Increasing the Density of Laboratory Measures for Machine Learning Applications
Abstract
1. Introduction
2. Methods
2.1. Study Cohort
2.2. Data Extraction
2.3. Data Processing
2.4. Data Abstraction and Imputation Strategy
2.5. Evaluation Strategy
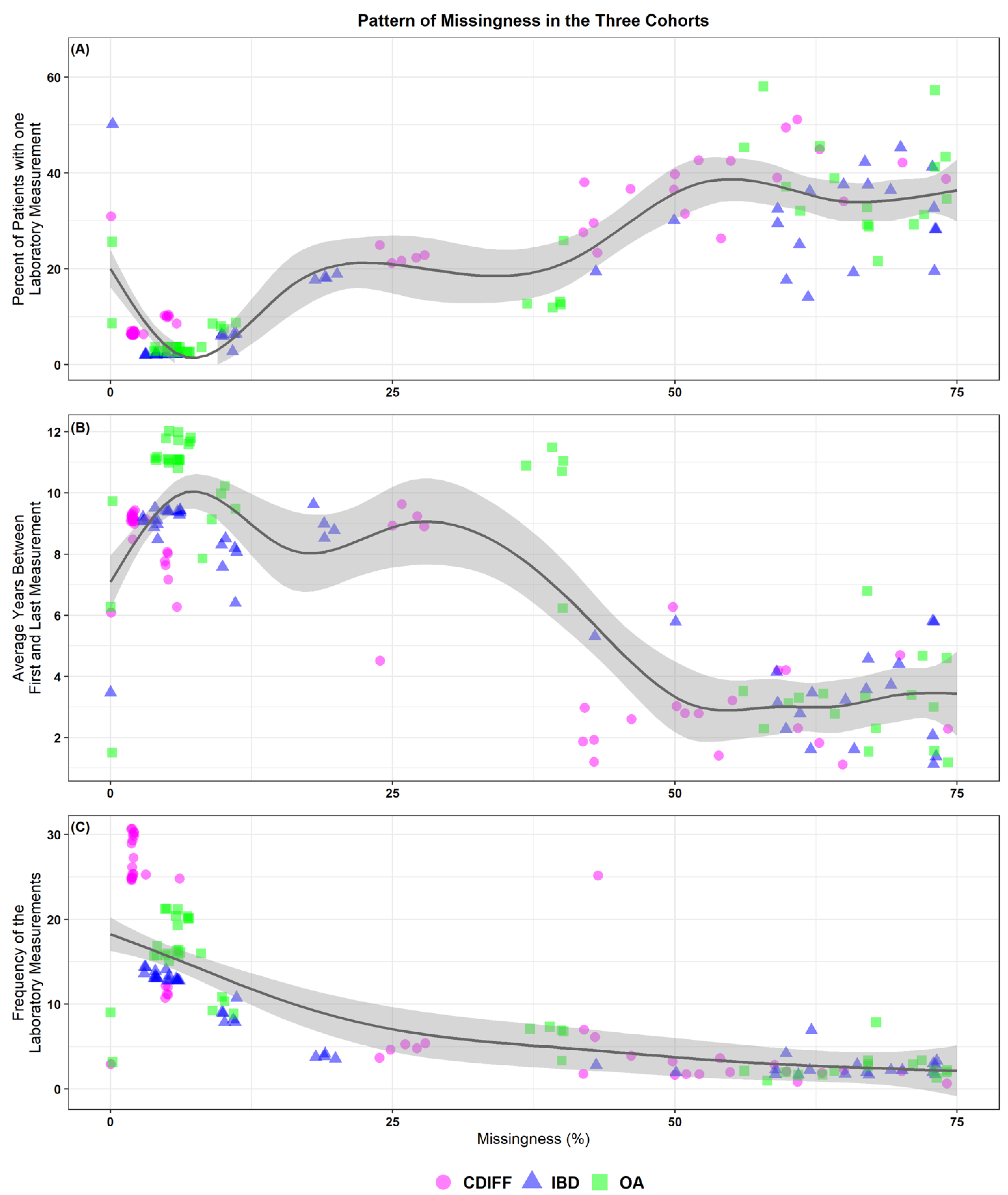
3. Results
3.1. Description of Laboratory Values for the Three Cohorts
3.2. Imputation Applied to Laboratory Values
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Diagnosis | Inclusion Criteria Using ICD Codes |
|---|---|
| ICD9 Diagnosis: Crohn’s and Ulcerative Colitis | 555, 55.0, 555.1, 555.2, 555.9, 556, 556.0, 556.1, 556.2, 556.3, 556.5, 556.6, 556.8, 556.9 |
| ICD10 Diagnosis: Crohn’s and Ulcerative Colitis | K50.00, K50.011, K50.012, K50.013, K50.014, K50.018, K50.019, K50.10, K50.111, K50.112, K50.113, K50.114, K50.118, K50.119, K50.80, K50.811, K50.812, K50.813, K50.814, K50.818, K50.819, K50.90, K50.911, K50.912, K50.913, K50.914, K50.918, K50.919, K51.80, K51.00, K51.011, K51.012, K51.014, K51.018, K51.019, K51.20, K51.211, K51.212, K51.213, K51.218, K51.219, K51.30, K51.311, K51.313, K51.314, K51.318, K51.319, K51.411, K51.414, K51.419, K51.50, K51.511, K51.513, K51.514, K51.518, K51.519, K51.80, K51.811, K51.812, K51.813, K51.814, K51.818, K51.819, K51.90, K51.911, K51.912, K51.913, K51.914, K51.918, K51.919 |
| ICD9 Diagnosis: Osteoarthritis | 715; 715.0; 715.00; 715.09; 715.1; 715.10; 715.15; 715.16; 715.30; 715.35; 715.36; 715.8; 715.80; 715.85; 715.86; 715.89; 715.9; 715.90; 715.95; 715.96; |
| ICD10 Diagnosis: Osteoarthritis | M15.0; M15.9; M16.0; M16.10; M16.11; M16.12; M16.2; M16.30; M16.31; M16.32; M16.9; M17.0; M17.10; M17.11; M17.12; M17.9; M19.91 |
| Percentage Missing | Percent of Patient with 1 Lab Value | Average Number of Years between First and Last Laboratory Measurement, for Patient with 2 or More Measurements ( in Years) | Frequency of the Laboratory Measurements Calculated for Patients with Two or More Measurements | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOINC ID | Short Description | Cdiff | IBD | OA | Cdiff | IBD | OA | Cdiff | IBD | OA | Cdiff | IBD | OA | ||||||||||||||||||
| Mean | Median | Q1 | Q3 | Mean | Median | Q1 | Q3 | Mean | Median | Q1 | Q3 | Mean | Median | Q1 | Q3 | Mean | Median | Q1 | Q3 | Mean | Median | Q1 | Q3 | ||||||||
| 14957-5 | Microalbumin in Urine | 75% | 73% | 31% | 28% | 26% | 7.1 | 6.0 | 2.8 | 10.6 | 6.8 | 5.8 | 2.6 | 10.1 | 7.4 | 6.3 | 3.0 | 11.0 | 5 | 3 | 1 | 8 | 5 | 3 | 1 | 7 | 6 | 4 | 1 | 8 | |
| 14959-1 | Microalbumin/Creatinine in Urine | 73% | 31% | 28% | 25% | 7.1 | 6.1 | 2.8 | 10.6 | 6.8 | 5.8 | 2.7 | 10.1 | 7.3 | 6.3 | 3.0 | 11.0 | 5 | 3 | 1 | 8 | 5 | 3 | 1 | 7 | 6 | 4 | 1 | 8 | ||
| 13969-1 | Creatine kinase.MB in Serum/Plasma | 54% | 73% | 26% | 20% | 24% | 3.6 | 1.4 | 0.0 | 5.9 | 3.6 | 1.1 | 0.0 | 6.0 | 3.9 | 1.5 | 0.0 | 6.7 | 7 | 4 | 2 | 9 | 6 | 3 | 1 | 7 | 6 | 3 | 2 | 7 | |
| 18262-6 | Cholesterol in LDL in Serum/Plasma (by direct assay) | 70% | 67% | 42% | 38% | 35% | 5.5 | 4.7 | 2.1 | 8.2 | 5.5 | 4.6 | 1.9 | 7.9 | 5.9 | 5.2 | 2.3 | 8.7 | 4 | 2 | 1 | 5 | 3 | 2 | 1 | 4 | 4 | 2 | 1 | 5 | |
| 6768-6 | Alkaline phosphatase in Serum/Plasma | 5% | 11% | 10% | 6% | 9% | 8.5 | 7.8 | 3.1 | 13.4 | 8.9 | 8.2 | 3.5 | 13.7 | 10.1 | 9.7 | 4.9 | 15.1 | 18 | 11 | 5 | 22 | 14 | 8 | 3 | 18 | 14 | 9 | 4 | 17 | |
| 2284-8 | Folate in Serum/Plasma | 65% | 73% | 74% | 34% | 33% | 35% | 3.2 | 1.1 | 0.0 | 5.1 | 3.5 | 1.4 | 0.0 | 5.6 | 3.4 | 1.2 | 0.0 | 5.5 | 3 | 2 | 1 | 3 | 3 | 2 | 1 | 3 | 3 | 2 | 1 | 3 |
