Protein-Based Graphene Biosensors: Optimizing Artificial Chemoreception in Bilayer Lipid Membranes
Abstract
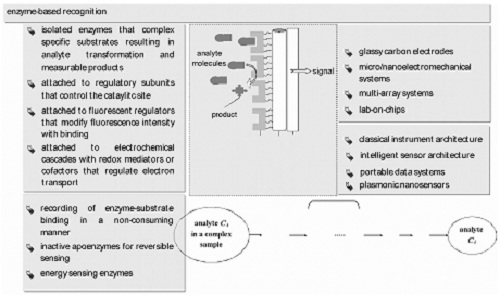
:1. Introduction
2. Materials and Methods
2.1. Reagents and Solutions
2.2. Procedures
2.3. Electrochemical Measurements
2.4. Sensor Validation for Biological Samples
3. Results and Discussion
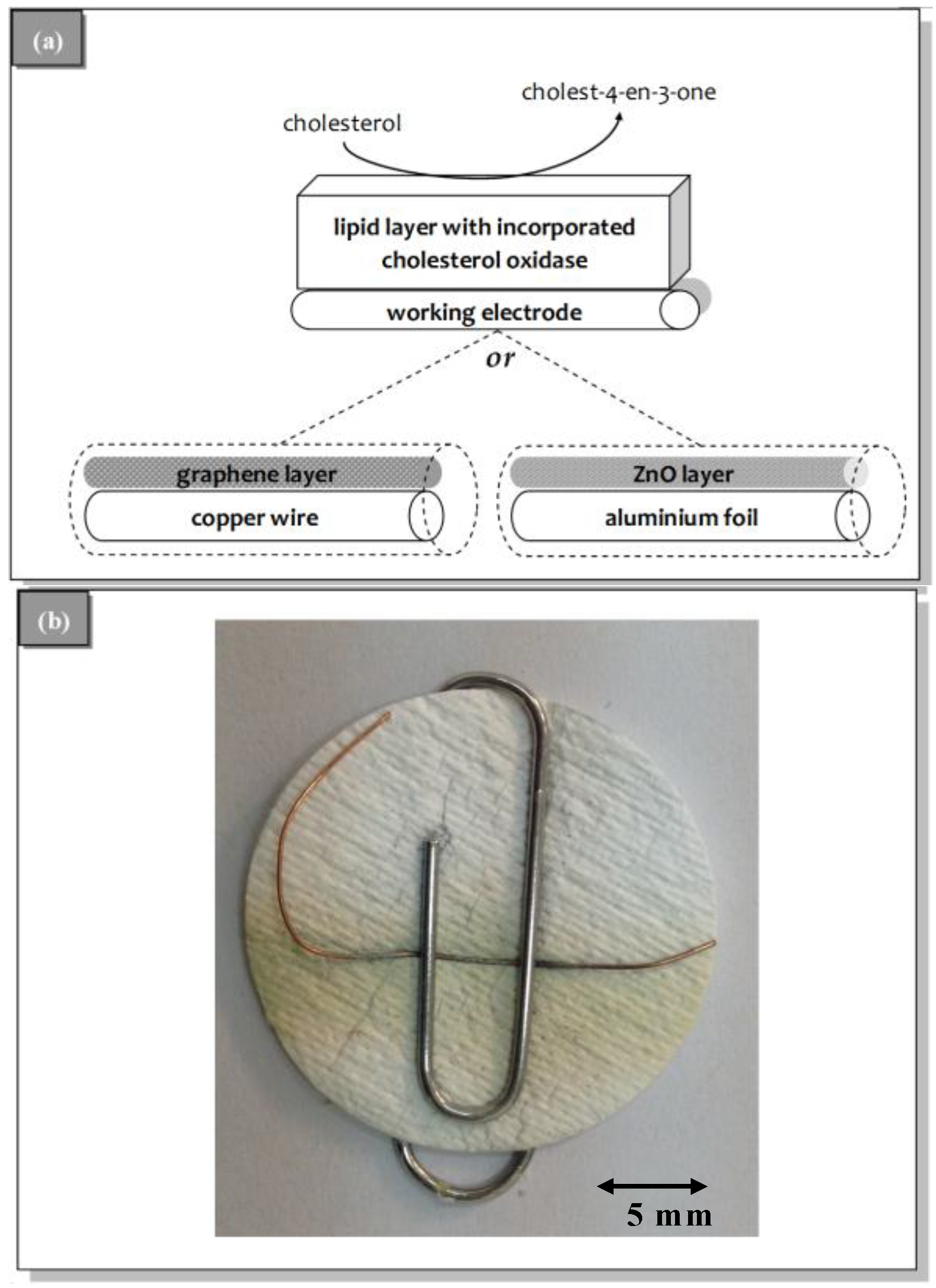
3.1. Sensor Construction
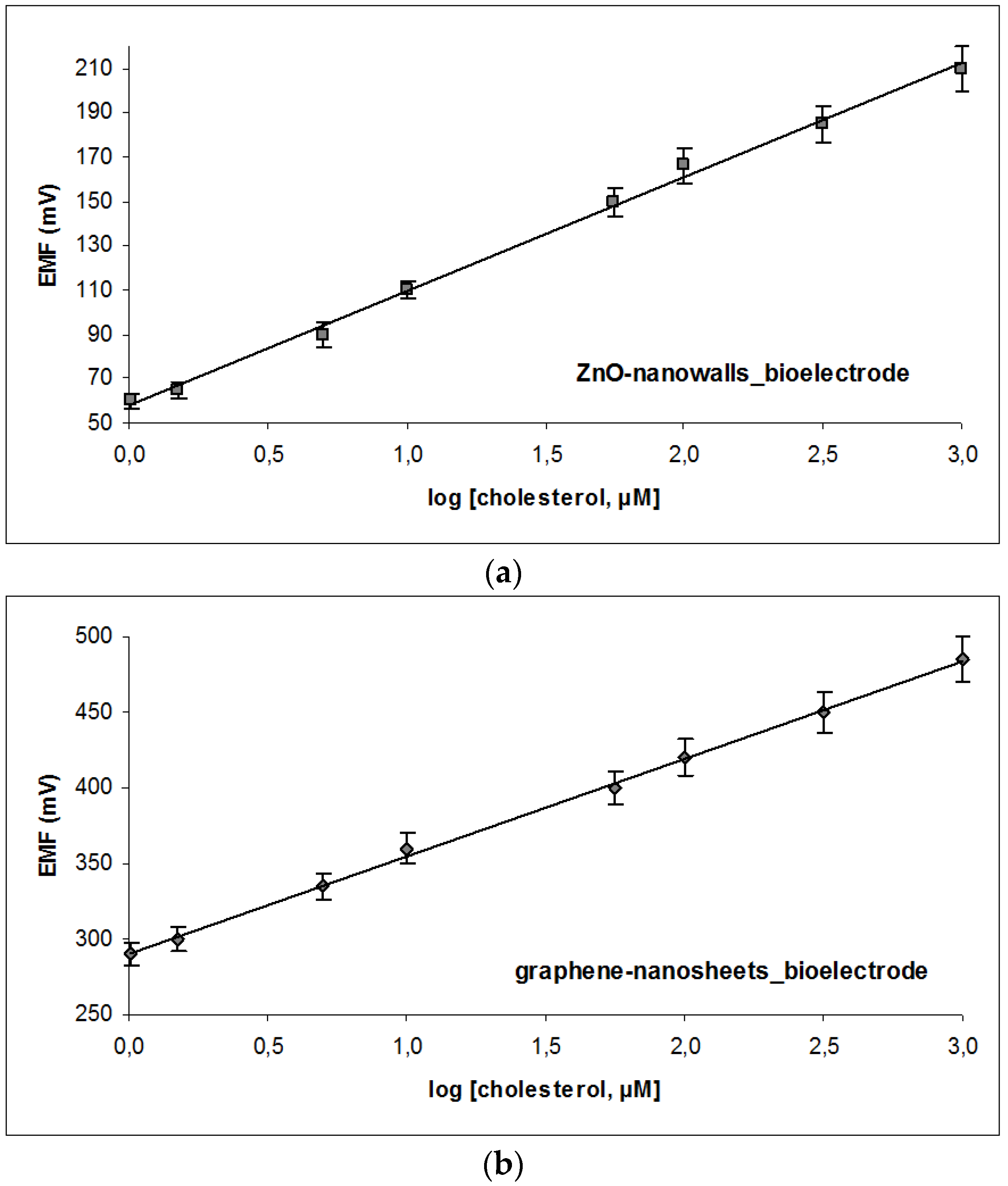
3.2. Sensor Characteristics and Performance
4. Conclusions
Author Contributions
Conflicts of Interest
References
- Siontorou, C.G.; Batzias, F.A. A methodological combined framework for roadmapping biosensor research: A fault tree analysis approach within a strategic technology evaluation frame. Crit. Rev. Biotechnol. 2013, 34, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Batzias, F.A. Determining the sources of measurement uncertainty in environmental cell-based biosensing. IEEE Trans. Instrum. Meas. 2014, 63, 794–804. [Google Scholar] [CrossRef]
- Jia, X.; Dong, S.; Wang, E. Engineering the bioelectrochemical interface using functional nanomaterials and microchip technique toward sensitive and portable electrochemical biosensors. Biosens. Bioelectron. 2016, 76, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Dong, J. From fluorescence polarization to Quenchbody: Recent progress in fluorescent reagentless biosensors based on antibody and other binding proteins. BBA Proteins Proteom. 2014, 1844, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Villaverde, A. Allosteric enzymes as biosensors for molecular diagnosis. FEBS Lett. 2003, 554, 169–172. [Google Scholar] [CrossRef]
- Razumiene, J.; Cirbaite, E.; Razumas, V.; Laurinavicius, V. New mediators for biosensors based on PQQ-dependent alcohol dehydrogenases. Sens. Actuators B Chem. 2015, 207, 1019–1025. [Google Scholar] [CrossRef]
