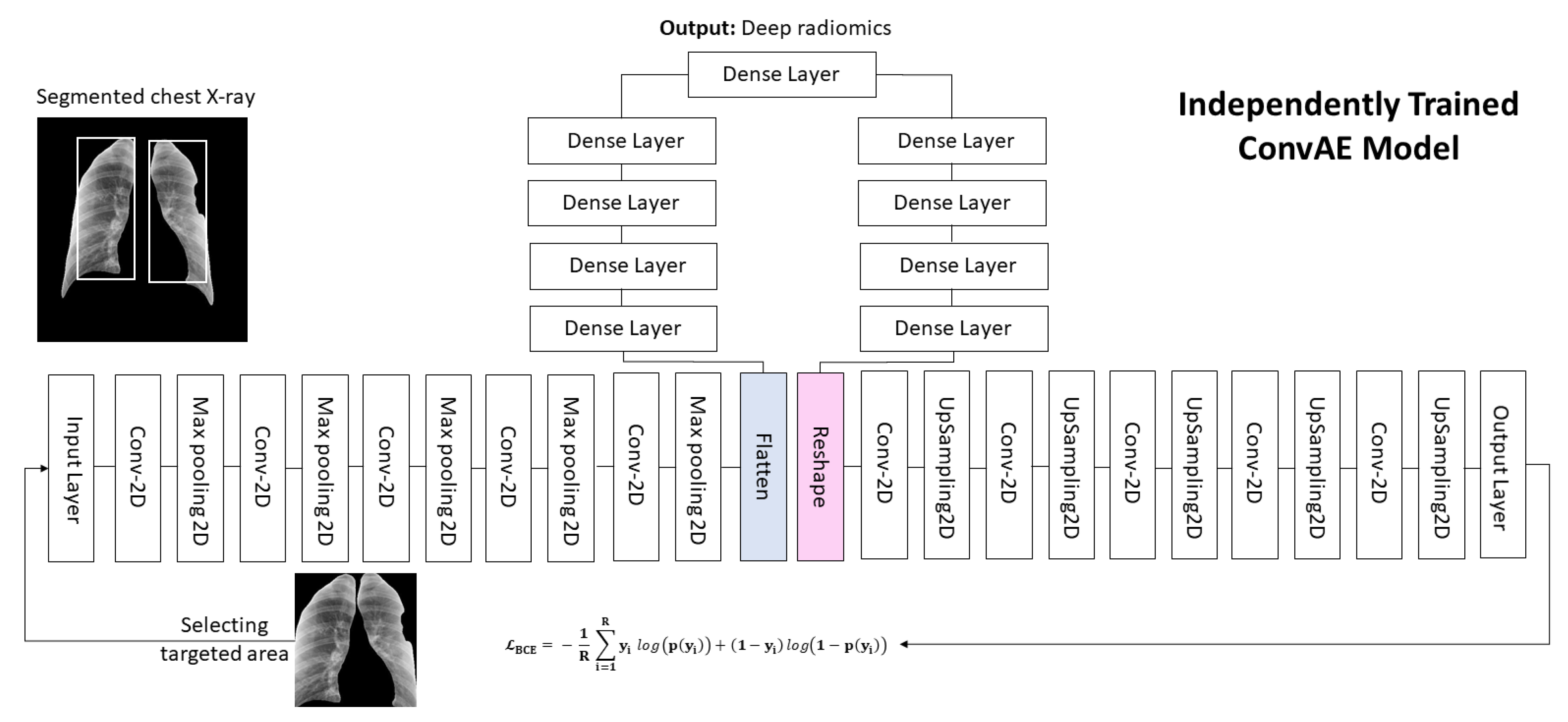
- We proposed a convolutional deep autoencoder (ConvAE) to extract low-dimensional deep-imaging features, called deep radiomics, from CXR images as potential diagnostic biomarkers for COVID-19;
- Our study addresses the curse of dimensionality problem using high-dimensional deep radiomics by utilizing a ConvAE to compress the feature space and combine them with conventional radiomics for diagnostic purposes;
- The proposed model successfully classifies subjects into healthy, pneumonia, and COVID-19 cases through binary- and multiclass classification, as validated with an independent cohort of patients. We also provided comparative analyses of the different combinations of feature categories.