| 3024-7 | Thyroxine (T4) free in Serum/Plasma | 60% | 70% | 74% | 49% | 45% | 43% | 5.7 | 4.2 | 1.4 | 8.9 | 6.0 | 4.4 | 1.7 | 9.4 | 6.3 | 4.6 | 1.8 | 9.8 | 3 | 2 | 1 | 4 | 3 | 2 | 1 | 3 | 4 | 2 | 1 | 4 |
| 1798-8 | Amylase in Serum/Plasma | 61% | 73% | 51% | 50% | 57% | 3.9 | 2.3 | 0.2 | 6.5 | 4.5 | 3.1 | 0.8 | 7.2 | 4.5 | 3.0 | 0.4 | 7.7 | 3 | 1 | 1 | 3 | 3 | 1 | 1 | 3 | 2 | 1 | 1 | 2 | |
| 2502-3 | Iron saturation in Serum/Plasma | 63% | 73% | 73% | 45% | 41% | 41% | 2.9 | 1.8 | 0.3 | 4.7 | 3.2 | 2.1 | 0.6 | 5.1 | 2.8 | 1.6 | 0.2 | 4.6 | 4 | 2 | 1 | 4 | 3 | 2 | 1 | 3 | 3 | 2 | 1 | 3 |
| 27353-2 | Glucose mean value in Blood Estimated from glycated hemoglobin | 59% | 59% | 72% | 39% | 33% | 31% | 4.6 | 4.2 | 1.8 | 7.2 | 4.5 | 4.1 | 1.8 | 7.3 | 4.9 | 4.7 | 2.1 | 7.8 | 7 | 3 | 1 | 9 | 6 | 2 | 1 | 7 | 7 | 3 | 1 | 10 |
| 2157-6 | Creatine kinase in Serum/Plasma | 46% | 61% | 71% | 37% | 25% | 29% | 4.4 | 2.6 | 0.1 | 7.3 | 4.5 | 2.8 | 0.2 | 7.5 | 4.9 | 3.4 | 0.2 | 8.0 | 7 | 4 | 1 | 8 | 5 | 2 | 1 | 5 | 5 | 3 | 1 | 6 |
| 2340-8 | Glucose in Blood by Automated test strip | 43% | 62% | 68% | 23% | 14% | 22% | 3.8 | 1.9 | 0.1 | 6.2 | 3.8 | 1.6 | 0.0 | 6.3 | 4.1 | 2.3 | 0.0 | 6.8 | 77 | 25 | 3 | 92 | 40 | 7 | 2 | 33 | 41 | 8 | 2 | 39 |
| 17856-6 | Hemoglobin A1c/Hemoglobin.total in Blood | 50% | 50% | 67% | 37% | 30% | 29% | 7.5 | 6.3 | 2.7 | 11.4 | 7.1 | 5.8 | 2.4 | 10.8 | 7.8 | 6.8 | 3.0 | 11.8 | 9 | 3 | 1 | 12 | 8 | 2 | 1 | 8 | 10 | 3 | 1 | 13 |
| 2777-1 | Phosphate in Serum/Plasma | 42% | 60% | 67% | 28% | 18% | 29% | 4.0 | 1.9 | 0.1 | 6.4 | 4.4 | 2.3 | 0.1 | 7.2 | 5.2 | 3.3 | 0.5 | 8.2 | 14 | 7 | 2 | 17 | 10 | 4 | 1 | 10 | 8 | 3 | 1 | 8 |
| 19123-9 | Magnesium in Serum / Plasma | 43% | 66% | 67% | 30% | 19% | 33% | 3.0 | 1.2 | 0.1 | 4.5 | 3.6 | 1.6 | 0.1 | 5.6 | 3.4 | 1.5 | 0.0 | 5.3 | 13 | 6 | 2 | 16 | 10 | 3 | 1 | 10 | 7 | 3 | 1 | 7 |
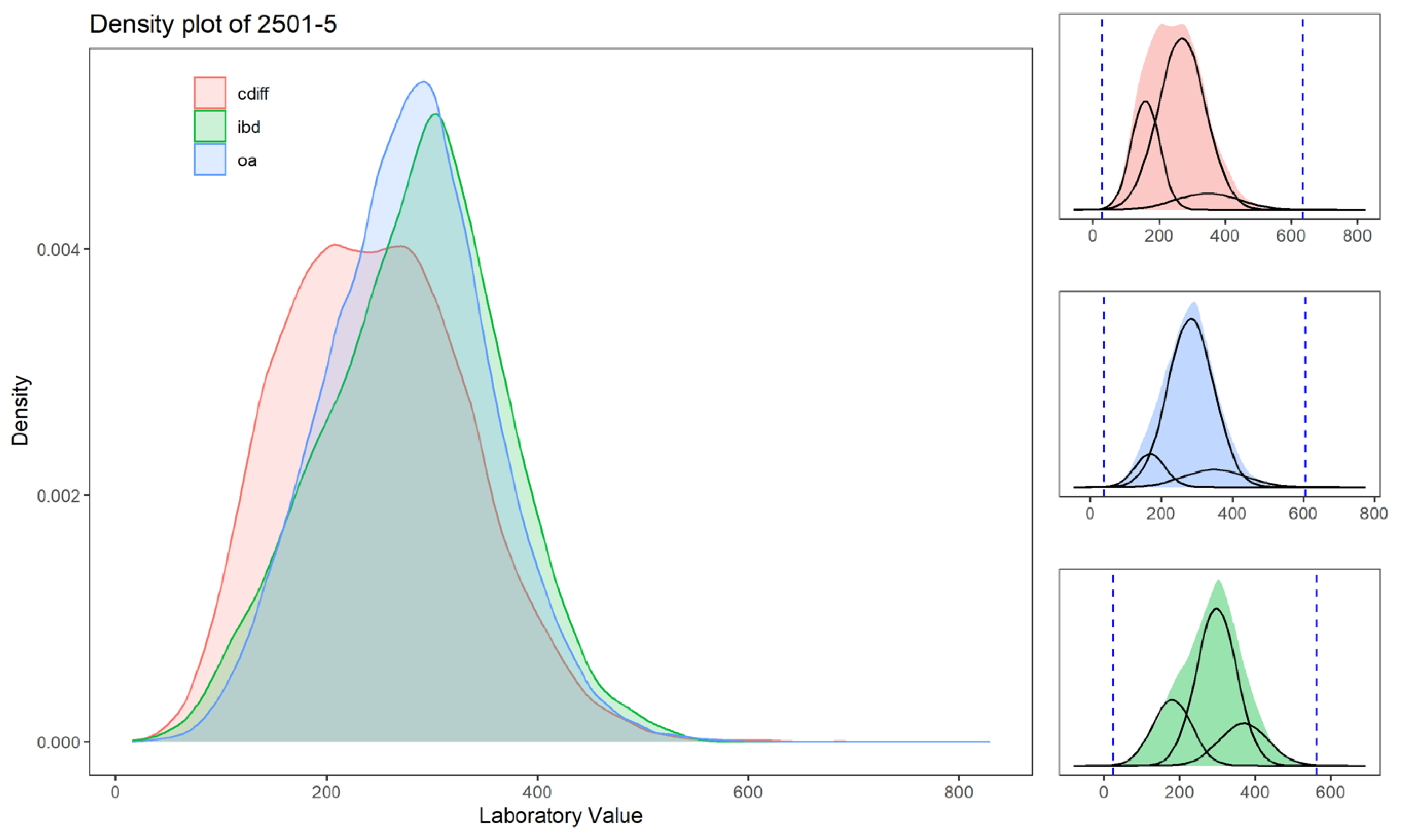
| 2501-5 | Iron binding capacity.unsaturated in Serum/Plasma | 52% | 65% | 64% | 43% | 38% | 39% | 4.2 | 2.8 | 0.6 | 6.5 | 4.8 | 3.2 | 1.1 | 7.3 | 4.3 | 2.8 | 0.6 | 6.5 | 4 | 2 | 1 | 4 | 3 | 2 | 1 | 4 | 3 | 2 | 1 | 4 |
| 2276-4 | Ferritin in Serum/Plasma | 55% | 67% | 63% | 42% | 42% | 46% | 4.5 | 3.2 | 0.9 | 6.8 | 4.9 | 3.6 | 1.3 | 7.3 | 4.7 | 3.4 | 1.1 | 7.1 | 4 | 2 | 1 | 4 | 4 | 2 | 1 | 4 | 3 | 2 | 1 | 3 |
| 2132-9 | Cobalamin (Vitamin B12) in Serum/Plasma | 51% | 59% | 61% | 31% | 29% | 32% | 4.5 | 2.8 | 0.2 | 7.2 | 5.1 | 3.1 | 0.4 | 8.3 | 4.9 | 3.3 | 0.2 | 7.8 | 3 | 2 | 1 | 4 | 4 | 2 | 1 | 4 | 3 | 2 | 1 | 4 |
| 2498-4 | Iron in Serum/Plasma | 50% | 62% | 60% | 40% | 36% | 37% | 4.4 | 3.0 | 0.7 | 6.7 | 5.0 | 3.5 | 1.2 | 7.5 | 4.5 | 3.1 | 0.6 | 6.9 | 4 | 2 | 1 | 4 | 4 | 2 | 1 | 4 | 3 | 2 | 1 | 4 |
| 1988-5 | C reactive protein in Serum/Plasma | 74% | 58% | 39% | 56% | 58% | 3.6 | 2.3 | 0.5 | 5.8 | 4.5 | 3.5 | 1.3 | 7.0 | 3.8 | 2.3 | 0.5 | 5.8 | 3 | 1 | 1 | 3 | 4 | 2 | 1 | 5 | 2 | 1 | 1 | 2 | |
| 3040-3 | Lipase in Serum/Plasma | 42% | 69% | 56% | 38% | 36% | 45% | 4.5 | 3.0 | 0.6 | 7.1 | 5.1 | 3.7 | 1.2 | 7.9 | 5.0 | 3.5 | 0.9 | 8.0 | 4 | 2 | 1 | 4 | 4 | 2 | 1 | 4 | 3 | 2 | 1 | 3 |
| 13457-7 | Cholesterol in LDL in Serum/Plasma (by calculation) | 28% | 20% | 40% | 23% | 19% | 13% | 9.5 | 8.9 | 4.2 | 14.5 | 9.4 | 8.8 | 4.0 | 14.7 | 10.7 | 10.7 | 5.5 | 16.0 | 8 | 5 | 2 | 11 | 7 | 4 | 2 | 9 | 10 | 7 | 3 | 14 |
