Comprehensive Insights into the Cholesterol-Mediated Modulation of Membrane Function Through Molecular Dynamics Simulations
Abstract
1. Introduction
2. Planar Bilayers
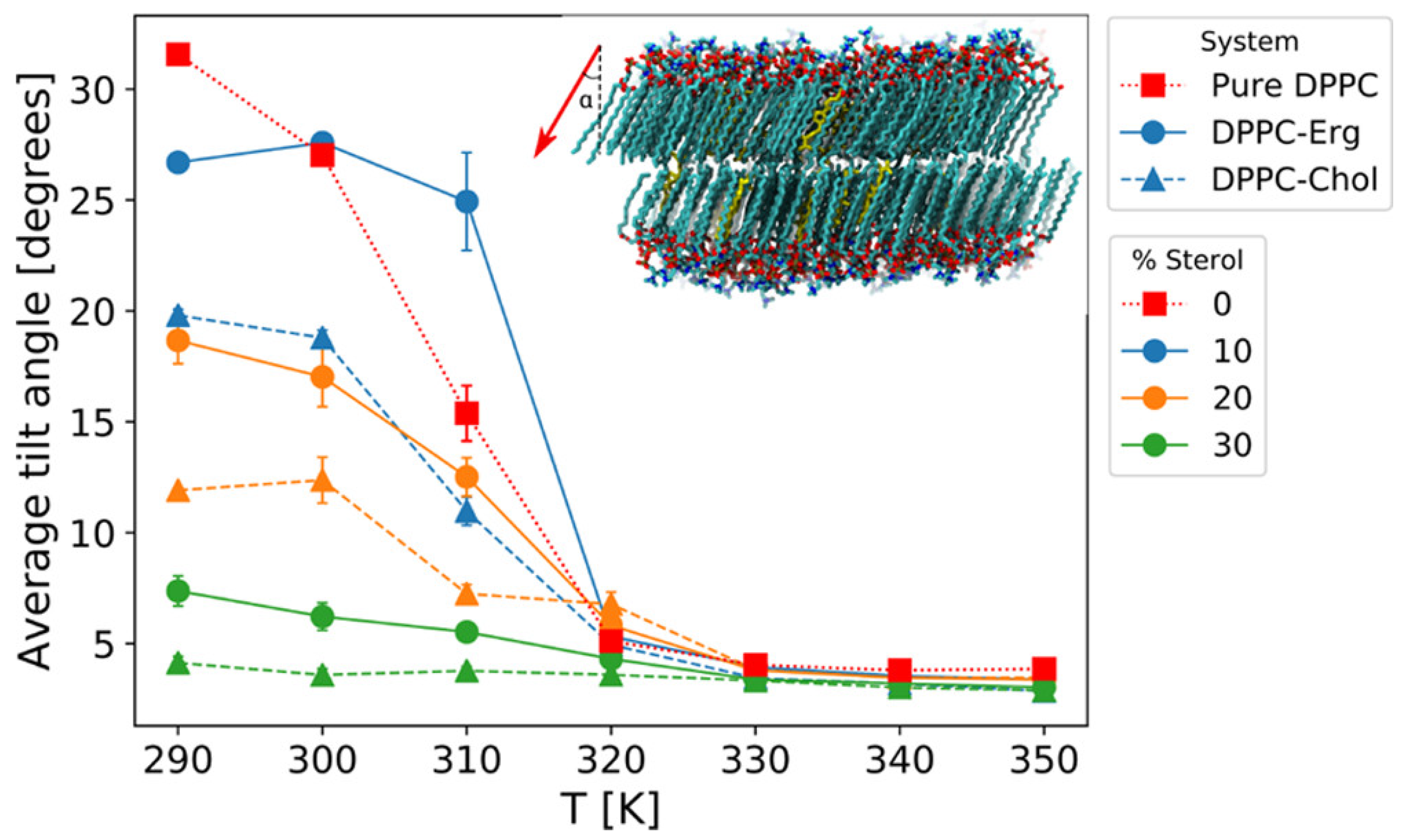
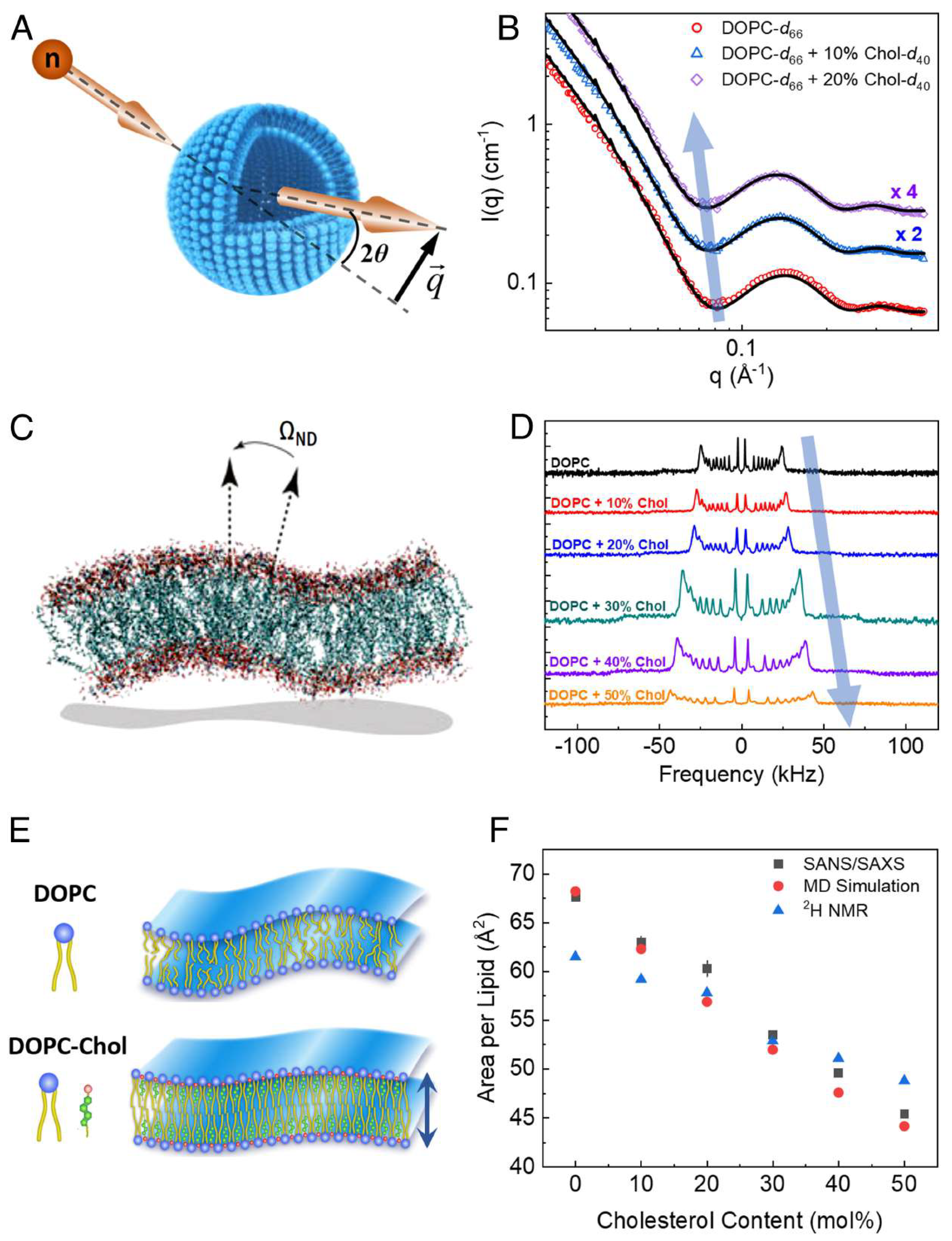
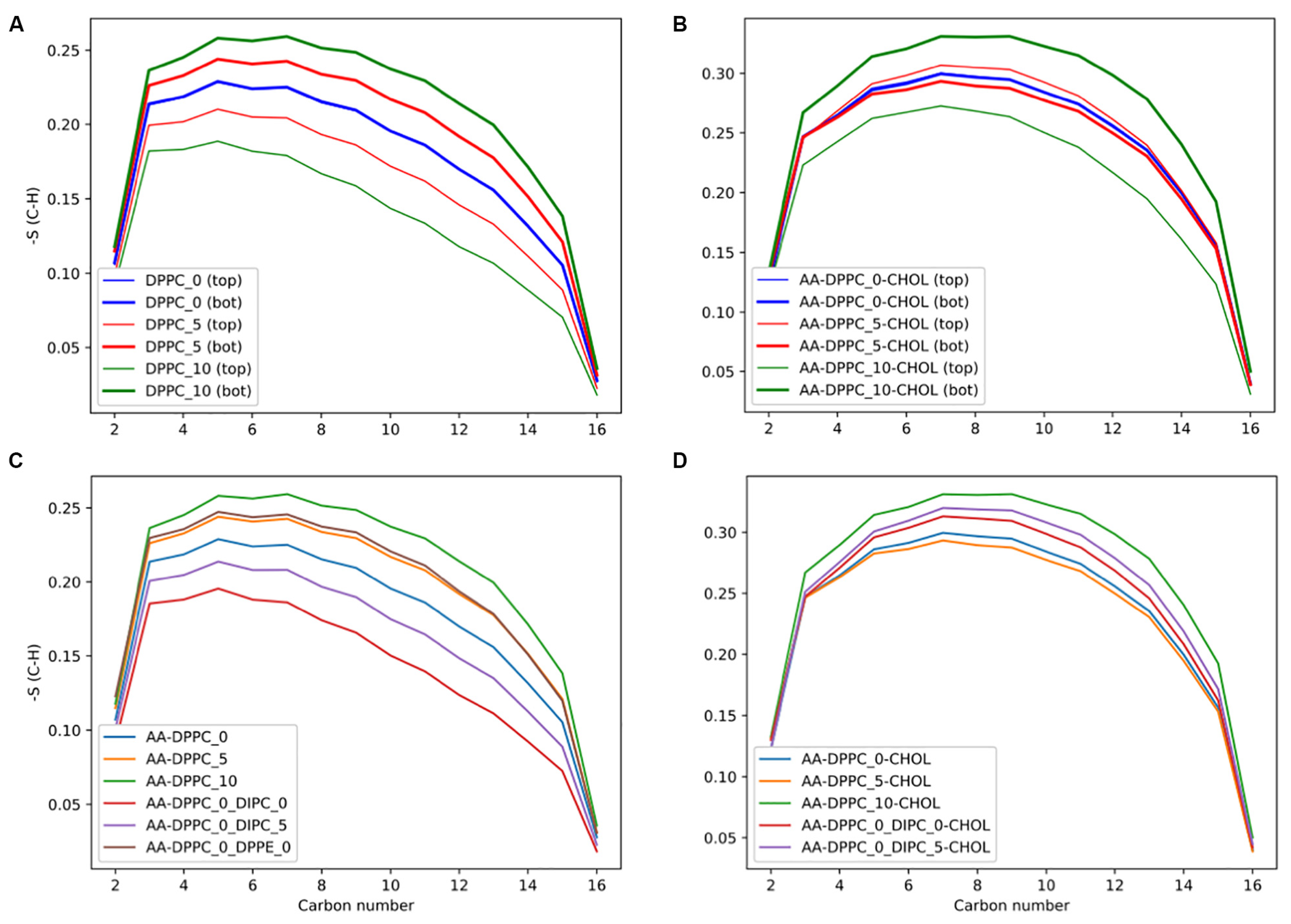
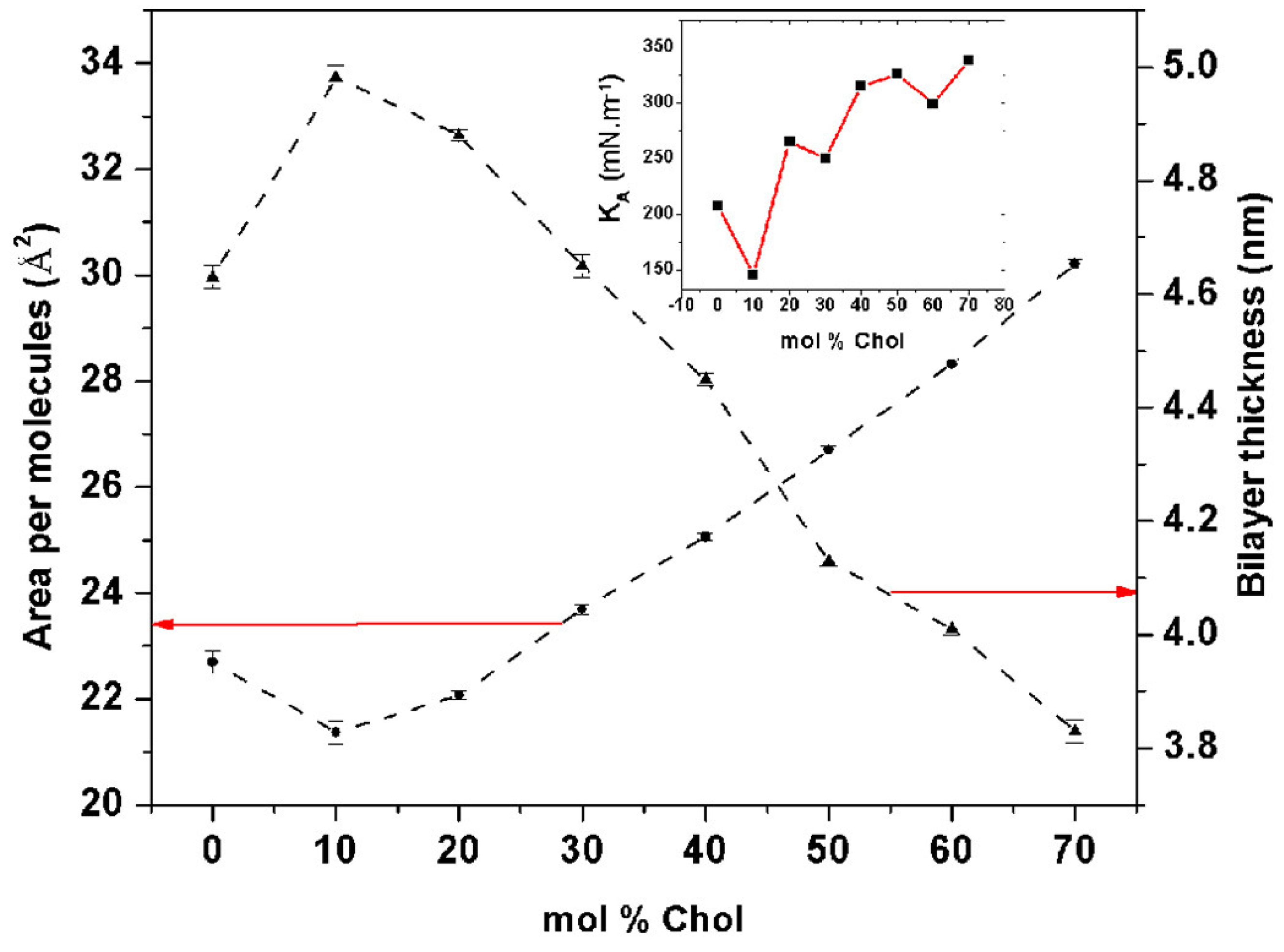
2.1. Membrane Thickness and Lipid Ordering
2.2. Mechanical Properties and Membrane Rigidity
2.3. Asymmetry and Leaflet Coupling
2.4. Phase Separation and Fluidity
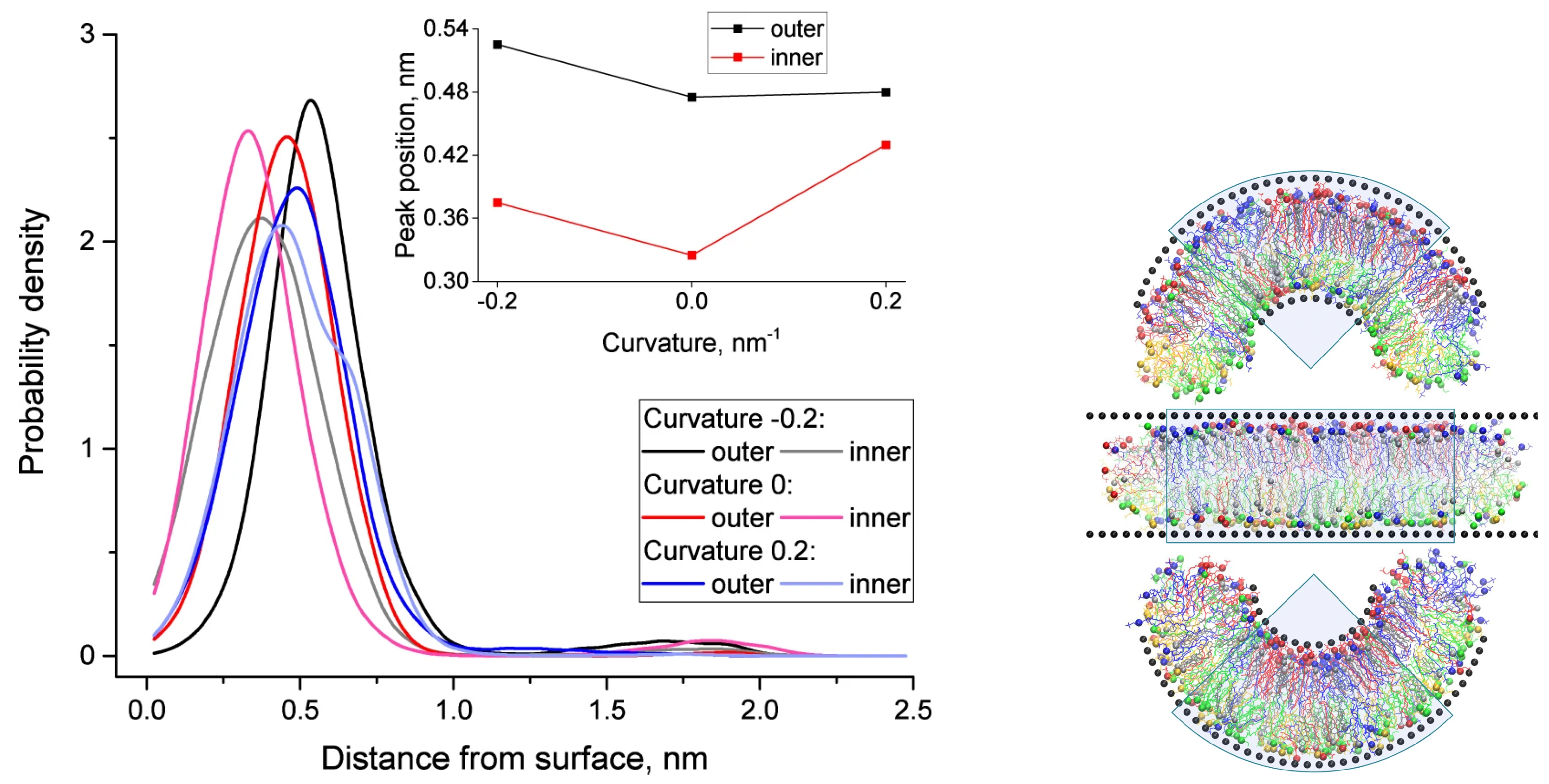
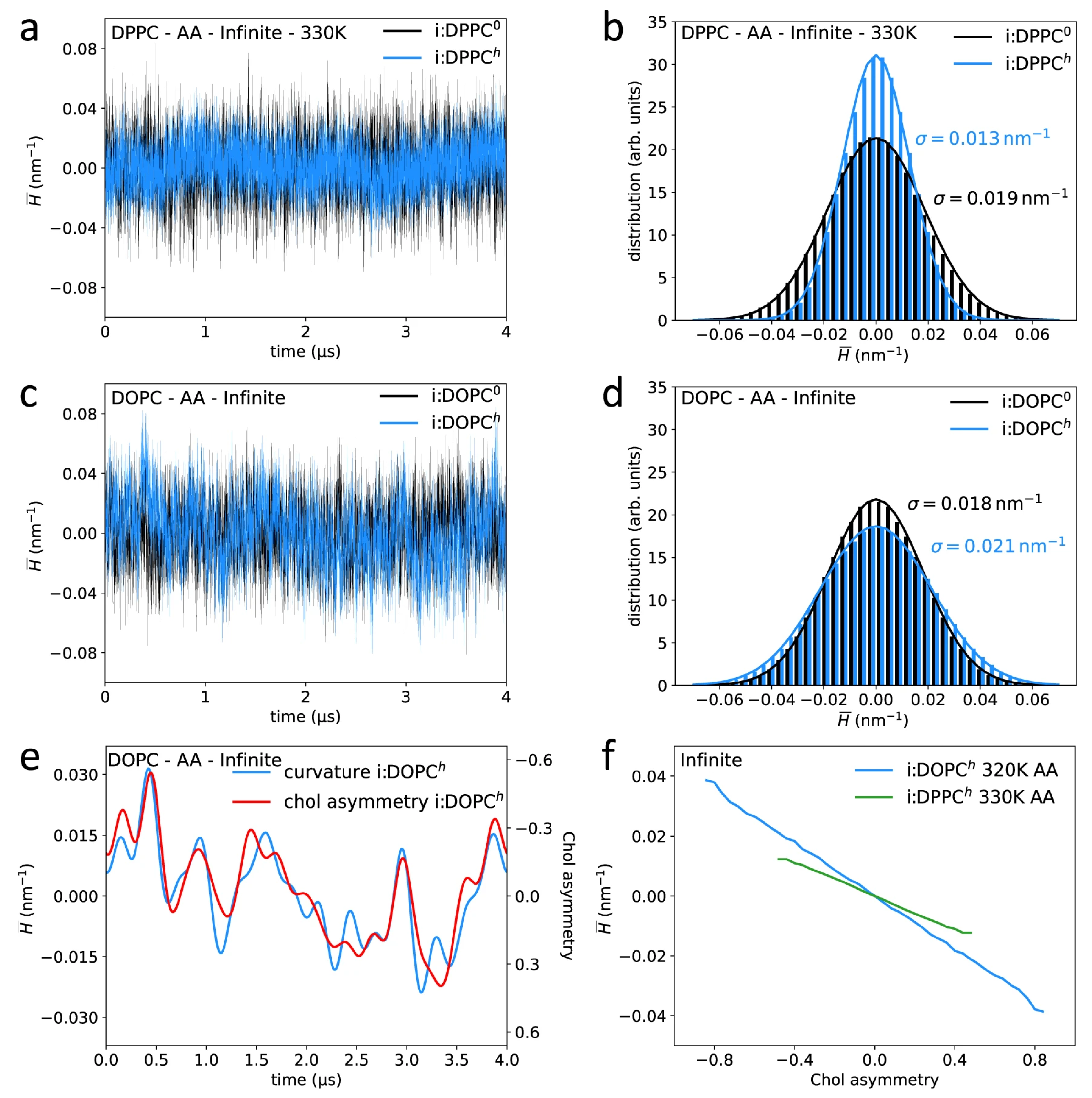
3. Curved Bilayers
3.1. Cholesterol-Induced Stability
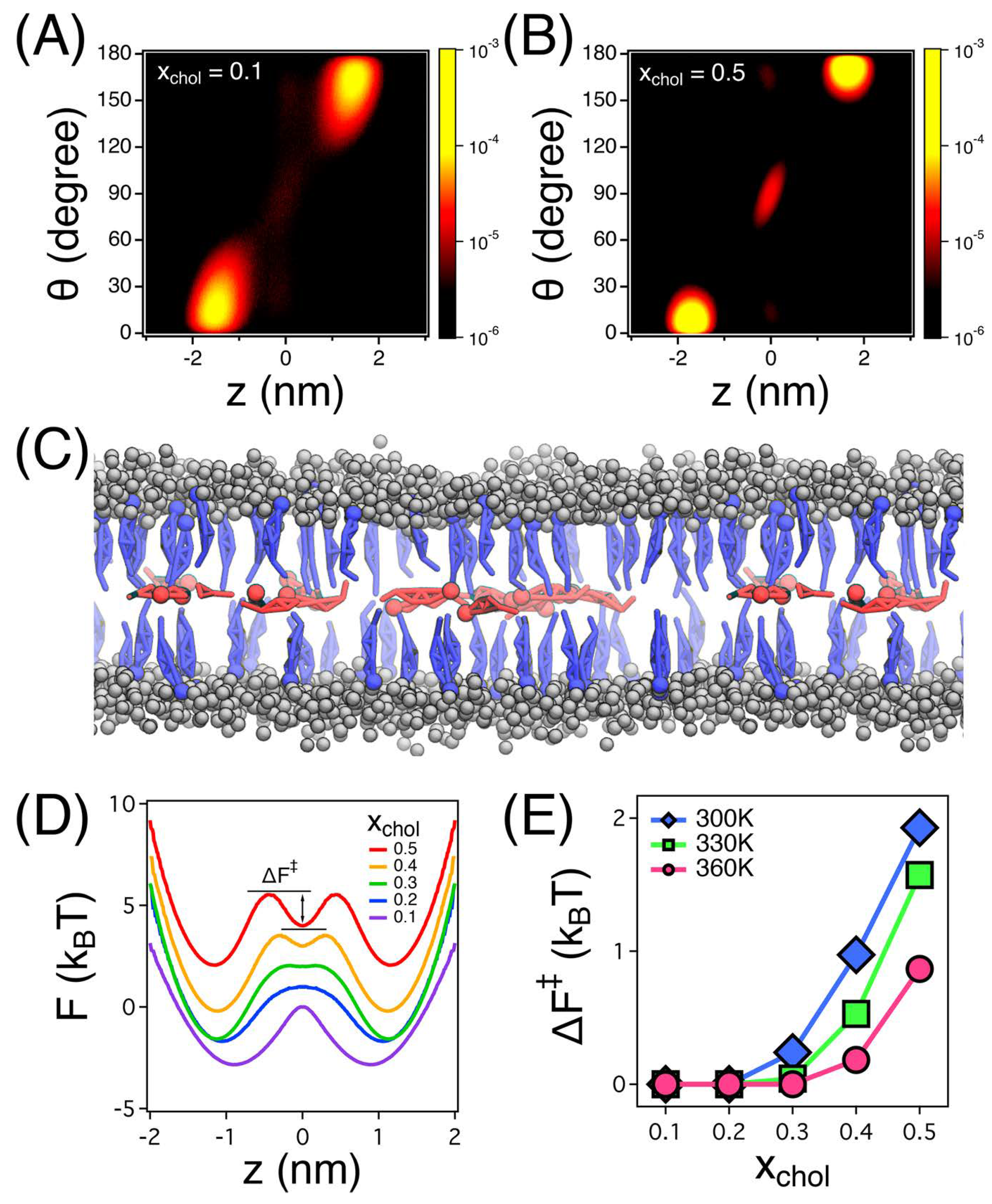
3.2. Flip-Flop Dynamics
4. Nanodisc
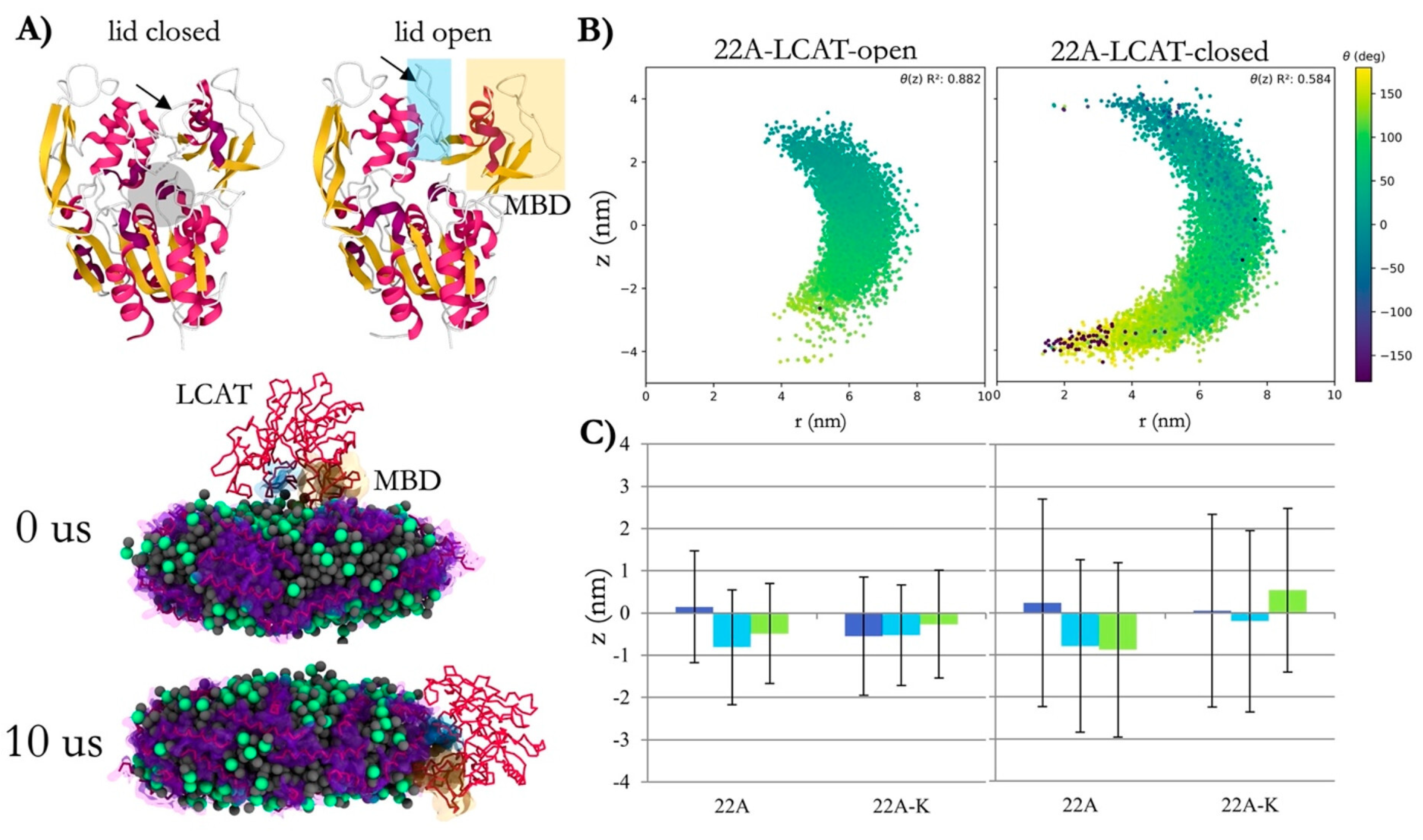
4.1. Role of Cholesterol in Reverse Cholesterol Transport (RCT)
4.2. Structural Adaptation and Stability in Synthetic Nanodiscs
4.3. Impact of Cholesterol on Protein Configuration and Nanodisc Shape
5. Liposome
5.1. Formation and Stability
5.2. Drug Loading and Release
5.3. Phase Behavior and Structural Organization
6. Proteoliposomes
6.1. Membrane Protein Stability
6.2. Structural Changes in Proteoliposomes
6.3. Conformational Dynamics
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MD | Molecular Dynamics |
| Lo | Liquid-Ordered |
| Ld | Liquid-Disordered |
| SCD | Deuterium Order Parameter |
| DLPE | Dioleoylphosphatidylethanolamine |
| DOPE | Dioleoylphosphatidylethanolamine |
| DLiPE | Dilinoleoylphosphatidylethanolamine |
| DOPS | Dioleoylphosphatidylserine |
| DLiPS | Dilinoleoylphosphatidylserine |
| DLiPC | Dilinoleoylphosphatidylcholine |
| DOPC | Dioleoylphosphatidylcholine |
| POPC | Palmitoyloleoylphosphatidylcholine |
| DSPC | Distearoylphosphatidylcholine |
| DSPE | Distearoylphosphatidylethanolamine |
| MSPC | Monostearoylphosphatidylcholine |
| AMT | Amantadine |
| A | Amyloid- |
| hIAPP | Human Islet Amyloid Polypeptide |
| LLO | Listeriolysin O |
| MI | Membrane-Inserted |
| MB | Membrane-Bound |
| PAMAM | Polyamidoamine |
| AFM | Atomic Force Microscopy |
| HSPC | Hydrogenated Soy Phosphatidylcholine |
| CDs | Cyclodextrins |
| MβCD | Methyl-β-Cyclodextrin |
| SASA | Solvent Accessible Surface Area |
| APOA1 | Apolipoprotein A-I |
| RCT | Reverse cholesterol transport |
| MSPs | Membrane scaffold proteinsMSPs |
| LCAT | Lecithin–Cholesterol Acyltransferase |
| HDL | High-density lipoprotein |
References
- Zakany, F.; Kovacs, T.; Panyi, G.; Varga, Z. Direct and indirect cholesterol effects on membrane proteins with special focus on potassium channels. Biochim. Biophys. Acta-(BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158706. [Google Scholar] [CrossRef] [PubMed]
- Somjid, S.; Krongsuk, S.; Johns, J.R. Cholesterol concentration effect on the bilayer properties and phase formation of niosome bilayers: A molecular dynamics simulation study. J. Mol. Liq. 2018, 256, 591–598. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Dong, M.; Han, X. Effect of cholesterol on the fluidity of supported lipid bilayers. Colloids Surf. Biointerfaces 2020, 196, 111353. [Google Scholar] [CrossRef]
- Ramos-Martín, F.; D’Amelio, N. Biomembrane lipids: When physics and chemistry join to shape biological activity. Biochimie 2022, 203, 118–138. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kaur, N.; Hitaishi, P.; Ghosh, S.K.; Mithu, V.S.; Scheidt, H.A. Role of Cholesterol in Interaction of Ionic Liquids with Model Lipid Membranes and Associated Permeability. J. Phys. Chem. B 2024, 128, 5407–5418. [Google Scholar] [CrossRef]
- Giraldo-Lorza, J.M.; Leidy, C.; Manrique-Moreno, M. The Influence of Cholesterol on Membrane Targeted Bioactive Peptides: Modulating Peptide Activity Through Changes in Bilayer Biophysical Properties. Membranes 2024, 14, 220. [Google Scholar] [CrossRef]