2. Methods
2.1. U-Net for Segmentation of Lung Lobes
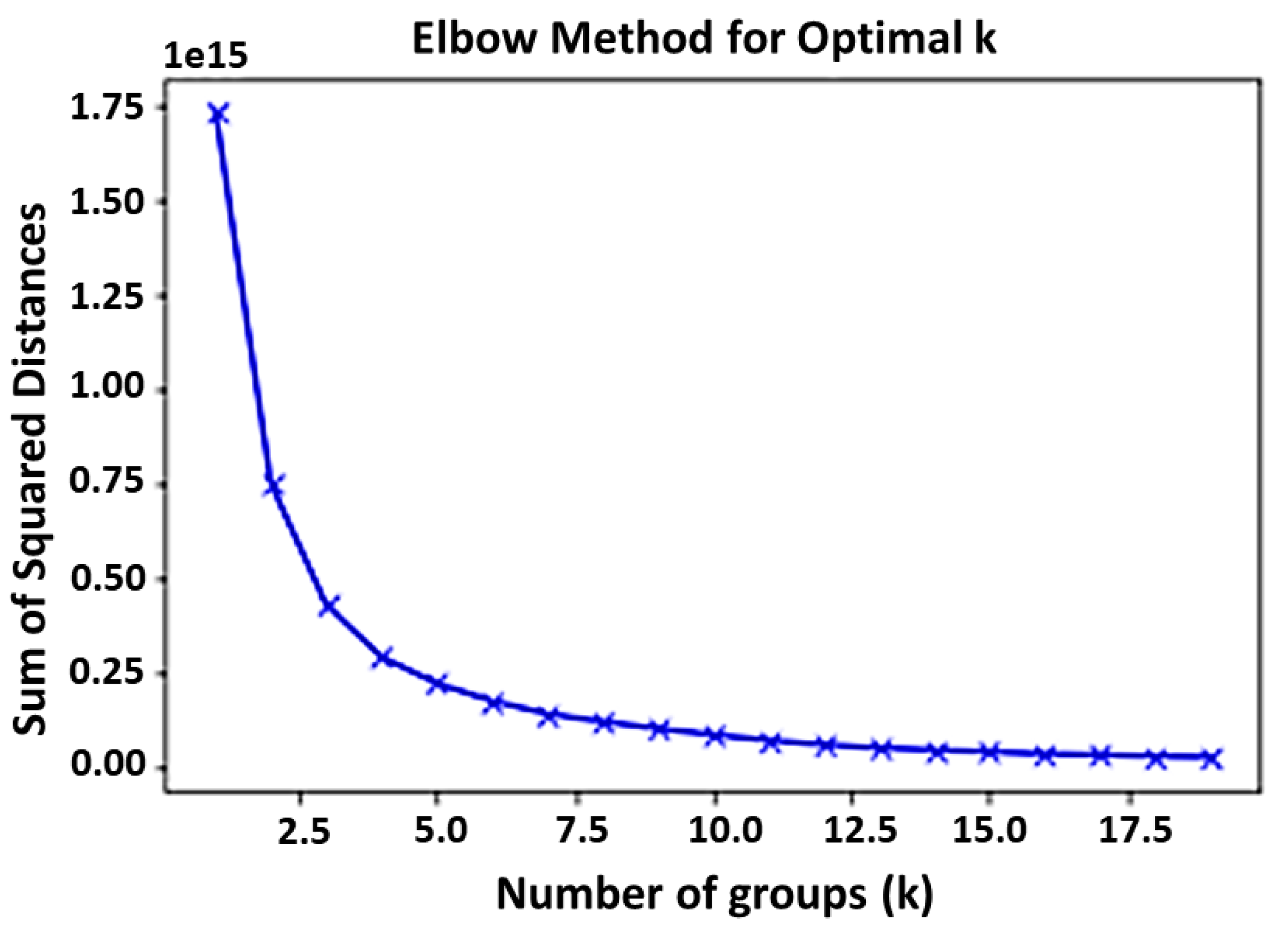
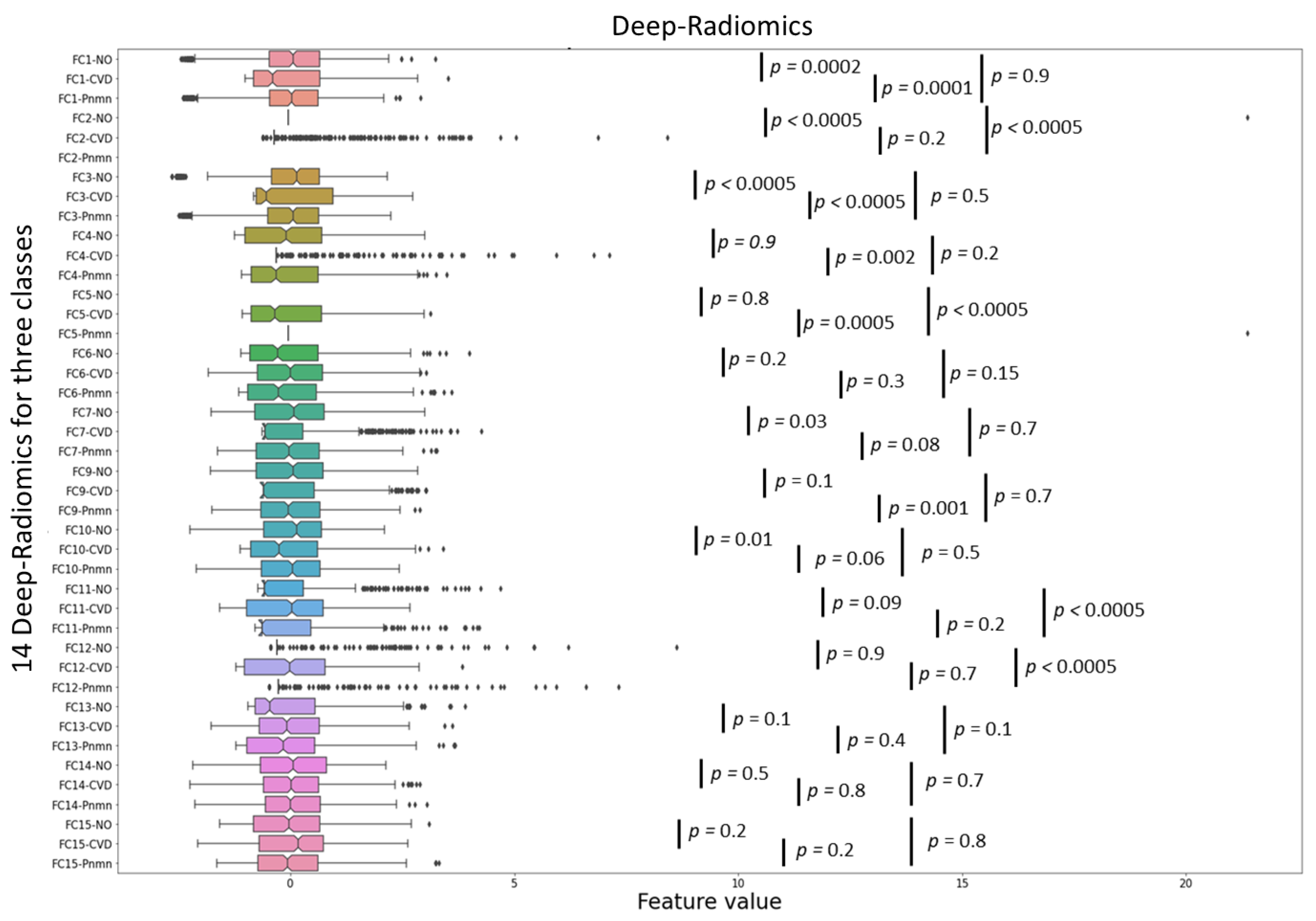
2.2. Deep Radiomics
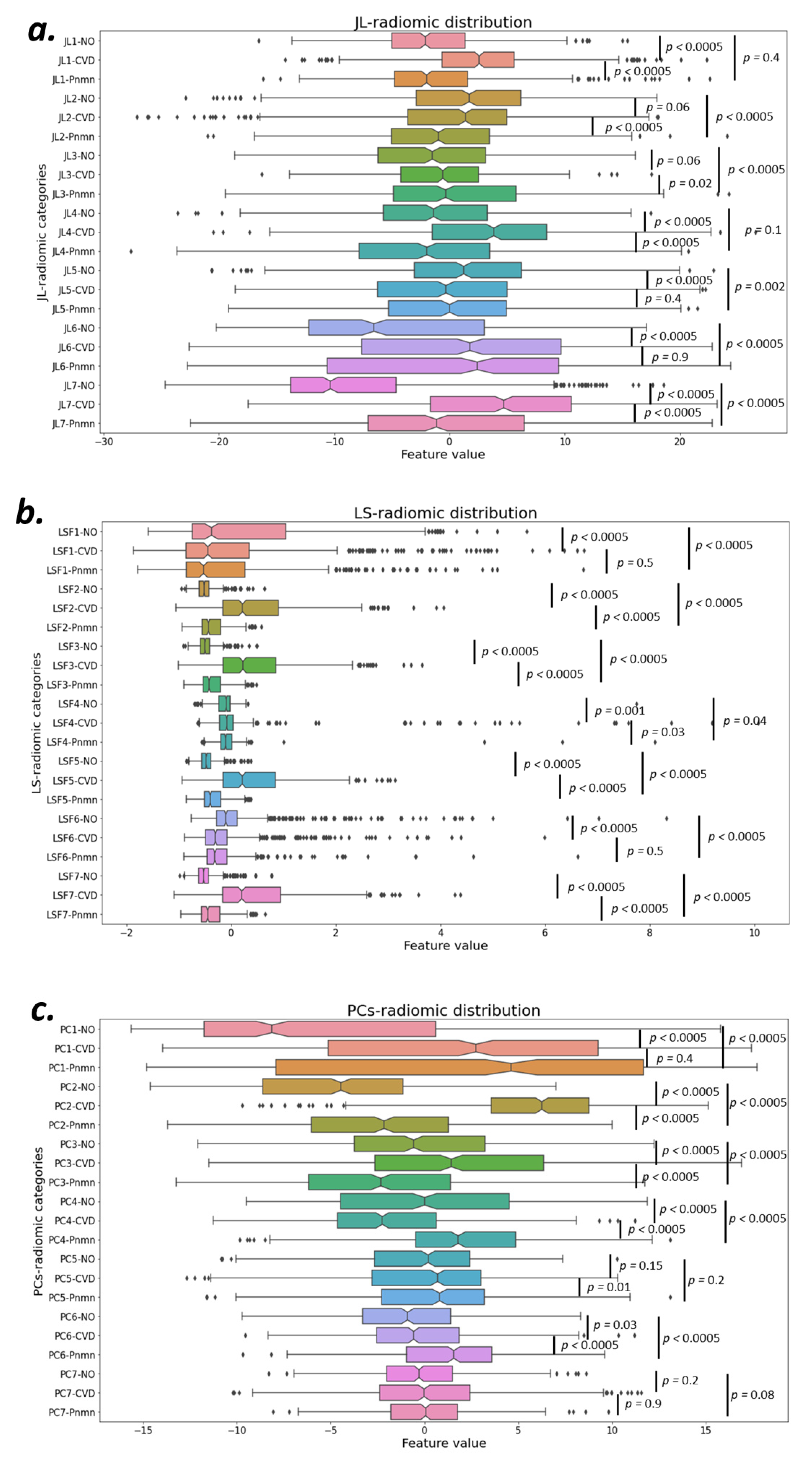
2.3. Conventional Radiomics Features
3. Results
3.1. Study Population
3.1.1. Pulmonary Chest X-ray Disease Detection
3.1.2. COVID-19 Cases Collected from Multiple Sources
3.1.3. Pneumonia, Healthy Controls, and COVID-19
3.1.4. Figure 1 COVID-19 Chest X-ray Dataset Initiative
3.1.5. U-Net-Based Segmentation of the Lung Lobes
3.1.6. Conventional and Deep Latent Space Radiomics
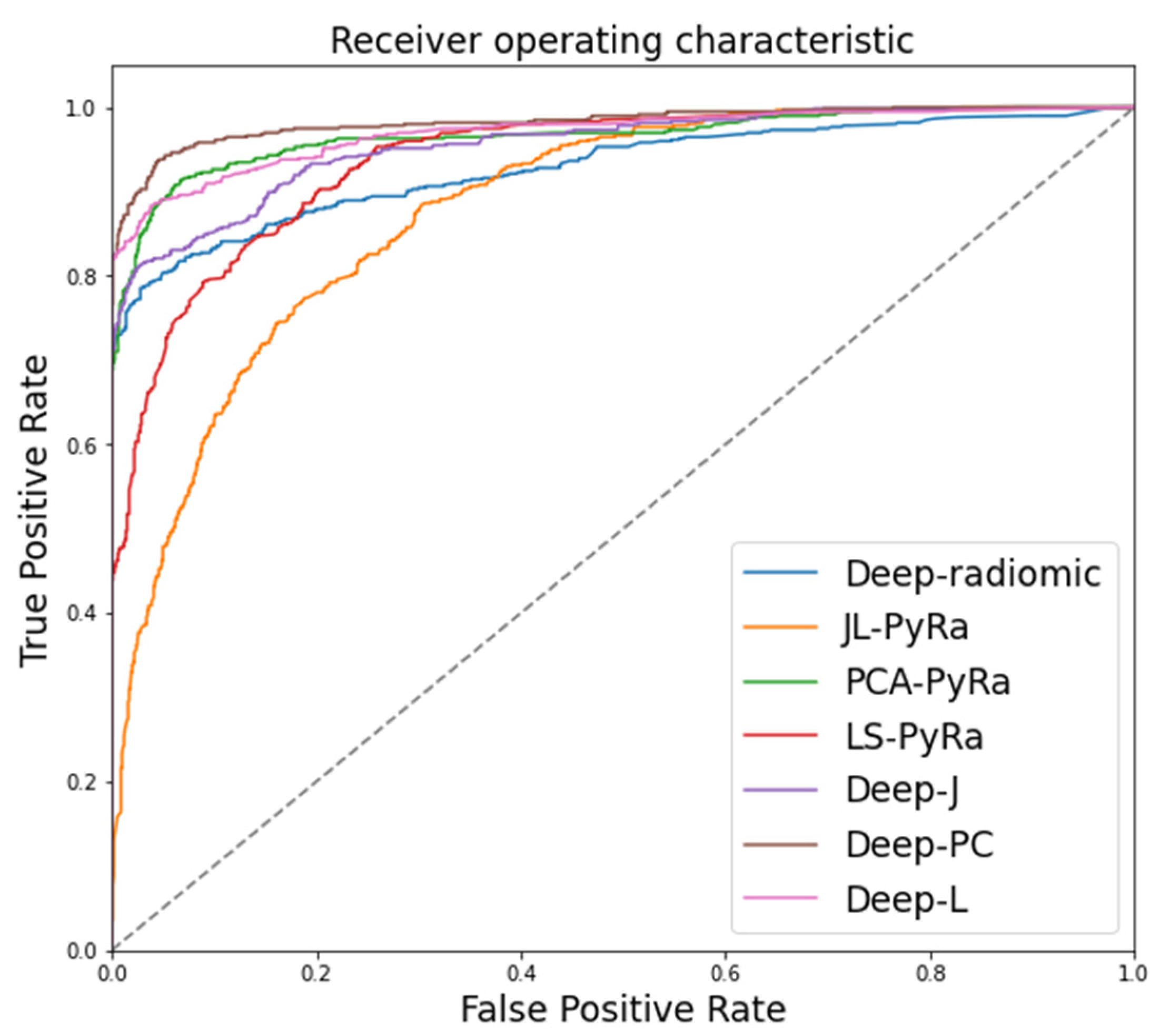
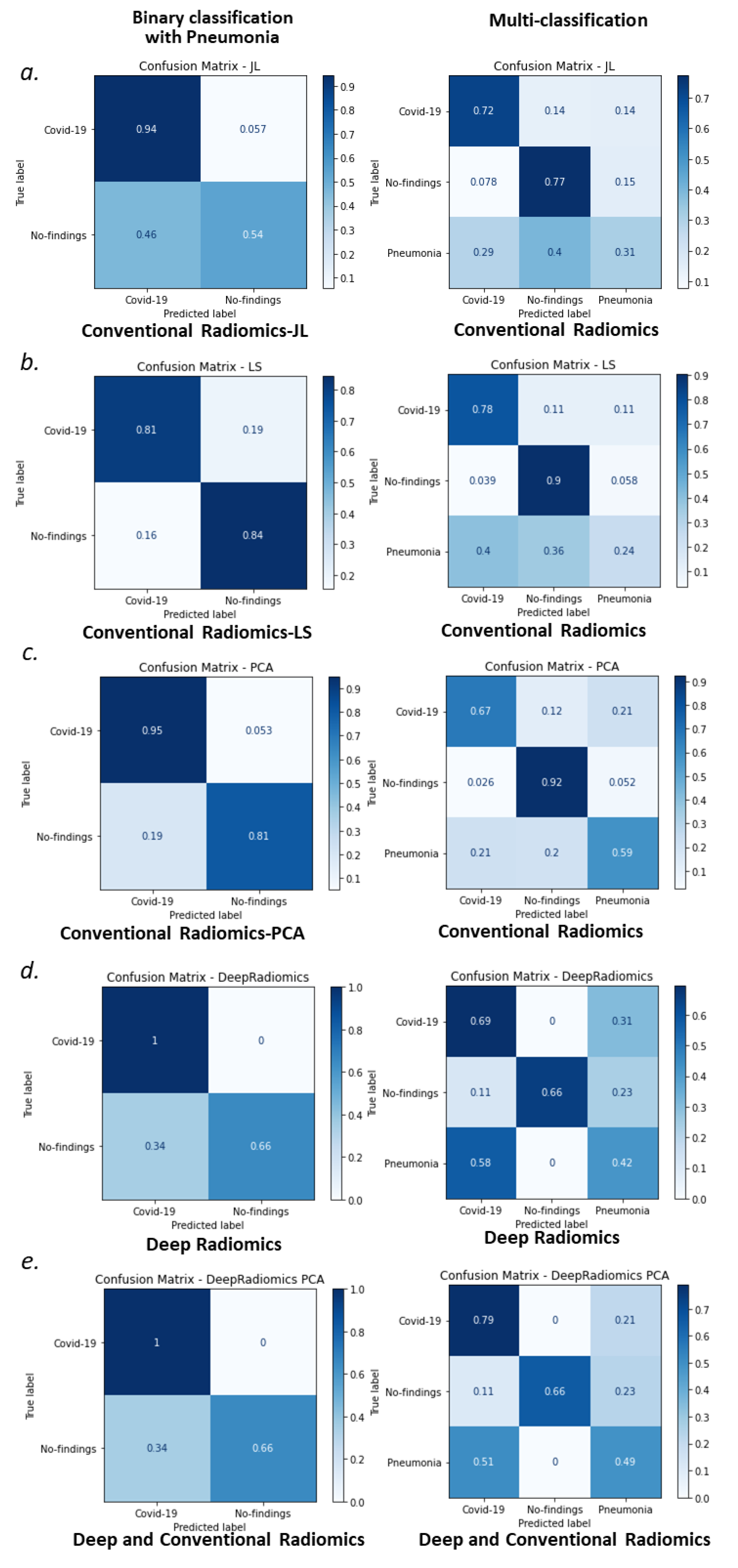
3.2. Random Forest Classification of COVID-19
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- World Health Organization (WHO). Coronavirus Disease 2019 (COVID-19) Situation Report—80; WHO: Geneva, Switzerland, 2020; Available online: https://www.who.int/docs/default-source/coronaviruse/weekly-updates/wou-16oct-cleared.pdf?sfvrsn=c8f87df8_0&download=true (accessed on 16 October 2020).
- World Health Organization (WHO). Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV); WHO: Geneva, Switzerland, 2020. [Google Scholar]
- World Health Organization (WHO). WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Kanne, J.P. Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: Key points for the radiologist. Radiology 2020, 295, 16–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernheim, A.; Mei, X.; Huang, M.; Yang, Y.; Fayad, Z.A.; Zhang, N.; Diao, K.; Lin, B.; Zhu, X.; Li, K.; et al. Chest CT findings in coronavirus disease-19 (COVID-19): Relationship to duration of infection. Radiology 2020, 295, 200463. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J.J.R. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology 2020, 296, E41–E45. [Google Scholar] [CrossRef] [Green Version]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv 2020, arXiv:2003.10849. [Google Scholar]
- Apostolopoulos, I.D.; Bessiana, T. COVID-19: Automatic detection from X-Ray images utilizing transfer learning with convolutional neural networks. arXiv 2020, arXiv:2003.11617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunraj, H.; Wang, L.; Wong, A. COVIDNet-CT: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest CT images. arXiv 2020, arXiv:2009.05383. [Google Scholar]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of chest CT and RT-PCR testing for Coronavirus Disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef]