- Scognamiglio, V. Nanotechnology in glucose monitoring: Advances and challenges in the last 10 years. Biosens. Bioelectron. 2013, 47, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors—Sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Chambers, J.P.; Arulanandam, B.P.; Matta, L.L.; Weis, A.; Valdes, J.J. Biosensor recognition elements. Curr. Issues Mol. Biol. 2008, 10, 1–12. [Google Scholar] [PubMed]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Immobilization strategies to develop enzymatic biosensors. Biotechnol. Adv. 2012, 30, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Polanka, M.; Jun, D.; Kalasz, H.; Kuca, K. Cholinesterase biosensor construction: A review. Protein Pept. Lett. 2008, 15, 795–798. [Google Scholar] [CrossRef]
- Nikolelis, D.P.; Raftopoulou, G.; Nikoleli, G.P.; Simantiraki, M. Stabilized lipid membrane based biosensors with incorporated enzyme for repetitive uses. Electroanalysis 2006, 18, 2467–2474. [Google Scholar] [CrossRef]
- Nikolelis, D.P.; Hianik, T.; Nikoleli, G.-P. Stabilized lipid films in electrochemical biosensors. Electroanalysis 2010, 22, 2747–2763. [Google Scholar] [CrossRef]
- Cordeiro, C.A.; de Vries, M.G.; Cremers, T.I.F.H.; Westerink, B.H.C. The role of surface availability in membrane-induced selectivity for amperometric enzyme-based biosensors. Sens. Actuators B Chem. 2016, 223, 679–688. [Google Scholar] [CrossRef]
- Campitelli, A.; Bartic, C.; Friedt, J.-M.; de Keersmaecker, K.; Laureyn, W.; Francis, L.; Frederix, F.; Reekmans, G.; Angelova, A.; Suls, J.; et al. Development of microelectronic based biosensors. In Proceedings of the IEEE Custom Integrated Circuits Conference, San Jose, CA, USA, 21–24 September 2003; IEEE: New York, NY, USA, 2003; pp. 505–512. [Google Scholar]
- Zhou, C.; Friedt, J.-M.; Angelova, A.; Choi, K.-H.; Laureyn, W.; Frederix, F.; Francis, L.A.; Campitelli, A.; Engelborghs, Y.; Borghs, G. Human immunoglobulin adsorption investigated by means of quartz crystal microbalance dissipation, atomic force microscopy, surface acoustic wave, and surface plasmon resonance techniques. Langmuir 2004, 20, 5870–5878. [Google Scholar] [CrossRef] [PubMed]