| 2085-9 | Cholesterol in HDL in Serum/Plasma | 27% | 19% | 40% | 22% | 18% | 13% | 9.6 | 9.2 | 4.3 | 14.8 | 9.6 | 9.0 | 4.1 | 15.0 | 11.0 | 11.0 | 5.7 | 16.3 | 8 | 5 | 2 | 12 | 7 | 4 | 2 | 10 | 10 | 7 | 3 | 14 |
| 1968-7 | Bilirubin.direct in Serum/Plasma | 24% | 43% | 40% | 25% | 19% | 26% | 6.0 | 4.5 | 1.0 | 9.8 | 6.6 | 5.3 | 1.8 | 10.4 | 7.2 | 6.2 | 2.1 | 11.6 | 8 | 4 | 2 | 9 | 7 | 3 | 2 | 8 | 6 | 3 | 1 | 7 |
| 2093-3 | Cholesterol in Serum/Plasma | 26% | 18% | 39% | 22% | 18% | 12% | 10.0 | 9.6 | 4.5 | 15.5 | 10.0 | 9.6 | 4.2 | 15.7 | 11.4 | 11.5 | 5.9 | 16.9 | 9 | 5 | 2 | 12 | 8 | 4 | 2 | 10 | 10 | 7 | 3 | 15 |
| 2571-8 | Triglyceride in Serum/Plasma | 25% | 19% | 37% | 21% | 18% | 13% | 9.4 | 8.9 | 4.1 | 14.6 | 9.2 | 8.5 | 3.5 | 14.7 | 10.8 | 10.9 | 5.6 | 16.2 | 8 | 5 | 2 | 12 | 7 | 4 | 2 | 10 | 10 | 7 | 3 | 14 |
| 1975-2 | Bilirubin.total in Serum/Plasma | 5% | 11% | 11% | 10% | 6% | 9% | 8.4 | 7.6 | 3.0 | 13.1 | 8.8 | 8.1 | 3.5 | 13.5 | 9.9 | 9.5 | 4.8 | 14.8 | 17 | 11 | 5 | 22 | 14 | 8 | 3 | 17 | 14 | 9 | 4 | 17 |
| 30239-8 | Aspartate aminotransferase in Serum/Plasma | 5% | 10% | 10% | 10% | 6% | 8% | 8.8 | 8.1 | 3.2 | 13.9 | 9.1 | 8.5 | 3.7 | 14.1 | 10.4 | 10.2 | 5.2 | 15.7 | 19 | 12 | 5 | 24 | 15 | 9 | 3 | 19 | 15 | 10 | 4 | 20 |
| 1743-4 | Alanine aminotransferase in Serum/Plasma | 5% | 10% | 10% | 10% | 6% | 8% | 8.6 | 8.0 | 3.2 | 13.5 | 9.0 | 8.3 | 3.8 | 13.8 | 10.2 | 10.0 | 5.2 | 15.2 | 19 | 12 | 5 | 25 | 15 | 9 | 4 | 20 | 16 | 11 | 5 | 22 |
| 2885-2 | Protein in Serum/Plasma | 5% | 10% | 9% | 10% | 6% | 9% | 7.9 | 7.2 | 2.9 | 12.4 | 8.3 | 7.6 | 3.3 | 12.8 | 9.3 | 9.1 | 4.6 | 14.0 | 17 | 11 | 5 | 22 | 14 | 8 | 3 | 17 | 14 | 9 | 4 | 17 |
| 10466-1 | Anion gap 3 in Serum/Plasma | 6% | 11% | 8% | 9% | 3% | 4% | 6.3 | 6.3 | 2.5 | 10.2 | 6.4 | 6.4 | 2.7 | 10.4 | 7.3 | 7.9 | 3.9 | 11.1 | 39 | 25 | 10 | 51 | 22 | 11 | 4 | 27 | 26 | 16 | 7 | 32 |
| 2028-9 | Carbon dioxide, total in Serum/Plasma | 2% | 4% | 7% | 7% | 2% | 3% | 9.6 | 9.2 | 3.8 | 15.1 | 9.5 | 8.9 | 3.7 | 15.0 | 11.3 | 11.6 | 6.0 | 16.9 | 45 | 29 | 12 | 59 | 26 | 13 | 5 | 31 | 31 | 20 | 9 | 40 |
| 2951-2 | Sodium in Serum/Plasma | 2% | 4% | 7% | 7% | 2% | 3% | 9.6 | 9.2 | 3.8 | 15.2 | 9.6 | 9.1 | 3.8 | 15.1 | 11.3 | 11.7 | 6.0 | 16.9 | 45 | 30 | 12 | 60 | 26 | 13 | 5 | 32 | 31 | 20 | 9 | 40 |
| 3094-0 | Urea nitrogen in Serum/Plasma | 2% | 4% | 7% | 7% | 2% | 3% | 9.7 | 9.3 | 3.9 | 15.3 | 9.6 | 9.1 | 3.7 | 15.2 | 11.4 | 11.8 | 6.1 | 17.1 | 45 | 30 | 12 | 60 | 26 | 13 | 5 | 32 | 32 | 20 | 9 | 41 |
| 17861-6 | Calcium in Serum/Plasma | 2% | 4% | 6% | 7% | 2% | 3% | 8.8 | 8.5 | 3.5 | 14.0 | 8.9 | 8.5 | 3.5 | 14.0 | 10.4 | 10.8 | 5.5 | 15.6 | 44 | 29 | 12 | 58 | 25 | 13 | 5 | 31 | 30 | 19 | 9 | 38 |
| 777-3 | Platelets in Blood | 2% | 6% | 6% | 6% | 2% | 4% | 9.5 | 9.1 | 3.7 | 15.1 | 9.8 | 9.4 | 3.9 | 15.4 | 11.0 | 11.1 | 5.6 | 16.6 | 40 | 25 | 11 | 52 | 25 | 13 | 5 | 31 | 27 | 16 | 7 | 33 |
| 789-8 | Erythrocytes in Blood | 2% | 6% | 6% | 6% | 2% | 4% | 9.5 | 9.1 | 3.7 | 15.1 | 9.8 | 9.4 | 3.9 | 15.4 | 11.0 | 11.1 | 5.6 | 16.7 | 40 | 25 | 11 | 52 | 25 | 13 | 5 | 31 | 27 | 16 | 7 | 33 |
| 788-0 | Erythrocyte distribution width | 3% | 6% | 6% | 6% | 2% | 4% | 9.5 | 9.1 | 3.7 | 15.0 | 9.8 | 9.4 | 3.9 | 15.4 | 11.0 | 11.1 | 5.6 | 16.6 | 40 | 25 | 11 | 52 | 25 | 13 | 5 | 31 | 27 | 16 | 7 | 33 |
| 6690-2 | Leukocytes in Blood | 2% | 6% | 6% | 6% | 2% | 4% | 9.5 | 9.1 | 3.7 | 15.1 | 9.8 | 9.4 | 3.9 | 15.4 | 11.0 | 11.1 | 5.6 | 16.7 | 41 | 25 | 11 | 53 | 25 | 13 | 5 | 31 | 27 | 16 | 7 | 33 |