- Salehi, S.; Abedi, A.; Balakrishnan, S.; Gholamrezanezhad, A. Coronavirus Disease 2019 (COVID-19): A systematic review of imaging findings in 919 patients. Am. J. Roentgenol. 2020, 215, 87–93. [Google Scholar] [CrossRef]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2021, 14, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Bullock, J.; Luccioni, A.; Pham, K.H.; Lam, C.S.N.; Luengo-Oroz, M. Mapping the landscape of artificial intelligence applications against COVID-19. arXiv 2020, arXiv:2003.11336. [Google Scholar]
- Zheng, C.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Wang, X. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Xu, Z.; Feng, J.; Jin, C.; Han, X.; Wu, H.; Shi, H. Longitudinal assessment of COVID-19 using a deep learning-based quantitative CT pipeline: Illustration of two cases. Radiol. Cardiothorac. Imaging 2020, 2, e200082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Han, R.; Ai, T.; Yu, P.; Kang, H.; Tao, Q.; Xia, L. Serial quantitative chest CT assessment of COVID-19: A deep learning approach. Radiol. Cardiothorac. Imaging 2020, 2, e200075. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Jiang, Z.; Yu, Q.; Shao, C.; Zhang, H.; Yue, H.; Ma, B.; Wang, Y.; Liu, C.; Meng, X.; et al. Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study. MedRxiv 2020. [Google Scholar] [CrossRef]
- Gozes, O.; Frid-Adar, M.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid AI development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection and patient monitoring using deep learning CT image analysis. arXiv 2020, arXiv:2003.05037, 2020. [Google Scholar]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy. Radiology 2020, 296, E65–E71. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, L.; Zhang, J.; Zhang, L.; Gong, D.; Zhao, Y.; Chen, Q.; Huang, S.; Yang, M.; Yang, X.; et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: A prospective study. MedRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Jin, S.; Yan, Q.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Xu, Z.; et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system. MedRxiv 2020. [Google Scholar] [CrossRef]