- Angelova, A.; Ollivon, M.; Campitelli, A.; Bourgaux, C. Lipid cubic phases as stable nanochannel network structures for protein biochip development: X-ray diffraction study. Langmuir 2003, 19, 6928–6935. [Google Scholar] [CrossRef]
- Pumera, M. Graphene in biosensing. Mater. Today 2011, 14, 308–315. [Google Scholar] [CrossRef]
- Yang, Y.; Asiri, A.M.; Tang, Z.; Du, D.; Lin, Y. Graphene based materials for biomedical applications. Mater. Today 2013, 16, 365–373. [Google Scholar] [CrossRef]
- Viswanathan, S.; Narayanan, T.N.; Aran, K.; Fink, K.D.; Paredes, J.; Ajayan, P.M.; Filipek, S.; Miszta, P.; Tekin, H.C.; Inci, F.; et al. Graphene–protein field effect biosensors: Glucose sensing. Mater. Today 2015, 18, 513–522. [Google Scholar] [CrossRef]
- Walcarius, A.; Shelley, D.; Minteer, J.W.; Yuehe, L.; Arben, M. Nanomaterials for bio-functionalized electrodes: Recent trends. J. Mater. Chem. B 2013, 1, 4878–4908. [Google Scholar] [CrossRef]
- Du, D.; Wang, L.; Shao, Y.; Wang, J.; Engelhard, M.H.; Lin, Y. Functionalized graphene oxide as a nanocarrier in a multienzyme labeling amplification strategy for ultrasensitive electrochemical immunoassay of phosphorylated p53 (S392). Anal. Chem. 2011, 83, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Arya, S.K.; Pandey, P.; Malhotra, B.D.; Saha, S.; Sreenivas, K.; Gupta, V. Cholesterol biosensor based on RF sputtered zinc oxide nanoporous thin film. Appl. Phys. Lett. 2007, 91. [Google Scholar] [CrossRef]