| 2345-7 | Glucose in Serum/Plasma | 2% | 3% | 6% | 7% | 2% | 3% | 9.7 | 9.4 | 3.9 | 15.4 | 9.7 | 9.2 | 3.8 | 15.4 | 11.5 | 12.0 | 6.1 | 17.3 | 46 | 30 | 13 | 61 | 27 | 14 | 5 | 32 | 32 | 21 | 9 | 41 |
| 2075-0 | Chloride in Serum/Plasma | 2% | 4% | 6% | 7% | 2% | 3% | 9.6 | 9.2 | 3.8 | 15.1 | 9.5 | 9.0 | 3.7 | 15.0 | 11.3 | 11.7 | 6.0 | 16.9 | 45 | 30 | 12 | 59 | 26 | 13 | 5 | 31 | 31 | 20 | 9 | 40 |
| 32623-1 | Platelet mean volume in Blood | 2% | 6% | 5% | 7% | 2% | 4% | 9.4 | 9.0 | 3.6 | 15.0 | 9.7 | 9.3 | 3.9 | 15.4 | 11.0 | 11.0 | 5.5 | 16.6 | 39 | 25 | 11 | 51 | 25 | 13 | 5 | 31 | 26 | 15 | 7 | 32 |
| 2823-3 | Potassium in Serum/Plasma | 2% | 3% | 5% | 6% | 2% | 3% | 9.7 | 9.3 | 3.9 | 15.4 | 9.6 | 9.1 | 3.7 | 15.2 | 11.5 | 11.8 | 6.1 | 17.2 | 47 | 31 | 13 | 62 | 27 | 14 | 5 | 33 | 32 | 21 | 9 | 41 |
| 785-6 | MCH | 2% | 5% | 5% | 6% | 2% | 4% | 9.5 | 9.1 | 3.7 | 15.0 | 9.8 | 9.4 | 3.9 | 15.4 | 11.0 | 11.1 | 5.6 | 16.6 | 40 | 25 | 11 | 52 | 25 | 13 | 5 | 31 | 27 | 16 | 7 | 33 |
| 786-4 | MCHC | 2% | 5% | 5% | 6% | 2% | 4% | 9.5 | 9.1 | 3.7 | 15.0 | 9.8 | 9.4 | 3.9 | 15.4 | 11.0 | 11.1 | 5.6 | 16.6 | 40 | 25 | 11 | 52 | 25 | 13 | 5 | 31 | 27 | 16 | 7 | 33 |
| 2160-0 | Creatinine in Serum/Plasma | 2% | 3% | 5% | 6% | 2% | 3% | 9.8 | 9.4 | 3.9 | 15.4 | 9.6 | 9.1 | 3.8 | 15.2 | 11.6 | 12.0 | 6.4 | 17.2 | 46 | 31 | 13 | 62 | 27 | 14 | 5 | 33 | 33 | 21 | 9 | 42 |
| 718-7 | Hemoglobin in Blood | 2% | 4% | 4% | 6% | 2% | 3% | 9.6 | 9.2 | 3.7 | 15.3 | 9.9 | 9.5 | 4.0 | 15.7 | 11.0 | 11.2 | 5.5 | 16.8 | 43 | 27 | 11 | 56 | 27 | 14 | 6 | 32 | 29 | 17 | 8 | 35 |
| 4544-3 | Hematocrit of Blood by Automated count | 2% | 5% | 4% | 6% | 2% | 3% | 9.5 | 9.2 | 3.7 | 15.2 | 9.8 | 9.4 | 3.9 | 15.5 | 11.0 | 11.1 | 5.5 | 16.7 | 42 | 26 | 11 | 55 | 26 | 14 | 5 | 32 | 28 | 16 | 8 | 34 |
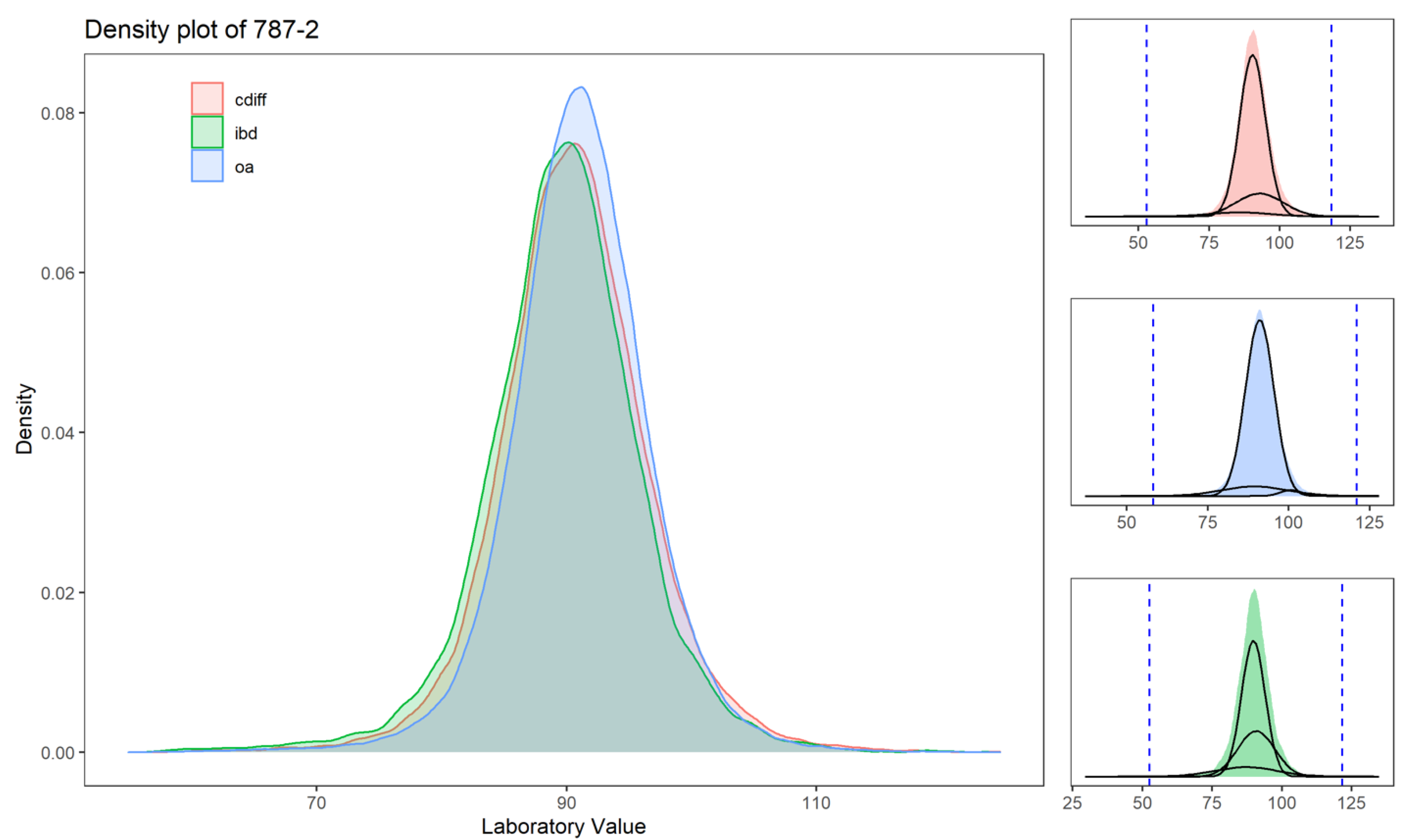
| 787-2 | Mean corpuscular volume, or MCV | 2% | 5% | 4% | 6% | 2% | 4% | 9.5 | 9.1 | 3.7 | 15.1 | 9.8 | 9.4 | 3.9 | 15.4 | 11.0 | 11.1 | 5.6 | 16.7 | 40 | 25 | 11 | 52 | 25 | 13 | 5 | 31 | 27 | 16 | 7 | 33 |
| Cdiff-PMM | Cdiff-RF | |||||
|---|---|---|---|---|---|---|
| Missingness Level | Dimensionality Level (g) | Cluster Number | RMSE Difference | p-Value | RMSE Difference | p-Value |
| 25% | 100 | 4 | −0.774 | 0.376 | 0.349 | 0.625 |
| 25% | 100 | 8 | 0.110 | 0.739 | 1.402 | 0.532 |
| 25% | 100 | 16 | −2.121 | 0.306 | −0.189 | 0.629 |
| 25% | 1000 | 4 | 7.417 | 0.456 | 2.066 | 0.391 |
| 25% | 1000 | 8 | 0.141 | 0.584 | 1.238 | 0.581 |
| 25% | 1000 | 16 | 5.916 | 0.139 | −0.035 | 0.233 |
| 25% | 8160 | 4 | −3.088 | 0.419 | −4.397 | 0.582 |
| 25% | 8160 | 8 | 4.628 | 0.150 | −0.882 | 0.868 |
| 25% | 8160 | 16 | 4.910 | 0.493 | 0.631 | 0.594 |
| 50% | 100 | 4 | 7.117 | 0.459 | −1.470 | 0.789 |
| 50% | 100 | 8 | 9.189 | 0.759 | 11.064 | 0.796 |
| 50% | 100 | 16 | −3.005 | 0.351 | 14.731 | 0.472 |
| 50% | 1000 | 4 | 6.934 | 0.920 | 0.503 | 0.675 |
| 50% | 1000 | 8 | 6.695 | 0.230 | 4.044 | 0.432 |
| 50% | 1000 | 16 | 16.207 | 0.087 | 5.976 | 0.196 |
| 50% | 8160 | 4 | 2.060 | 0.481 | −3.279 | 0.865 |
| 50% | 8160 | 8 | 10.087 | 0.435 | −7.323 | 0.502 |
| 50% | 8160 | 16 | 12.366 | 0.190 | −19.655 | 0.476 |
| 75% | 100 | 4 | −8.756 | 0.386 | −4.916 | 0.662 |
| 75% | 100 | 8 | 12.386 | 0.174 | −16.748 | 0.487 |
| 75% | 100 | 16 | 5.026 | 0.392 | −2.362 | 0.513 |
| 75% | 1000 | 4 | −31.468 | 0.017 | −12.722 | 0.982 |
| 75% | 1000 | 8 | 4.024 | 0.266 | 9.729 | 0.405 |
| 75% | 1000 | 16 | 23.333 | 0.139 | −9.162 | 0.258 |