- Zhao, W.; Prijic, S.; Urban, B.C.; Tisza, M.J.; Zuo, Y.; Li, L.; Tan, Z.; Chen, X.; Mani, S.A.; Chang, J.T. Candidate antimetastasis drugs suppress the metastatic capacity of breast cancer cells by reducing membrane fluidity. Cancer Res. 2016, 76, 2037–2049. [Google Scholar] [CrossRef]
- Levental, K.R.; Lorent, J.H.; Lin, X.; Skinkle, A.D.; Surma, M.A.; Stockenbojer, E.A.; Gorfe, A.A.; Levental, I. Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys. J. 2016, 110, 1800–1810. [Google Scholar] [CrossRef] [PubMed]
- Ghysels, A.; Krämer, A.; Venable, R.M.; Teague, W.E., Jr.; Lyman, E.; Gawrisch, K.; Pastor, R.W. Permeability of membranes in the liquid ordered and liquid disordered phases. Nat. Commun. 2019, 10, 5616. [Google Scholar] [CrossRef]
- Schachter, I.; Paananen, R.O.; Fábián, B.; Jurkiewicz, P.; Javanainen, M. The two faces of the liquid ordered phase. J. Phys. Chem. Lett. 2022, 13, 1307–1313. [Google Scholar] [CrossRef]
- Zhang, X.; Barraza, K.M.; Beauchamp, J. Cholesterol provides nonsacrificial protection of membrane lipids from chemical damage at air–water interface. Proc. Natl. Acad. Sci. USA 2018, 115, 3255–3260. [Google Scholar] [CrossRef] [PubMed]
- Silindir-Gunay, M.; Ozer, A.Y. Liposomes and micelles as nanocarriers for diagnostic and imaging purposes. In Design of Nanostructures for Theranostics Applications; Elsevier: Amsterdam, The Netherlands, 2018; pp. 305–340. [Google Scholar]
- Sternberg, B. Liposomes as a model for membrane structures and structural transformations: A liposome album. In Handbook of Nonmedical Applications of Liposomes; CRC Press: Boca Raton, FL, USA, 2019; pp. 271–298. [Google Scholar]
- Lanze, C.E.; Gandra, R.M.; Foderaro, J.E.; Swenson, K.A.; Douglas, L.M.; Konopka, J.B. Plasma membrane MCC/eisosome domains promote stress resistance in fungi. Microbiol. Mol. Biol. Rev. 2020, 84, 10–1128. [Google Scholar] [CrossRef]
- Kahli, H.; Béven, L.; Grauby-Heywang, C.; Debez, N.; Gammoudi, I.; Moroté, F.; Sbartai, H.; Cohen-Bouhacina, T. Impact of growth conditions on Pseudomonas fluorescens morphology characterized by atomic force microscopy. Int. J. Mol. Sci. 2022, 23, 9579. [Google Scholar] [CrossRef]
- Nicolson, G.L.; Ferreira de Mattos, G. Fifty years of the fluid–mosaic model of biomembrane structure and organization and its importance in biomedicine with particular emphasis on membrane lipid replacement. Biomedicines 2022, 10, 1711. [Google Scholar] [CrossRef] [PubMed]
- van Deventer, S.; Arp, A.B.; van Spriel, A.B. Dynamic plasma membrane organization: A complex symphony. Trends Cell Biol. 2021, 31, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Knittelfelder, O.; Geffard, O.; Clément, Y.; Testet, E.; Elie, N.; Touboul, D.; Abbaci, K.; Shevchenko, A.; Lemoine, J.; et al. Shotgun lipidomics and mass spectrometry imaging unveil diversity and dynamics in Gammarus fossarum lipid composition. IScience 2021, 24, 102115. [Google Scholar] [CrossRef]
- Melero, A.; Jiménez-Rojo, N. Cracking the membrane lipid code. Curr. Opin. Cell Biol. 2023, 83, 102203. [Google Scholar] [CrossRef]
- Lee, T.H.; Charchar, P.; Separovic, F.; Reid, G.E.; Yarovsky, I.; Aguilar, M.I. The intricate link between membrane lipid structure and composition and membrane structural properties in bacterial membranes. Chem. Sci. 2024, 15, 3408–3427. [Google Scholar] [CrossRef]
- Horne, J.E.; Brockwell, D.J.; Radford, S.E. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J. Biol. Chem. 2020, 295, 10340–10367. [Google Scholar] [CrossRef]