- Liu, B.; Zeng, H.C. Hydrothermal synthesis of ZnO nanorods in the diameter regime of 50 nm. J. Am. Chem. Soc. 2003, 125, 4430–4431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.F.; Wang, X.L.; Ai, S.Y.; Sun, Z.D.; Wan, Q.; Zhu, Z.Q.; Xian, Y.Z.; Jin, L.T.; Yamamoto, K. Immobilization of uricase on ZnO nanorods for a reagent less uric acid biosensor. Anal. Chim. Acta 2004, 519, 155–160. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, X.D.; Wang, Z.L. Microfibre–nanowire hybrid structure for energy scavenging. Nature 2008, 451, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.D.; Mulato, M. ZnO extended-gate field-effect transistors as pH sensors. Appl. Phys. Lett. 2007, 87. [Google Scholar] [CrossRef]
- Wei, A.; Sun, X.W.; Wang, J.X.; Lie, Y.; Cai, X.P.; Li, C.M.; Dong, Z.L.; Huang, W. Enzymatic glucose biosensor based on ZnO nanorod array grown by hydrothermal decomposition. Appl. Phys. Lett. 2006, 89. [Google Scholar] [CrossRef]
- Kumar, N.; Dorfman, A.; Hahm, J. Ultrasensitive DNA sequence detection using nanoscale ZnO sensor arrays. Nanotechnology 2006, 17, 2875–2881. [Google Scholar] [CrossRef]
- Wang, Z.L. Nanostructures of zinc oxide. Mater. Today 2004, 7, 26–33. [Google Scholar] [CrossRef]
- Nayak, A.P.; Katzenmeyer, A.M.; Gosho, Y.; Tekin, B.; Islam, M.S. Sonochemical approach for rapid growth of zinc oxide nanowalls. Appl. Phys. A 2012, 107, 661–667. [Google Scholar] [CrossRef]
- Dorfman, A.; Kumar, N.; Hahm, J. Nanoscale ZnO-enhanced fluorescence detection of protein interactions. Adv. Mater. 2006, 18, 2685–2690. [Google Scholar] [CrossRef]
- Asif, M.H.; Nur, O.; Willander, M.; Danielsson, B. Selective calcium ion detection with functionalized ZnO nanorods-extended gate MOSFET. Biosens. Bioelectron. 2006, 24, 3379–3382. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Shao, G.; Hong, J.; Bao, J.; Shen, J. Immobilization and direct electrochemistry of glucose oxidase on a tetragonal pyramid-shaped porous ZnO nanostructure for a glucose biosensor. Biosens. Bioelectron. 2009, 24, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Umar, A.; Rahman, M.M.; Vaseem, M.; Hahn, Y.B. Ultra-sensitive cholesterol biosensor based on low-temperature grown ZnO nanoparticles. Electrochem. Commun. 