| 75% | 8160 | 4 | 8.368 | 0.569 | 0.488 | 0.787 |
| 75% | 8160 | 8 | 6.993 | 0.515 | −5.113 | 0.631 |
| 75% | 8160 | 16 | 2.414 | 0.957 | −9.496 | 0.979 |
| IBD-PMM | IBD-RF | |||||
|---|---|---|---|---|---|---|
| Missingness Level | Dimensionality Level (g) | Cluster Number | RMSE Difference | p-Value | RMSE Difference | p-Value |
| 25% | 100 | 2 | 0.938 | 0.565 | 0.756 | 0.759 |
| 25% | 100 | 4 | 1.264 | 0.948 | 0.200 | 0.695 |
| 25% | 100 | 8 | −0.359 | 0.273 | 0.969 | 0.339 |
| 25% | 1000 | 2 | 1.284 | 0.583 | −1.145 | 0.425 |
| 25% | 1000 | 4 | 1.134 | 0.234 | −0.526 | 0.733 |
| 25% | 1000 | 8 | −2.696 | 0.196 | 1.083 | 0.132 |
| 25% | 7916 | 2 | −0.886 | 0.974 | 0.176 | 0.944 |
| 25% | 7916 | 4 | 0.313 | 0.210 | −0.906 | 0.249 |
| 25% | 7916 | 8 | 0.005 | 0.307 | 0.264 | 0.177 |
| 50% | 100 | 2 | 0.218 | 0.336 | 0.682 | 0.448 |
| 50% | 100 | 4 | 0.168 | 0.196 | 2.094 | 0.281 |
| 50% | 100 | 8 | 2.851 | 0.072 | −0.057 | 0.428 |
| 50% | 1000 | 2 | 0.080 | 0.411 | 0.230 | 0.561 |
| 50% | 1000 | 4 | 1.465 | 0.601 | 2.246 | 0.569 |
| 50% | 1000 | 8 | −0.745 | 0.609 | 2.145 | 0.604 |
| 50% | 7916 | 2 | 1.973 | 0.338 | 1.165 | 0.912 |
| 50% | 7916 | 4 | 1.922 | 0.188 | 1.973 | 0.676 |
| 50% | 7916 | 8 | 4.401 | 0.078 | 3.309 | 0.288 |
| 75% | 100 | 2 | −6.485 | 0.256 | −3.192 | 0.447 |
| 75% | 100 | 4 | −3.428 | 0.632 | 0.756 | 0.580 |
| 75% | 100 | 8 | 6.598 | 0.825 | −4.165 | 0.624 |
| 75% | 1000 | 2 | 5.436 | 0.721 | −4.835 | 0.306 |
| 75% | 1000 | 4 | 1.664 | 0.511 | 0.329 | 0.584 |
| 75% | 1000 | 8 | −7.031 | 0.581 | 1.175 | 0.771 |
| 75% | 7916 | 2 | 0.239 | 0.378 | −8.353 | 0.175 |
| 75% | 7916 | 4 | −4.155 | 0.470 | −4.033 | 0.689 |
| 75% | 7916 | 8 | 3.760 | 0.468 | −8.244 | 0.096 |
| OA-PMM | OA-RF | |||||
|---|---|---|---|---|---|---|
| Missingness Level | Dimensionality Level (g) | Cluster Number | RMSE Difference | p-Value | RMSE Difference | p-Value |
| 25% | 100 | 4 | 0.035 | 0.317 | 2.449 | 0.245 |
| 25% | 100 | 8 | −0.074 | 0.444 | 4.734 | 0.385 |
| 25% | 100 | 16 | −0.017 | 0.375 | −0.518 | 0.525 |
| 25% | 1000 | 4 | 0.035 | 0.687 | 3.351 | 0.247 |
| 25% | 1000 | 8 | −0.066 | 0.363 | 3.859 | 0.183 |
| 25% | 1000 | 16 | 0.085 | 0.706 | 1.414 | 0.172 |
| 25% | 2042 | 4 | 0.081 | 0.889 | 1.705 | 0.161 |
| 25% | 2042 | 8 | 0.004 | 0.595 | 4.417 | 0.460 |
| 25% | 2042 | 16 | −0.019 | 0.202 | 1.602 | 0.810 |
| 50% | 100 | 4 | 0.081 | 0.700 | −4.229 | 0.199 |
| 50% | 100 | 8 | 0.218 | 0.079 | 1.132 | 0.970 |
| 50% | 100 | 16 | 0.101 | 0.087 | 3.082 | 0.357 |
| 50% | 1000 | 4 | 0.106 | 0.653 | 10.161 | 0.843 |
| 50% | 1000 | 8 | −0.066 | 0.577 | −1.271 | 0.480 |
| 50% | 1000 | 16 | 0.147 | 0.620 | −0.328 | 0.891 |
| 50% | 2042 | 4 | 0.178 | 0.252 | −2.703 | 0.946 |
| 50% | 2042 | 8 | −0.013 | 0.216 | −11.300 | 0.409 |
| 50% | 2042 | 16 | 0.092 | 0.643 | 3.229 | 0.376 |
| 75% | 100 | 4 | −0.131 | 0.186 | 6.828 | 0.213 |
| 75% | 100 | 8 | 0.118 | 0.507 | −0.098 | 0.434 |
| 75% | 100 | 16 | 0.197 | 0.142 | −2.326 | 0.889 |
| 75% | 1000 | 4 | −0.077 | 0.092 | −4.702 | 0.222 |
| 75% | 1000 | 8 | 0.157 | 0.428 | −0.343 | 0.653 |
| 75% | 1000 | 16 | −0.053 | 0.508 | −6.447 | 0.651 |
| 75% | 2042 | 4 | 0.055 | 0.649 | −0.749 | 0.430 |
| 75% | 2042 | 8 | −0.089 | 0.549 | 1.865 | 0.768 |
| 75% | 2042 | 16 | 0.237 | 0.014 | 10.926 | 0.061 |
References
- Noorbakhsh-Sabet, N.; Zand, R.; Zhang, Y.; Abedi, V. Artificial Intelligence Transforms the Future of Health Care. Am. J. Med. 2019, 132, 795–801. [Google Scholar] [CrossRef]