- Sych, T.; Mély, Y.; Römer, W. Lipid self-assembly and lectin-induced reorganization of the plasma membrane. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170117. [Google Scholar] [CrossRef]
- Rowlett, V.W.; Mallampalli, V.K.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Levental, K.R.; Malmberg, E.; Symons, J.L.; Fan, Y.Y.; Chapkin, R.S.; Ernst, R.; Levental, I. Lipidomic and biophysical homeostasis of mammalian membranes counteracts dietary lipid perturbations to maintain cellular fitness. Nat. Commun. 2020, 11, 1339. [Google Scholar] [CrossRef] [PubMed]
- Preta, G. New insights into targeting membrane lipids for cancer therapy. Front. Cell Dev. Biol. 2020, 8, 571237. [Google Scholar] [CrossRef]
- Piper, S.J.; Johnson, R.M.; Wootten, D.; Sexton, P.M. Membranes under the magnetic lens: A dive into the diverse world of membrane protein structures using Cryo-EM. Chem. Rev. 2022, 122, 13989–14017. [Google Scholar] [CrossRef] [PubMed]
- Soubias, O.; Sodt, A.J.; Teague, W.E.; Hines, K.G.; Gawrisch, K. Physiological changes in bilayer thickness induced by cholesterol control GPCR rhodopsin function. Biophys. J. 2023, 122, 973–983. [Google Scholar] [CrossRef]
- Yesylevskyy, S.O.; Rivel, T.; Ramseyer, C. The influence of curvature on the properties of the plasma membrane. Insights from atomistic molecular dynamics simulations. Sci. Rep. 2017, 7, 16078. [Google Scholar] [CrossRef]
- Kaddah, S.; Khreich, N.; Kaddah, F.; Charcosset, C.; Greige-Gerges, H. Cholesterol modulates the liposome membrane fluidity and permeability for a hydrophilic molecule. Food Chem. Toxicol. 2018, 113, 40–48. [Google Scholar] [CrossRef]
- Briuglia, M.L.; Rotella, C.; McFarlane, A.; Lamprou, D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015, 5, 231–242. [Google Scholar] [CrossRef]
- Yang, S.T.; Kreutzberger, A.J.; Lee, J.; Kiessling, V.; Tamm, L.K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids 2016, 199, 136–143. [Google Scholar] [CrossRef]
- Gu, R.X.; Baoukina, S.; Tieleman, D.P. Cholesterol flip-flop in heterogeneous membranes. J. Chem. Theory Comput. 2019, 15, 2064–2070. [Google Scholar] [CrossRef]
- Paukner, K.; Králová Lesná, I.; Poledne, R. Cholesterol in the cell membrane—An emerging player in atherogenesis. Int. J. Mol. Sci. 2022, 23, 533. [Google Scholar] [CrossRef] [PubMed]
- Varma, M.; Deserno, M. Distribution of cholesterol in asymmetric membranes driven by composition and differential stress. Biophys. J. 2022, 121, 4001–4018. [Google Scholar] [CrossRef]
- Ikonen, E.; Zhou, X. Cholesterol transport between cellular membranes: A balancing act between interconnected lipid fluxes. Dev. Cell 2021, 56, 1430–1436. [Google Scholar] [CrossRef]
- van der Koog, L.; Gandek, T.B.; Nagelkerke, A. Liposomes and extracellular vesicles as drug delivery systems: A comparison of composition, pharmacokinetics, and functionalization. Adv. Healthc. Mater. 2022, 11, 2100639. [Google Scholar] [CrossRef] [PubMed]
- Hudiyanti, D.; Fawrin, H.; Siahaan, P. Simultant encapsulation of vitamin C and beta-carotene in sesame (Sesamum indicum L.) liposomes. IOP Conf. Ser. Mater. Sci. Eng. 2018, 349, 012014. [Google Scholar] [CrossRef]
- Hudiyanti, D.; Christa, S.M.; Mardhiyyah, N.H.; Anugrah, D.S.B.; Widiarih, T.; Siahaan, P. Dynamics insights into aggregation of phospholipid species with cholesterol and vitamin C. Pharmacia 2022, 69, 385–391. [Google Scholar] [CrossRef]
- Liu, X.; Wang, P.; Zou, Y.X.; Luo, Z.G.; Tamer, T.M. Co-encapsulation of Vitamin C and β-Carotene in liposomes: Storage stability, antioxidant activity, and in vitro gastrointestinal digestion. Food Res. Int. 2020, 136, 109587. [Google Scholar] [CrossRef]