2009, 11, 118–121. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, D.; Zhi, J. A novel tyrosinase biosensor based on biofunctional ZnO nanorod microarrays on the nanocrystalline diamond electrode for detection of phenolic compounds. Bioelectrochemistry 2009, 75, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Nikolelis, D.P.; Ntanos, N.; Nikoleli, G.-P.; Tampouris, K. Development of an Electrochemical biosensor for the rapid detection of naphthalene acetic acid in fruits by using air stable lipid films with incorporated auxin-bin ding protein 1 receptor. Protein Pept. Lett. 2008, 15, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Nikolelis, D.P.; Chaloulakos, T.-I.; Nikoleli, G.-P.; Psaroudakis, N. A portable sensor for the rapid detection of naphthalene acetic acid in fruits and vegetables using stabilized in air lipid films with incorporated auxin-binding protein 1 receptor. Talanta 2008, 77, 786–792. [Google Scholar] [CrossRef]
- Bratakou, S.; Nikoleli, G.-P.; Nikolelis, D.P.; Psaroudakis, N. Development of a potentiometric chemical sensor for the rapid detection of carbofuran based on air stable lipid films with incorporated calix[4]arene phosphoryl receptor using graphene electrodes. Electroanalysis 2015, 27, 2608–2613. [Google Scholar] [CrossRef]
- Psychoyios, V.N.; Nikoleli, G.-P.; Tzamtzis, N.; Nikolelis, D.P.; Psaroudakis, N.; Danielsson, B.; Israr, M.Q.; Willander, M. Potentiometric cholesterol biosensor based on ZnO nanowalls and stabilized polymerized lipid film. Electroanalysis 2013, 25, 367–372. [Google Scholar] [CrossRef]
- Nikoleli, G.-P.; Israr, M.Q.; Tzamtzis, N.; Nikolelis, D.P.; Willander, M.; Psaroudakis, N. Structural characterization of graphene nanosheets for miniaturization of potentiometric urea lipid film based biosensors. Electroanalysis 2012, 24, 1285–1295. [Google Scholar] [CrossRef]