- Botsis, T.; Hartvigsen, G.; Chen, F.; Weng, C. Secondary Use of EHR: Data Quality Issues and Informatics Opportunities. AMIA Jt. Summits Transl. Sci. 2010, 1, 1–5. [Google Scholar]
- Sterne, J.; White, I.R.; Carlin, J.B.; Spratt, M.; Royston, P.; Kenward, M.G.; Wood, A.M.; Carpenter, J.R. Multiple imputation for missing data in epidemiological and clinical research: Potential and pitfalls. BMJ 2009, 338, b2393. [Google Scholar] [CrossRef]
- Netten, A.P.; Dekker, F.W.; Rieffe, C.; Soede, W.; Briaire, J.J.; Frijns, J.H.M. Missing Data in the Field of Otorhinolaryngology and Head & Neck Surgery. Ear Hear. 2017, 38, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu-Jones, B.K.; Lavage, D.R.; Snyder, J.W.; Moore, J.H.; Pendergrass, S.A.; Bauer, C.R. Characterizing and Managing Missing Structured Data in Electronic Health Records: Data Analysis. JMIR Med. Inform. 2018, 6, e11. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu-Jones, B.K.; Moore, J.H. Missing data imputation in the electronic health record using deeply learned autoencoders. Biocomputing 2017, 207–218. [Google Scholar] [CrossRef]
- Troyanskaya, O.G.; Cantor, M.; Sherlock, G.; Brown, P.O.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing value estimation methods for DNA microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef]
- Kuppusamy, V.; Paramasivam, I. Integrating WLI fuzzy clustering with grey neural network for missing data imputation. Int. J. Intell. Enterp. 2017, 4, 103. [Google Scholar] [CrossRef]
- Lee, K.J.; Carlin, J.B. Multiple imputation in the presence of non-normal data. Stat. Med. 2017, 36, 606–617. [Google Scholar] [CrossRef]
- Liu, Y.; Gopalakrishnan, V. An Overview and Evaluation of Recent Machine Learning Imputation Methods Using Cardiac Imaging Data. Data 2017, 2, 8. [Google Scholar] [CrossRef]
- Ford, E.; Rooney, P.; Hurley, P.; Oliver, S.; Bremner, S.; Cassell, J. Can the Use of Bayesian Analysis Methods Correct for Incompleteness in Electronic Health Records Diagnosis Data? Development of a Novel Method Using Simulated and Real-Life Clinical Data. Front. Public Health 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Wells, B.J.; Nowacki, A.S.; Chagin, K.M.; Kattan, M.W. Strategies for Handling Missing Data in Electronic Health Record Derived Data. eGEMs Gener. Évid. Methods Improv. Patient Outcomes 2013, 1, 1035. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, Y.; Moore, J.H. Integration of genetic and clinical information to improve imputation of data missing from electronic health records. J. Am. Med. Inform. Assoc. 2019, 26, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- White, I.R.; Royston, P.; Wood, A.M. Multiple imputation using chained equations: Issues and guidance for practice. Stat. Med. 2010, 30, 377–399. [Google Scholar] [CrossRef] [PubMed]
- van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. Available online: https://www.jstatsoft.org/v45/i03/ (accessed on 5 October 2020). [CrossRef]
- Luo, Y.; Szolovits, P.; Dighe, A.S.; Baron, J.M. 3D-MICE: Integration of cross-sectional and longitudinal imputation for multi-analyte longitudinal clinical data. J. Am. Med. Inform. Assoc. 2017, 25, 645–653. [Google Scholar] [CrossRef]
- Abt, M.C.; McKenney, P.T.; Pamer, E.G. Clostridium difficile colitis: Pathogenesis and host defence. Nat. Rev. Genet. 2016, 14, 609–620. [Google Scholar] [CrossRef]
- Carrell, D.; Denny, J. Group Health and Vanderbilt. In Clostridium Difficile Colitis; PheKB: Nashville, TN, USA, 2012. [Google Scholar]