- Fantini, J.; Epand, R.M.; Barrantes, F.J. Cholesterol-recognition motifs in membrane proteins. In Direct Mechanisms in Cholesterol Modulation of Protein Function; Springer: Berlin/Heidelberg, Germany, 2019; pp. 3–25. [Google Scholar]
- Shahane, G.; Ding, W.; Palaiokostas, M.; Orsi, M. Physical properties of model biological lipid bilayers: Insights from all-atom molecular dynamics simulations. J. Mol. Model. 2019, 25, 76. [Google Scholar] [CrossRef] [PubMed]
- Tieleman, D.; Sejdiu, B.; Cino, E.; Smith, P.; Barreto-Ojeda, E.; Khan, H.; Corradi, V. Insights into lipid-protein interactions from computer simulations. Biophys. Rev. 2021, 13, 1019–1027. [Google Scholar] [CrossRef]
- Chaisson, E.H.; Heberle, F.A.; Doktorova, M. Building asymmetric lipid bilayers for molecular dynamics simulations: What methods exist and how to choose one? Membranes 2023, 13, 629. [Google Scholar] [CrossRef]
- Cho, I.; Jackson, M.R.; Swift, J. Roles of cross-membrane transport and signaling in the maintenance of cellular homeostasis. Cell. Mol. Bioeng. 2016, 9, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S. Mammalian lipids: Structure, synthesis and function. Essays Biochem. 2021, 65, 813–845. [Google Scholar] [CrossRef] [PubMed]
- Risselada, H.J. Cholesterol: The plasma membrane’s constituent that chooses sides. Biophys. J. 2019, 116, 2235–2236. [Google Scholar] [CrossRef]
- Zakany, F.; Mándity, I.M.; Varga, Z.; Panyi, G.; Nagy, P.; Kovacs, T. Effect of the Lipid Landscape on the Efficacy of Cell-Penetrating Peptides. Cells 2023, 12, 1700. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, L.M.; Yoon, B.K.; Jackman, J.A.; Knoll, W.; Weiss, P.S.; Cho, N.J. Understanding how sterols regulate membrane remodeling in supported lipid bilayers. Langmuir 2017, 33, 14756–14765. [Google Scholar] [CrossRef]
- Santamaria, A.; Batchu, K.C.; Fragneto, G.; Laux, V.; Haertlein, M.; Darwish, T.A.; Russell, R.A.; Zaccai, N.R.; Guzmán, E.; Maestro, A. Investigation on the relationship between lipid composition and structure in model membranes composed of extracted natural phospholipids. J. Colloid Interface Sci. 2023, 637, 55–66. [Google Scholar] [CrossRef]
- Subczynski, W.K.; Pasenkiewicz-Gierula, M.; Widomska, J.; Mainali, L.; Raguz, M. High cholesterol/low cholesterol: Effects in biological membranes: A review. Cell Biochem. Biophys. 2017, 75, 369–385. [Google Scholar] [CrossRef]
- Hryniewicz-Jankowska, A.; Augoff, K.; Sikorski, A.F. The role of cholesterol and cholesterol-driven membrane raft domains in prostate cancer. Exp. Biol. Med. 2019, 244, 1053–1061. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef]
- Ivanova, N.; Genova, J.; Chamati, H. Physical properties of SOPC lipid membranes containing cholesterol by molecular dynamics simulation. In Advances in Biomembranes and Lipid Self-Assembly; Elsevier: Amsterdam, The Netherlands, 2021; Volume 34, pp. 1–30. [Google Scholar]
- Ivanova, N.; Chamati, H. The effect of cholesterol in SOPC lipid bilayers at low temperatures. Membranes 2023, 13, 275. [Google Scholar] [CrossRef]
- Saito, H.; Morishita, T.; Mizukami, T.; Nishiyama, K.i.; Kawaguchi, K.; Nagao, H. Molecular dynamics study of binary POPC bilayers: Molecular condensing effects on membrane structure and dynamics. J. Physics Conf. Ser. 2018, 1136, 012022. [Google Scholar] [CrossRef]
- Elkins, M.R.; Bandara, A.; Pantelopulos, G.A.; Straub, J.E.; Hong, M. Direct observation of cholesterol dimers and tetramers in lipid bilayers. J. Phys. Chem. B 2021, 125, 1825–1837. [Google Scholar] [CrossRef]
- Kondela, T.; Dushanov, E.; Vorobyeva, M.; Mamatkulov, K.; Drolle, E.; Soloviov, D.; Hrubovčák, P.; Kholmurodov, K.; Arzumanyan, G.; Leonenko, Z.; et al. Investigating the competitive effects of cholesterol and melatonin in model lipid membranes. Biochim. Biophys. Acta-(BBA)-Biomembr. 2021, 1863, 183651. [Google Scholar] [CrossRef]