- Israr, M.Q.; Sadaf, J.R.; Asif, M.H.; Nur, O.; Willander, M.; Danielsson, B. Potentiometric cholesterol biosensor based on ZnO nanorods chemically grown on Ag wire. Thin Solid Films 2010, 519, 1106–1109. [Google Scholar] [CrossRef]
- Ibupoto, Z.H.; Khun, K.; Liu, X.; Willander, M. Low temperature synthesis of seed mediated CuO bundle of nanowires, their structural characterisation and cholesterol detection. Mater. Sci. Eng. C 2013, 33, 3889–3898. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Nia, Y.; Kokot, S. Electrochemical cholesterol sensor based on cholesterol oxidase and MoS2-AuNPs modified glassy carbon electrode. Sens. Actuators B Chem. 2016, 233, 100–106. [Google Scholar] [CrossRef]
- Aravamudhan, S.; Kumar, A.; Mohapatra, S.; Bhansali, S. Sensitive estimation of total cholesterol in blood using Au nanowires based micro-fluidic platform. Biosens. Bioelectron. 2007, 22, 2289–2294. [Google Scholar] [CrossRef] [PubMed]
- Nikoleli, G.-P.; Ibupoto, Z.H.; Nikolelis, D.P.; Likodimos, V.; Psaroudakis, N.; Tzamtzis, N.; Willander, M.; Hianik, T. Potentiometric cholesterol biosensing application of graphene electrode with stabilized polymeric lipid membrane. Cent. Eur. J. Chem. 2013, 11, 1554–1561. [Google Scholar] [CrossRef]
- Angelova, A.; Angelov, B.; Garamus, V.M.; Couvreur, P.; Lesieur, S. Small-angle X-ray scattering investigations of biomolecular confinement, loading, and release from liquid-crystalline nanochannel assemblies. J. Phys. Chem. Lett. 2012, 3, 445–457. [Google Scholar] [CrossRef] [PubMed]
| Graphene-Based Sensor | ZnO-Based Sensor | |
|---|---|---|
| Noise level | 29 ± 1.4 mV | 34 mV ± 2.3 mV |
| Equation of calibration | EVolts = 64.003 × log [cholesterol] + 84.861 | EVolts = 54.560 × log [cholesterol] + 61.348 |
| r2 = 0.9987, [cholesterol] in μM | r2 = 0.9989, [cholesterol] in μM | |
| Sensitivity | 64 mV/decade of concentration | 57 mV/decade of concentration |