- Abedi, V.; Shivakumar, M.K.; Lu, P.; Hontecillas, R.; Leber, A.; Ahuja, M.; Ulloa, A.E.; Shellenberger, M.J.; Bassaganya-Riera, J. Latent-Based Imputation of Laboratory Measures from Electronic Health Records: Case for Complex Diseas-es. bioRxiv 2018, 275743. [Google Scholar] [CrossRef]
- Landauer, T.K.; Dumais, S.T. A solution to Plato’s problem: The latent semantic analysis theory of acquisition, induction, and representation of knowledge. Psychol. Rev. 1997, 104, 211–240. [Google Scholar] [CrossRef]
- Aspects of Automatic Text Analysis; Mehler, A., Köhler, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 209. [Google Scholar]
- Breiman, L. Manual on Setting Up, Using, and Understanding Random Forests v3.1; Tech. Report; Statistics Department University of California Berkeley: Berkeley, CA, USA, 2002; Available online: https://www.stat.berkeley.edu/~breiman/Using_random_forests_V3.1.pdf (accessed on 29 December 2020).
- Leber, A.; Hontecillas, R.; Tubau-Juni, N.; Zoccoli-Rodriguez, V.; Hulver, M.; McMillan, R.; Eden, K.; Allen, I.C.; Bassaganya-Riera, J. NLRX1 Regulates Effector and Metabolic Functions of CD4+ T Cells. J. Immunol. 2017, 198, 2260–2268. [Google Scholar] [CrossRef]
- Burgette, L.F.; Reiter, J.P. Multiple Imputation for Missing Data via Sequential Regression Trees. Am. J. Epidemiol. 2010, 172, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.D.; Bartlett, J.W.; Carpenter, J.; Nicholas, O.; Hemingway, H. Comparison of Random Forest and Parametric Imputation Models for Imputing Missing Data Using MICE: A CALIBER Study. Am. J. Epidemiol. 2014, 179, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.J.; Shlens, J.; Szegedy, C. Explaining and harnessing adversarial examples. arXiv 2014, arXiv:1412.6572. [Google Scholar]
- Yoon, J.; Jordon, J.; van der Schaar, M. GAIN: Missing data imputation using generative adversarial nets. arXiv 2018, arXiv:1806.02920. [Google Scholar]
- Breiman, L. Using Iterated Bagging to Debias Regressions. Mach. Learn. 2001, 45, 261–277. [Google Scholar] [CrossRef]
- Bühlmann, P.; Yu, B. Analyzing bagging. Ann. Stat. 2002, 30, 927–961. [Google Scholar] [CrossRef]
- Chen, R.; Stewart, W.F.; Sun, J.; Ng, K.; Yan, X. Recurrent Neural Networks for Early Detection of Heart Failure from Longitudinal Electronic Health Record Data: Implications for Temporal Modeling with Respect to Time Before Diagnosis, Data Density, Data Quantity, and Data Type. Circ. Cardiovasc. Qual. Outcomes 2019, 12, e005114. [Google Scholar] [CrossRef]
- Ng, K.; Steinhubl, S.R.; Defilippi, C.; Dey, S.; Stewart, W.F. Early Detection of Heart Failure Using Electronic Health Records: Practical Implications for Time before Diagnosis, Data Diversity, Data Quantity, and Data Density. Circ. Cardiovasc. Qual. Outcomes 2016, 9, 649–658. [Google Scholar] [CrossRef]
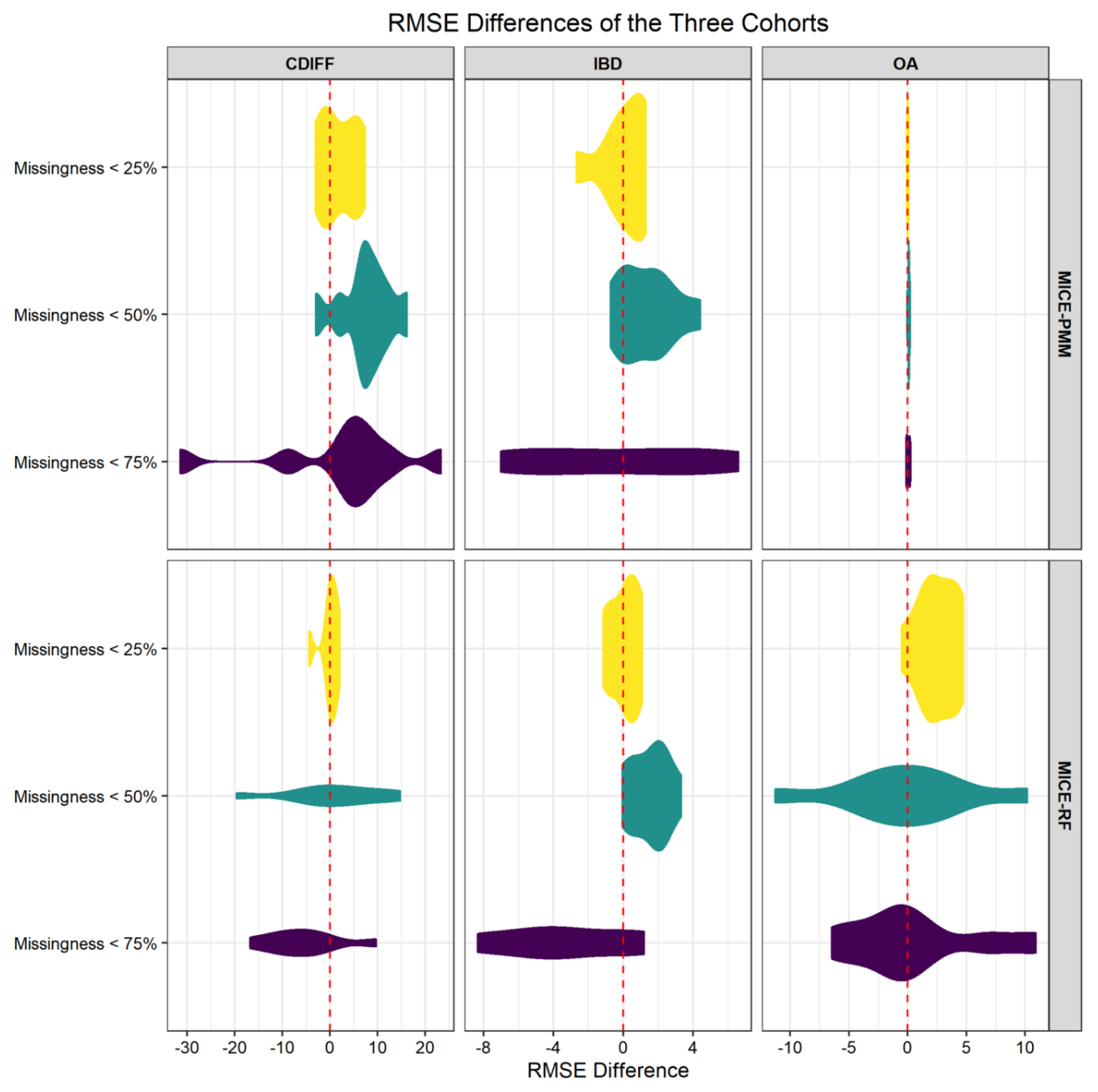
| C. difficile (Cdiff) Infection | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MICE-PMM | MICE-RF | ||||||||
| Cluster Number | Dimensionality Level (g) | Missingness < 25% | Missingness < 50% | Missingness < 75% | Cluster Number | Dimensionality Level (g) | Missingness < 25% | Missingness < 50% | Missingness < 75% |
| 4 | 100 | −0.77 | 7.12 | −8.76 | 4 | 100 | 0.35 | −1.47 | −4.92 |
| 1000 | 7.42 | 6.93 | −31.47 | 1000 | 2.07 | 0.50 | −12.72 | ||
| 8160 | −3.09 | 2.06 | 8.37 | 8160 | −4.40 | −3.28 | 0.49 | ||