- Li, Z.B.; Fang, B.; Cui, X.X.; Zhang, C.Z.; Meng, Q.T. Physical mechanism analysis of cholesterol concentration effect on asymmetric phospholipid membrane. Eur. Phys. J. Plus 2021, 136, 136. [Google Scholar] [CrossRef]
- Alavizargar, A.; Keller, F.; Wedlich-Soldner, R.; Heuer, A. Effect of cholesterol versus ergosterol on DPPC bilayer properties: Insights from atomistic simulations. J. Phys. Chem. B 2021, 125, 7679–7690. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Doktorova, M.; Molugu, T.R.; Heberle, F.A.; Scott, H.L.; Dzikovski, B.; Nagao, M.; Stingaciu, L.R.; Standaert, R.F.; Barrera, F.N.; et al. How cholesterol stiffens unsaturated lipid membranes. Proc. Natl. Acad. Sci. USA 2020, 117, 21896–21905. [Google Scholar] [CrossRef] [PubMed]
- Doktorova, M.; LeVine, M.V.; Khelashvili, G.; Weinstein, H. A new computational method for membrane compressibility: Bilayer mechanical thickness revisited. Biophys. J. 2019, 116, 487–502. [Google Scholar] [CrossRef]
- Saeedimasine, M.; Montanino, A.; Kleiven, S.; Villa, A. Role of lipid composition on the structural and mechanical features of axonal membranes: A molecular simulation study. Sci. Rep. 2019, 9, 8000. [Google Scholar] [CrossRef]
- Blumer, M.; Harris, S.; Li, M.; Martinez, L.; Untereiner, M.; Saeta, P.N.; Carpenter, T.S.; Ingólfsson, H.I.; Bennett, W.D. Simulations of asymmetric membranes illustrate cooperative leaflet coupling and lipid adaptability. Front. Cell Dev. Biol. 2020, 8, 575. [Google Scholar] [CrossRef]
- Lu, S.M.; Fairn, G.D. Mesoscale organization of domains in the plasma membrane–beyond the lipid raft. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 192–207. [Google Scholar] [CrossRef]
- Aghaaminiha, M.; Farnoud, A.M.; Sharma, S. Quantitative relationship between cholesterol distribution and ordering of lipids in asymmetric lipid bilayers. Soft Matter 2021, 17, 2742–2752. [Google Scholar] [CrossRef] [PubMed]
- Olzynska, A.; Kulig, W.; Mikkolainen, H.; Czerniak, T.; Jurkiewicz, P.; Cwiklik, L.; Rog, T.; Hof, M.; Jungwirth, P.; Vattulainen, I. Tail-oxidized cholesterol enhances membrane permeability for small solutes. Langmuir 2020, 36, 10438–10447. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Sung, B.J. Facilitated and non-Gaussian diffusion of cholesterol in liquid ordered phase bilayers depends on the flip-flop and spatial arrangement of cholesterol. J. Phys. Chem. Lett. 2018, 9, 6529–6535. [Google Scholar] [CrossRef]
- Ermilova, I.; Lyubartsev, A.P. Cholesterol in phospholipid bilayers: Positions and orientations inside membranes with different unsaturation degrees. Soft Matter 2019, 15, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Matos, A.L.; Keller, F.; Wegner, T.; Del Castillo, C.E.C.; Grill, D.; Kudruk, S.; Spang, A.; Glorius, F.; Heuer, A.; Gerke, V. CHIMs are versatile cholesterol analogs mimicking and visualizing cholesterol behavior in lipid bilayers and cells. Commun. Biol. 2021, 4, 720. [Google Scholar]
- Kremkow, J.; Luck, M.; Huster, D.; Müller, P.; Scheidt, H.A. Membrane interaction of ibuprofen with cholesterol-containing lipid membranes. Biomolecules 2020, 10, 1384. [Google Scholar] [CrossRef]
- Pedrosa, M.; Moncho-Jordá, A.; Gálvez-Ruiz, M.J.; Kanduč, M. Impact of cholesterol on the structure and phase separation of DPPC Langmuir monolayers: Experiments and simulations. Surf. Interfaces 2024, 51, 104757. [Google Scholar] [CrossRef]
- Bennett, W.F.D.; Tieleman, D.P. Computer simulations of lipid membrane domains. Biochim. Biophys. Acta-(BBA)-Biomembr. 2013, 1828, 1765–1776. [Google Scholar] [CrossRef]