| Variability of response | 2.5%–3.1% | 2.1%–4.8% |
| Working range | 1 × 10−6–1 × 10−3 M | 1 × 10−6–1 × 10−3 M |
| Detection limit | 1.08 × 10−6 M | 5.56 × 10−6 M |
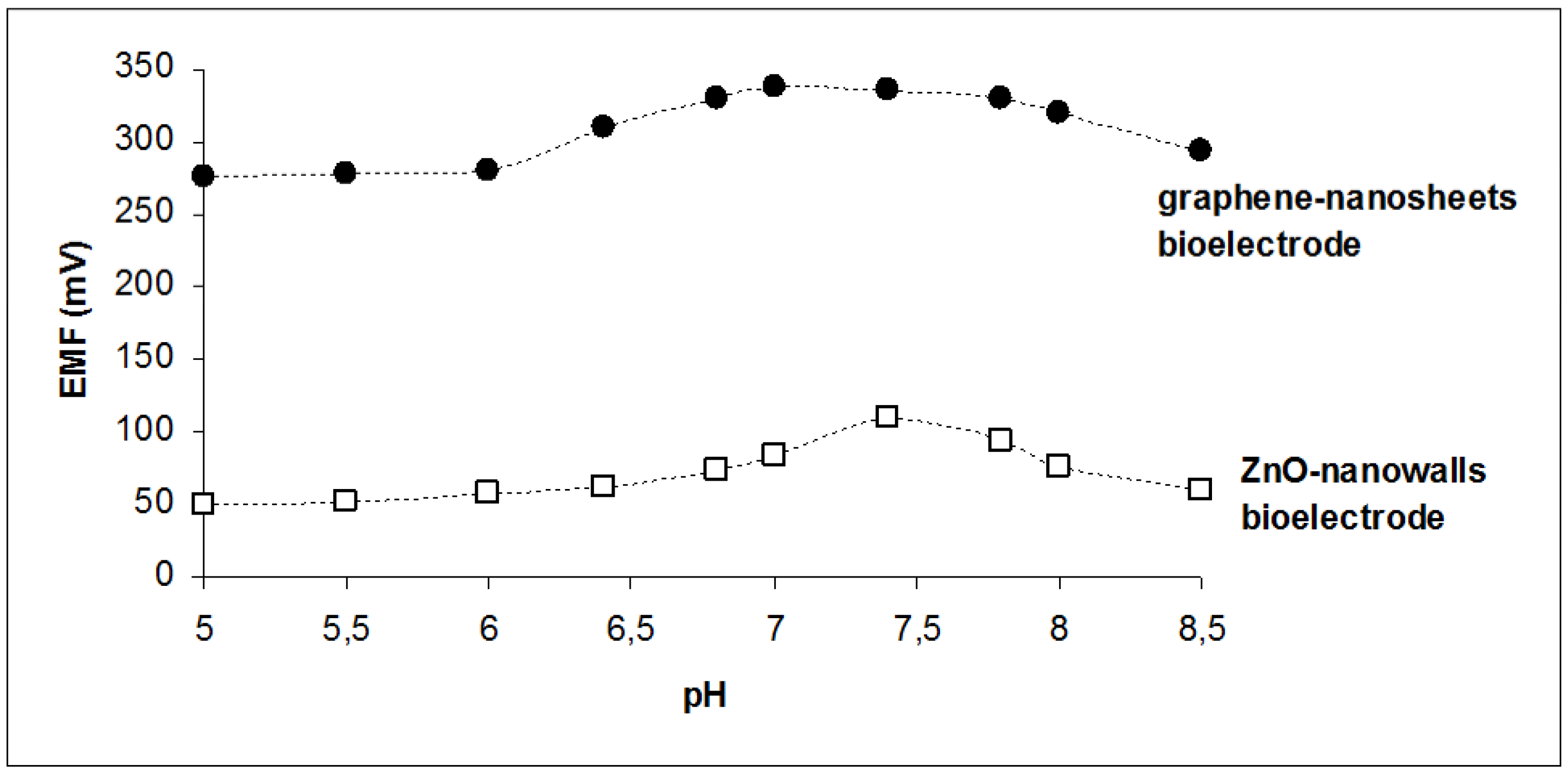
| pH opt | 6.8–7.8 | 7.4 |
| Sample Nr. | Hospital Analyser Cholesterol Value | Graphene-Nanosheet Enzyme Sensor | ZnO-Nanowalls Enzyme Sensor | ||
|---|---|---|---|---|---|
| mg/dL | mg/dL | % rel. Error | mg/dL | % rel. Error | |
| 1 | 100 | 98 | –2.00% | 104 | +4.00% |
| 2 | 116 | 120 | +3.45% | 118 | +1.72% |
| 3 | 155 | 150 | –3.23% | 153 | –1.29% |
| 4 | 180 | 185 | +2.77% | 189 | +5.00% |
| 5 | 201 | 208 | +3.48% | 190 | –5.47% |
| 6 | 220 | 214 | –2.72% | 230 | +4.55% |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siontorou, C.G.; Georgopoulos, K.N.; Nikoleli, G.-P.; Nikolelis, D.P.; Karapetis, S.K.; Bratakou, S. Protein-Based Graphene Biosensors: Optimizing Artificial Chemoreception in Bilayer Lipid Membranes. Membranes 2016, 6, 43. https://doi.org/10.3390/membranes6030043
Siontorou CG, Georgopoulos KN, Nikoleli G-P, Nikolelis DP, Karapetis SK, Bratakou S. Protein-Based Graphene Biosensors: Optimizing Artificial Chemoreception in Bilayer Lipid Membranes. Membranes. 2016; 6(3):43. https://doi.org/10.3390/membranes6030043
Chicago/Turabian StyleSiontorou, Christina G., Konstantinos N. Georgopoulos, Georgia-Paraskevi Nikoleli, Dimitrios P. Nikolelis, Stefanos K. Karapetis, and Spyridoula Bratakou. 2016. "Protein-Based Graphene Biosensors: Optimizing Artificial Chemoreception in Bilayer Lipid Membranes" Membranes 6, no. 3: 43. https://doi.org/10.3390/membranes6030043
APA StyleSiontorou, C. G., Georgopoulos, K. N., Nikoleli, G.-P., Nikolelis, D. P., Karapetis, S. K., & Bratakou, S. (2016). Protein-Based Graphene Biosensors: Optimizing Artificial Chemoreception in Bilayer Lipid Membranes. Membranes, 6(3), 43. https://doi.org/10.3390/membranes6030043