- Shan, F.; Gao, Y.; Wang, J.; Shi, W.; Shi, N.; Han, M.; Xue, Z.; Shen, D.; Shi, Y. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv 2020, arXiv:2003.04655. [Google Scholar]
- Ghoshal, B.; Tucker, A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv 2020, arXiv:2003.10769. [Google Scholar]
- Zhang, J.; Xie, Y.; Li, Y.; Shen, C.; Xia, Y. COVID-19 screening on Chest X-ray images using deep learning based anomaly detection. arXiv 2020, arXiv:2003.12338. [Google Scholar]
- Wong, H.Y.F.; Lam, H.Y.S.; Fong, A.H.-T.; Leung, S.T.; Chin, T.W.-Y.; Lo, C.S.Y.; Lui, M.M.-S.; Lee, J.C.Y.; Chiu, K.W.-H.; Chung, T.W.-H.; et al. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology 2020, 296, E72–E78. [Google Scholar] [CrossRef] [Green Version]
- Heidari, M.; Mirniaharikandehei, S.; Khuzani, A.Z.; Danala, G.; Qiu, Y.; Zheng, B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inform. 2020, 144, 104284. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Park, S.; Ye, J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.H.; Geng, H.; Chiu, T.L.; Yu, S.K.; Cho, D.C.; Heo, J.; Choi, M.S.; Choi, I.H.; Van, C.C.; Nhung, N.V.; et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front. Med. 2020, 7, 427. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Mitra, S.; Saha, N. Deep learning for screening COVID-19 using chest X-Ray images. In Proceedings of the 2020 IEEE Symposium Series on Computational Intelligence (SSCI), Canberra, ACT, Australia, 1–4 December 2020; pp. 2521–2527. [Google Scholar]
- Sethy, P.K.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Preprints 2020, 2020030300. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Prog. Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Wang, D.; Mo, J.; Zhou, G.; Xu, L.; Liu, Y. An efficient mixture of deep and machine learning models for COVID-19 diagnosis in chest X-ray images. PLoS ONE 2020, 15, e0242535. [Google Scholar] [CrossRef]
- Rahimzadeh, M.; Attar, A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform. Med. Unlocked 2020, 19, 100360. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- El-Din, H.E.; Shouman, M.A.; Karar, M.E. COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Chowdhury, N.K.; Rahman, M.; Kabir, M.A. PDCOVIDNet: A parallel-dilated convolutional neural network architecture for detecting COVID-19 from chest X-ray images. Health Inf. Sci. Syst. 2020, 8, 27. [Google Scholar] [CrossRef]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the 2015 International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Sorensen, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. K. Dan. Vidensk. Selsk. Biol. Skr. 1948, 5, 1–34. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- LeCun, Y. LeNet-5, Convolutional Neural Networks. Available online: http://yann.lecun.com/exdb/lenet (accessed on 1 July 2020).
- Aghdam, H.H.; Heravi, E.J. Guide to Convolutional Neural Networks: A Practical Application to Traffic-Sign Detection and Classification; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Convolutional Neural Networks (LeNet)—DeepLearning 0.1 Documentation; LISA Lab.: Online, 31 August 2013.
- Ciresan, D.C.; Meier, U.; Masci, J.; Maria Gambardella, L.; Schmidhuber, J. Flexible, high performance convolutional neural networks for image classification. In Proceedings of the International Joint Conference on Artificial Intelligence, Barcelona, Spain, 16–22 July 2011; Volume 22, p. 1237. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2012; Volume 25, pp. 1097–1105. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Homayounieh, F.; Ebrahimian, S.; Babaei, R.; Mobin, H.K.; Zhang, E.; Bizzo, B.C.; Mohseni, I.; Digumarthy, S.R.; Kalra, M.K. CT radiomics, radiologists, and clinical information in predicting outcome of patients with COVID-19 pneumonia. Radiol. Cardiothorac. Imaging 2020, 2, e200322. [Google Scholar] [CrossRef]
- Fang, X.; Li, X.; Bian, Y.; Ji, X.; Lu, J. Radiomics nomogram for the prediction of 2019 novel coronavirus pneumonia caused by SARS-CoV-2. Eur. Radiol. 2020, 30, 6888–6901. [Google Scholar] [CrossRef]
- Yousefi, B.; Akbari, H.; Maldague, X. Detecting vasodilation as potential diagnostic biomarker in breast cancer using deep learning-driven thermomics. Biosensors 2020, 10, 164. [Google Scholar] [CrossRef]
- Cavallari, G.B.; Ribeiro, L.S.; Ponti, M. Unsupervised representation learning using convolutional and stacked auto-encoders: A domain and cross-domain feature space analysis. In Proceedings of the 2018 31st SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Paraná, Brazil, 29 October–1 November 2018; pp. 440–446. [Google Scholar]