- Drabik, D.; Chodaczek, G.; Kraszewski, S.; Langner, M. Mechanical Properties Determination of DMPC, DPPC, DSPC, and HSPC Solid-Ordered Bilayers. Langmuir 2020, 36, 3826–3835. [Google Scholar] [CrossRef]
- Hung, W.C.; Lee, M.T.; Chung, H.; Sun, Y.T.; Chen, H.; Charron, N.E.; Huang, H.W. Comparative Study of the Condensing Effects of Ergosterol and Cholesterol. Biophys. J. 2016, 110, 2026–2033. [Google Scholar] [CrossRef][Green Version]
- Goicoechea, L.; de la Rosa, L.C.; Torres, S.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial cholesterol: Metabolism and impact on redox biology and disease. Redox Biol. 2023, 61, 102643. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.H. Molecular dynamics simulations of curved lipid membranes. Int. J. Mol. Sci. 2022, 23, 8098. [Google Scholar] [CrossRef]
- Wiczew, D.; Szulc, N.; Tarek, M. Molecular dynamics simulations of the effects of lipid oxidation on the permeability of cell membranes. Bioelectrochemistry 2021, 141, 107869. [Google Scholar] [CrossRef]
- Beaven, A.H.; Sapp, K.; Sodt, A.J. Simulated dynamic cholesterol redistribution favors membrane fusion pore constriction. Biophys. J. 2023, 122, 2162–2175. [Google Scholar] [CrossRef]
- Caparotta, M.; Masone, D. Cholesterol plays a decisive role in tetraspanin assemblies during bilayer deformations. Biosystems 2021, 209, 104505. [Google Scholar] [CrossRef]
- Jafari, M.; Mehrnejad, F.; Talandashti, R.; Doustdar, F.; Vakili, M.R.; Lavasanifar, A. Effect of the lipid composition and cholesterol on the membrane selectivity of low generations PAMAM dendrimers: A molecular dynamics simulation study. Appl. Surf. Sci. 2021, 540, 148274. [Google Scholar] [CrossRef]
- Yang, Y.; Dong, H.; Zhou, H.X. Effects of cholesterol on the partitioning of a drug molecule in lipid bilayers. J. Phys. Chem. B 2021, 125, 5338–5345. [Google Scholar] [CrossRef] [PubMed]
- Kramar, P.; Miklavčič, D. Effect of the cholesterol on electroporation of planar lipid bilayer. Bioelectrochemistry 2022, 144, 108004. [Google Scholar] [CrossRef] [PubMed]
- Canepa, E.; Bochicchio, D.; Gasbarri, M.; Odino, D.; Canale, C.; Ferrando, R.; Canepa, F.; Stellacci, F.; Rossi, G.; Dante, S.; et al. Cholesterol hinders the passive uptake of amphiphilic nanoparticles into fluid lipid membranes. J. Phys. Chem. Lett. 2021, 12, 8583–8590. [Google Scholar] [CrossRef]
- Pöhnl, M.; Trollmann, M.F.; Böckmann, R.A. Nonuniversal impact of cholesterol on membranes mobility, curvature sensing and elasticity. Nat. Commun. 2023, 14, 8038. [Google Scholar] [CrossRef]
- Li, M.J.; Atkins, W.M.; McClary, W.D. Preparation of lipid nanodiscs with lipid mixtures. Curr. Protoc. Protein Sci. 2019, 98, e100. [Google Scholar] [CrossRef] [PubMed]
- Parini, P. HDL, reverse cholesterol transport, and atherosclerosis: Unravelling the complexity or adding to the confusion? Atherosclerosis 2024, 397. [Google Scholar] [CrossRef] [PubMed]
- Georgila, K.; Vyrla, D.; Drakos, E. Apolipoprotein AI (ApoA-I), immunity, inflammation and cancer. Cancers 2019, 11, 1097. [Google Scholar] [CrossRef]
- Pourmousa, M.; Pastor, R.W. Molecular dynamics simulations of lipid nanodiscs. Biochim. Biophys. Acta-(BBA)-Biomembr. 2018, 1860, 2094–2107. [Google Scholar] [CrossRef]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef]
- Schachter, I.; Allolio, C.; Khelashvili, G.; Harries, D. Confinement in nanodiscs anisotropically modifies lipid bilayer elastic properties. J. Phys. Chem. B 2020, 124, 7166–7175. [Google Scholar] [CrossRef]
- Rouck, J.E.; Krapf, J.E.; Roy, J.; Huff, H.C.; Das, A. Recent advances in nanodisc technology for membrane protein studies (2012–2017). FEBS Lett. 2017, 591, 2057–2088. [Google Scholar] [CrossRef] [PubMed]