| 8 | 100 | 0.11 | 9.19 | 12.39 | 8 | 100 | 1.40 | 11.06 | −16.75 |
| 1000 | 0.14 | 6.69 | 4.02 | 1000 | 1.24 | 4.04 | 9.73 | ||
| 8160 | 4.63 | 10.09 | 6.99 | 8160 | −0.88 | −7.32 | −5.11 | ||
| 16 | 100 | −2.12 | −3.00 | 5.03 | 16 | 100 | −0.04 | 14.73 | −2.36 |
| 1000 | 5.92 | 16.21 | 23.33 | 1000 | −0.19 | 5.98 | −9.16 | ||
| 8160 | 4.91 | 12.37 | 2.41 | 8160 | 0.63 | −19.66 | −9.50 | ||
| Inflammatory Bowel Disease (IBD) | |||||||||
| MICE-PMM | MICE-RF | ||||||||
| Cluster Number | Dimensionality Level (g) | Missingness < 25% | Missingness< 50% | Missingness < 75% | Cluster Number | Dimensionality Level (g) | Missingness < 25% | Missingness < 50% | Missingness < 75% |
| 2 | 100 | 0.94 | 0.22 | −6.49 | 2 | 100 | 0.76 | 0.68 | −3.19 |
| 1000 | 1.28 | 0.08 | 5.44 | 1000 | −1.14 | 0.23 | −4.84 | ||
| 7916 | −0.89 | 1.97 | 0.24 | 7916 | 0.18 | 1.17 | −8.35 | ||
| 4 | 100 | 1.26 | 0.17 | −3.43 | 4 | 100 | 0.20 | 2.09 | 0.76 |
| 1000 | 1.13 | 1.46 | 1.66 | 1000 | −0.53 | 2.25 | 0.33 | ||
| 7916 | 0.31 | 1.92 | −4.15 | 7916 | −0.91 | 1.97 | −4.03 | ||
| 8 | 100 | −0.36 | 2.85 | 6.60 | 8 | 100 | 0.97 | −0.06 | −4.16 |
| 1000 | −2.70 | −0.74 | −7.03 | 1000 | 1.08 | 2.15 | 1.17 | ||
| 7916 | 0.01 | 4.40 | 3.76 | 7916 | 0.26 | 3.31 | −8.24 | ||
| Osteoarthritis (OA) | |||||||||
| MICE-PMM | MICE-RF | ||||||||
| Cluster Number | Dimensionality Level (g) | Missingness < 25% | Missingness < 50% | Missingness < 75% | Cluster Number | Dimensionality Level (g) | Missingness < 25% | Missingness < 50% | Missingness < 75% |
| 4 | 100 | 0.04 | 0.08 | −0.13 | 4 | 100 | 2.45 | −4.23 | 6.83 |
| 1000 | 0.03 | 0.11 | −0.08 | 1000 | 3.35 | 10.16 | −4.70 | ||
| 2042 | 0.08 | 0.18 | 0.05 | 2042 | 1.70 | −2.70 | −0.75 | ||
| 8 | 100 | −0.07 | 0.22 | 0.12 | 8 | 100 | 4.73 | 1.13 | −0.10 |
| 1000 | −0.07 | −0.07 | 0.16 | 1000 | 3.86 | −1.27 | −0.34 | ||
| 2042 | 0.00 | −0.01 | −0.09 | 2042 | 4.42 | −11.30 | 1.87 | ||
| 16 | 100 | −0.02 | 0.10 | 0.20 | 16 | 100 | −0.52 | 3.08 | −2.33 |
| 1000 | 0.08 | 0.15 | −0.05 | 1000 | 1.41 | −0.33 | −6.45 | ||
| 2042 | −0.02 | 0.09 | 0.24 | 2042 | 1.60 | 3.23 | 10.93 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abedi, V.; Li, J.; Shivakumar, M.K.; Avula, V.; Chaudhary, D.P.; Shellenberger, M.J.; Khara, H.S.; Zhang, Y.; Lee, M.T.M.; Wolk, D.M.; et al. Increasing the Density of Laboratory Measures for Machine Learning Applications. J. Clin. Med. 2021, 10, 103. https://doi.org/10.3390/jcm10010103
Abedi V, Li J, Shivakumar MK, Avula V, Chaudhary DP, Shellenberger MJ, Khara HS, Zhang Y, Lee MTM, Wolk DM, et al. Increasing the Density of Laboratory Measures for Machine Learning Applications. Journal of Clinical Medicine. 2021; 10(1):103. https://doi.org/10.3390/jcm10010103
Chicago/Turabian StyleAbedi, Vida, Jiang Li, Manu K. Shivakumar, Venkatesh Avula, Durgesh P. Chaudhary, Matthew J. Shellenberger, Harshit S. Khara, Yanfei Zhang, Ming Ta Michael Lee, Donna M. Wolk, and et al. 2021. "Increasing the Density of Laboratory Measures for Machine Learning Applications" Journal of Clinical Medicine 10, no. 1: 103. https://doi.org/10.3390/jcm10010103
APA StyleAbedi, V., Li, J., Shivakumar, M. K., Avula, V., Chaudhary, D. P., Shellenberger, M. J., Khara, H. S., Zhang, Y., Lee, M. T. M., Wolk, D. M., Yeasin, M., Hontecillas, R., Bassaganya-Riera, J., & Zand, R. (2021). Increasing the Density of Laboratory Measures for Machine Learning Applications. Journal of Clinical Medicine, 10(1), 103. https://doi.org/10.3390/jcm10010103