- Ponti, M.A.; Ribeiro, L.S.F.; Nazare, T.S.; Bui, T.; Collomosse, J. Everything you wanted to know about deep learning for computer vision but were afraid to ask. In Proceedings of the 2017 30th SIBGRAPI Conference on Graphics, Patterns and Images Tutorials (SIBGRAPI-T), Niteroi, Brazil, 17–20 October 2017; pp. 17–41. [Google Scholar]
- Shin, H.-C.; Orton, M.R.; Collins, D.; Doran, S.J.; Leach, M. Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1930–1943. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.J.; Balajewicz, M. Deep convolutional recurrent autoencoders for learning low-dimensional feature dynamics of fluid systems. arXiv 2018, arXiv:1808.01346. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Zeiler, M.D. ADADELTA: An adaptive learning rate method. arXiv 2012, arXiv:1212:5701. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Wilson, R.; Devaraj, A. Radiomics of pulmonary nodules and lung cancer. Transl. Lung Cancer Res. 2017, 6, 86–91. [Google Scholar] [CrossRef] [Green Version]
- Han, F.; Wang, H.; Zhang, G.; Han, H.; Song, B.; Li, L.; Moore, W.; Lu, H.; Zhao, H.; Liang, Z. Texture Feature Analysis for Computer-Aided Diagnosis on Pulmonary Nodules. J. Digit. Imaging 2015, 28, 99–115. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Johnson, W.B.; Lindenstrauss, J.; Beals, R.; Beck, A.; Bellow, A.; Hajian, A. Extensions of Lipschitz mappings into a Hilbert space. Contemp. Math. 1984, 26, 189–206. [Google Scholar] [CrossRef]
- Dasgupta, S.; Gupta, A. An Elementary Proof of the Johnson-Lindenstrauss Lemma; International Computer Science Institute: Berkeley, CA, USA, 1999. [Google Scholar]
- He, X.; Cai, D.; Niyogi, P. Laplacian score for feature selection. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2005; Volume 18, pp. 507–514. [Google Scholar]
- Malhi, A.; Gao, R. PCA-based feature selection scheme for machine defect classification. IEEE Trans. Instrum. Meas. 2004, 53, 1517–1525. [Google Scholar] [CrossRef]
- Jaeger, S.; Karargyris, A.; Candemir, S.; Folio, L.; Siegelman, J.; Callaghan, F.; Xue, Z.; Palaniappan, K.; Singh, R.K.; Antani, S.; et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging 2014, 33, 233–245. [Google Scholar] [CrossRef]
- Candemir, S.; Jaeger, S.; Palaniappan, K.; Musco, J.P.; Singh, R.K.; Xue, Z.; Karargyris, A.; Antani, S.; Thoma, G.; McDonald, C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 2014, 33, 577–590. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L.; Roth, K.; Duong, T.Q.; Ghassemi, M. COVID-19 image data collection: Prospective predictions are the future. arXiv 2020, arXiv:2006.11988. [Google Scholar]