- Bengtsen, T.; Holm, V.L.; Kjølbye, L.R.; Midtgaard, S.R.; Johansen, N.T.; Tesei, G.; Bottaro, S.; Schiøtt, B.; Arleth, L.; Lindorff-Larsen, K. Structure and dynamics of a nanodisc by integrating NMR, SAXS and SANS experiments with molecular dynamics simulations. Elife 2020, 9, e56518. [Google Scholar] [CrossRef]
- Ouimet, M.; Barrett, T.J.; Fisher, E.A. HDL and reverse cholesterol transport: Basic mechanisms and their roles in vascular health and disease. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef]
- Lau, S.; Middleton, D.A. Analysis of the orientation of cholesterol in high-density lipoprotein nanodiscs using solid-state NMR. Phys. Chem. Chem. Phys. 2022, 24, 23651–23660. [Google Scholar] [CrossRef]
- Denis, M.; Landry, Y.D.; Zha, X. ATP-binding cassette A1-mediated lipidation of apolipoprotein AI occurs at the plasma membrane and not in the endocytic compartments. J. Biol. Chem. 2008, 283, 16178–16186. [Google Scholar] [CrossRef] [PubMed]
- Norum, K.R. The function of lecithin: Cholesterol acyltransferase (LCAT). Scand. J. Clin. Lab. Investig. 2017, 77, 235–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiao, Q.; Wang, L.; Wang, Y.; Wang, D.; Ding, H. Role of ABCA1 in cardiovascular disease. J. Pers. Med. 2022, 12, 1010. [Google Scholar] [CrossRef]
- Jebari-Benslaiman, S.; Uribe, K.B.; Benito-Vicente, A.; Galicia-Garcia, U.; Larrea-Sebal, A.; Alloza, I.; Vandenbroeck, K.; Ostolaza, H.; Martín, C. Cholesterol efflux efficiency of reconstituted HDL is affected by nanoparticle lipid composition. Biomedicines 2020, 8, 373. [Google Scholar] [CrossRef]
- Schrijver, D.P.; de Dreu, A.; Hofstraat, S.R.; Kluza, E.; Zwolsman, R.; Deckers, J.; Anbergen, T.; de Bruin, K.; Trines, M.M.; Nugraha, E.G.; et al. Nanoengineering Apolipoprotein A1-Based Immunotherapeutics. Adv. Ther. 2021, 4, 2100083. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, Z.W.; Zeng, P.H.; Zhou, Y.J.; Yin, W.J. Molecular mechanisms for ABCA1-mediated cholesterol efflux. Cell Cycle 2022, 21, 1121–1139. [Google Scholar] [CrossRef] [PubMed]
- Jomard, A.; Osto, E. High density lipoproteins: Metabolism, function, and therapeutic potential. Front. Cardiovasc. Med. 2020, 7, 39. [Google Scholar] [CrossRef]
- Morvaridzadeh, M.; Zoubdane, N.; Heshmati, J.; Alami, M.; Berrougui, H.; Khalil, A. High-density lipoprotein metabolism and function in cardiovascular diseases: What about aging and diet effects? Nutrients 2024, 16, 653. [Google Scholar] [CrossRef]
- Islam, R.M.; Pourmousa, M.; Sviridov, D.; Gordon, S.M.; Neufeld, E.B.; Freeman, L.A.; Perrin, B.S., Jr.; Pastor, R.W.; Remaley, A.T. Structural properties of apolipoprotein AI mimetic peptides that promote ABCA1-dependent cholesterol efflux. Sci. Rep. 2018, 8, 2956. [Google Scholar] [CrossRef]
- Jaipuria, G.; Ukmar-Godec, T.; Zweckstetter, M. Challenges and approaches to understand cholesterol-binding impact on membrane protein function: An NMR view. Cell. Mol. Life Sci. 2018, 75, 2137–2151. [Google Scholar] [CrossRef]
- Giorgi, L.; Niemela, A.; Kumpula, E.P.; Natri, O.; Parkkila, P.; Huiskonen, J.T.; Koivuniemi, A. Mechanistic Insights into the Activation of Lecithin–Cholesterol Acyltransferase in Therapeutic Nanodiscs Composed of Apolipoprotein AI Mimetic Peptides and Phospholipids. Mol. Pharm. 2022, 19, 4135–4148. [Google Scholar] [CrossRef] [PubMed]
- Rosenson, R.S.; Brewer, H.B., Jr.; Davidson, W.S.; Fayad, Z.A.; Fuster, V.; Goldstein, J.; Hellerstein, M.; Jiang, X.C.; Phillips, M.C.; Rader, D.J.; et al. Cholesterol efflux and atheroprotection: Advancing the concept of reverse cholesterol transport. Circulation 2012, 125, 1905–1919. [Google Scholar] [CrossRef] [PubMed]