- Lung Bounding Boxes and Chest X-ray Segmentation. Available online: https://github.com/GeneralBlockchain/ (accessed on 30 June 2021).
- Pneumonia Severity Scores for 94 Images. Available online: https://github.com/ieee8023/covid-chestxray-dataset/blob/master/annotations/covid-severity-scores.csv (accessed on 30 May 2020).
- Cohen, J.P.; Dao, L.; Roth, K.; Morrison, P.; Bengio, Y.; Abbasi, A.F.; Shen, B.; Mahsa, H.K.; Ghassemi, M.; Li, H.; et al. Predicting COVID-19 pneumonia severity on chest X-ray with deep learning. arXiv 2020, arXiv:2005.11856. [Google Scholar] [CrossRef] [PubMed]
- Generated Lung Segmentations. Available online: https://github.com/ieee8023/covid-chestxray-dataset/blob/master/annotations/lungVAE-masks (accessed on 1 June 2020).
- Selvan, R.; Dam, E.B.; Rischel, S.; Sheng, K.; Nielsen, M.; Pai, A. Lung segmentation from chest X-rays using variational data imputation. arXiv 2020, arXiv:2005.10052. [Google Scholar]
- Brixia Score for 192 Images. Available online: https://github.com/BrixIA/Brixia-score-COVID-19 (accessed on 30 May 2020).
- Signoroni, A.; Savardi, M.; Benini, S.; Adami, N.; Leonardi, R.; Gibellini, P.; Vaccher, F.; Ravanelli, M.; Borghesi, A.; Farina, D. End-to-end learning for semiquantitative rating of COVID-19 severity on Chest X-rays. arXiv 2020, arXiv:2006.04603. [Google Scholar]
- Lung and Other Segmentations for 517 Images. Available online: https://github.com/v7labs/covid-19-xray-dataset/tree/master/annotations (accessed on 1 June 2020).
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 image data collection. arXiv 2020, arXiv:2003.11597. [Google Scholar]
- Cohen, J.P. COVID-19 Image Data Collection. 2020. Available online: https://github.com/ieee8023/covid-chestxray-dataset (accessed on 22 March 2020).
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chest X-ray 8: Hospital scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Wang, L.; Wong, A.; Lin, Z.Q.; Lee, J.; McInnis, P.; Chung, A.; Ross, M.; Van Berlo, B.; Ebadi, A. Figure 1 COVID-19 Chest X-ray Dataset Initiative. Available online: https://github.com/agchung/Figure1-COVID-chestxray-dataset/blob/master/README.md#figure-1-covid-19-chest-x-ray-dataset-initiative (accessed on 10 June 2021).
- COVID-19 Clinical Cases. Available online: https://www.figure1.com/covid-19-clinical-cases (accessed on 10 June 2021).
- COVID-19 DATABASE. Available online: https://www.sirm.org/category/senza-categoria/covid-19/ (accessed on 10 June 2021).
- Winther, H.B.; Laser, H.; Gerbel, S.; Maschke, S.K.; Hinrichs, J.B.; Vogel-Claussen, J.; Wacker, F.K.; Höper, M.M.; Meyer, B.C. COVID-19 Image Repository. 2020. Available online: https://data.uni-hannover.de/dataset/ca654807-033c-4650-a967-2d1641f49f0b (accessed on 11 June 2021).
- Poppler. Available online: https://poppler.freedesktop.org/dataset (accessed on 11 June 2021).
- Pandey, N. Lung Segmentation from Chest X-ray Dataset. Available online: https://www.kaggle.com/nikhilpandey360/lung-segmentation-from-chest-x-ray-dataset (accessed on 11 June 2021).
- Python 3, Google Compute Engine Backend, T4, and P100 GPU and 27.4 Gb RAM; Google 2020. Available online: https://colab.research.google.com/ (accessed on 11 June 2021).
- Abadi, M.; Barham, P.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; Zheng, X. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th {USENIX} Symposium on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Tartaglione, E.; Barbano, C.; Berzovini, C.; Calandri, M.; Grangetto, M. Unveiling COVID-19 from CHEST X-Ray with Deep Learning: A hurdles race with small data. Int. J. Environ. Res. Public Health 2020, 17, 6933. [Google Scholar] [CrossRef]


| Scheme of Model Training and Independent Validation | |||
|---|---|---|---|
| Purpose | Model | Learning Stage | Number of Cases and Dataset |
| Segmentation of Lung Lobes | U-Net 1 | Training | 704 The Shenzhen set |
| Independent Validation | 417 COVID-19 | ||
| 1125 ChestX-ray8 | |||
| 55 Figure 1 | |||
| COVID-19 Identification | ConvAE 2 | Training | 704 The Shenzhen set |
| Independent Validation | 417 COVID-19 | ||
| 1125 ChestX-ray8 | |||
| 55 Figure 1 | |||
| Accuracy of Multivariate Models | |||
|---|---|---|---|
| Methods | Classification Accuracy 1 (%) | t-Test 2; t-Statistic, Two-Tailed p-Value | |
| Binary Classification | Deep-radiomics | 88.7 (88.7–88.9) | 8.6, <0.0005 |
| JL-PyRad | 79.3 (77.2–80.3) | >90, <0.0005 | |
| PCA-PyRad | 89.6 (88.4–90.7) | - | |
| LS-PyRad | 83.6 (82.9–84.3) | 55.4, <0.0005 | |
| Deep-JL | 88.7 (88.7–88.8) | 8.6, <0.0005 | |
| Deep-PCA | 88.8 (88.7–89.0) | 7.9, <0.0005 | |
| Deep-LS | 88.9 (88.7–93.2) | 1.05, 0.29 | |
| Multiclass Classification | Deep-radiomics | 63.5 (61.3–65.1) | >90, <0.0005 |
| JL-PyRad | 63.9 (62.5–65.2) | >90, <0.0005 | |
| PCA-PyRad | 72.6 (69.4–74.4) | - | |
| LS-PyRad | 65.7 (65.1–66.2) | 72.6, <0.0005 | |
| Deep-JL | 67.7 (64.9–71.4) | 42.1, <0.0005 | |
| Deep-PCA | 72.5 (71.2–73.4) | 2.3, 0.02 | |
| Deep-LS | 66.8 (65.9–73.4) | 32.3, <0.0005 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousefi, B.; Kawakita, S.; Amini, A.; Akbari, H.; Advani, S.M.; Akhloufi, M.; Maldague, X.P.V.; Ahadian, S. Impartially Validated Multiple Deep-Chain Models to Detect COVID-19 in Chest X-ray Using Latent Space Radiomics. J. Clin. Med. 2021, 10, 3100. https://doi.org/10.3390/jcm10143100
Yousefi B, Kawakita S, Amini A, Akbari H, Advani SM, Akhloufi M, Maldague XPV, Ahadian S. Impartially Validated Multiple Deep-Chain Models to Detect COVID-19 in Chest X-ray Using Latent Space Radiomics. Journal of Clinical Medicine. 2021; 10(14):3100. https://doi.org/10.3390/jcm10143100
Chicago/Turabian StyleYousefi, Bardia, Satoru Kawakita, Arya Amini, Hamed Akbari, Shailesh M. Advani, Moulay Akhloufi, Xavier P. V. Maldague, and Samad Ahadian. 2021. "Impartially Validated Multiple Deep-Chain Models to Detect COVID-19 in Chest X-ray Using Latent Space Radiomics" Journal of Clinical Medicine 10, no. 14: 3100. https://doi.org/10.3390/jcm10143100
APA StyleYousefi, B., Kawakita, S., Amini, A., Akbari, H., Advani, S. M., Akhloufi, M., Maldague, X. P. V., & Ahadian, S. (2021). Impartially Validated Multiple Deep-Chain Models to Detect COVID-19 in Chest X-ray Using Latent Space Radiomics. Journal of Clinical Medicine, 10(14), 3100. https://doi.org/10.3390/jcm10143100