- Cang, X.; Du, Y.; Mao, Y.; Wang, Y.; Yang, H.; Jiang, H. Mapping the functional binding sites of cholesterol in β2-adrenergic receptor by long-time molecular dynamics simulations. J. Phys. Chem. B 2013, 117, 1085–1094. [Google Scholar] [CrossRef]
- Pluhackova, K.; Gahbauer, S.; Kranz, F.; Wassenaar, T.A.; Böckmann, R.A. Dynamic cholesterol-conditioned dimerization of the G protein coupled chemokine receptor type 4. PLoS Comput. Biol. 2016, 12, e1005169. [Google Scholar] [CrossRef]
- Corradi, V.; Mendez-Villuendas, E.; Ingólfsson, H.I.; Gu, R.X.; Siuda, I.; Melo, M.N.; Moussatova, A.; DeGagné, L.J.; Sejdiu, B.I.; Singh, G.; et al. Lipid–protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 2018, 4, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Moon, H.; Kim, H.R. Effects of lipid shape and interactions on the conformation, dynamics, and curvature of ultrasound-responsive liposomes. Pharmaceutics 2022, 14, 1512. [Google Scholar] [CrossRef]
- López, C.A.; de Vries, A.H.; Marrink, S.J. Molecular mechanism of cyclodextrin mediated cholesterol extraction. PLoS Comput. Biol. 2011, 7, e1002020. [Google Scholar] [CrossRef]
- Hammouda, Z.; Khreich, N.; Auezova, L.; Fourmentin, S.; Elaissari, A.; Greige-Gerges, H. Cyclodextrin-membrane interaction in drug delivery and membrane structure maintenance. Int. J. Pharm. 2019, 564, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Parchekani, J.; Allahverdi, A.; Taghdir, M.; Naderi-Manesh, H. Design and simulation of the liposomal model by using a coarse-grained molecular dynamics approach towards drug delivery goals. Sci. Rep. 2022, 12, 2371. [Google Scholar] [CrossRef]
- Wu, H.; Yu, M.; Miao, Y.; He, S.; Dai, Z.; Song, W.; Liu, Y.; Song, S.; Ahmad, E.; Wang, D.; et al. Cholesterol-tuned liposomal membrane rigidity directs tumor penetration and anti-tumor effect. Acta Pharm. Sin. B 2019, 9, 858–870. [Google Scholar] [CrossRef]
- Costa, R.R.; Domínguez-Arca, V.; Velasco, B.; Reis, R.L.; Rodríguez-Cabello, J.C.; Pashkuleva, I.; Taboada, P.; Prieto, G. Cholesterol Conjugated Elastin-like Recombinamers: Molecular Dynamics Simulations, Conformational Changes, and Bioactivity. ACS Appl. Mater. Interfaces 2024, 16, 66327–66340. [Google Scholar] [CrossRef] [PubMed]
- Ingram, T.; Storm, S.; Kloss, L.; Mehling, T.; Jakobtorweihen, S.; Smirnova, I. Prediction of micelle/water and liposome/water partition coefficients based on molecular dynamics simulations, COSMO-RS, and COSMOmic. Langmuir 2013, 29, 3527–3537. [Google Scholar] [CrossRef] [PubMed]
- Yoshiaki, Y.; Kotaro, K.; Ryota, K.; Arisa, Y.; Katsumi, M. Cholesterol-Induced Lipophobic Interaction between Transmembrane Helices Using Ensemble and Single-Molecule Fluorescence Resonance Energy Transfer. Biochemistry 2015, 54, 1371–1379. [Google Scholar]
- Liu, Y.; Zhang, D.; Zhang, Y.; Tang, Y.; Xu, L.; He, H.; Wu, J.; Zheng, J. Molecular dynamics simulations of cholesterol effects on the interaction of hIAPP with lipid bilayer. J. Phys. Chem. B 2020, 124, 7830–7841. [Google Scholar] [CrossRef]
- Qian, S.; Heller, W.T. Melittin-induced cholesterol reorganization in lipid bilayer membranes. Biochim. Biophys. Acta-(BBA)-Biomembr. 2015, 1848, 2253–2260. [Google Scholar] [CrossRef]
- Cheerla, R.; Ayappa, K.G. Molecular dynamics study of lipid and cholesterol reorganization due to membrane binding and pore formation by Listeriolysin O. J. Membr. Biol. 2020, 253, 535–550. [Google Scholar] [CrossRef]
- Hashemi, M.; Banerjee, S.; Lyubchenko, Y.L. Free cholesterol accelerates Aβ self-assembly on membranes at physiological concentration. Int. J. Mol. Sci. 2022, 23, 2803. [Google Scholar] [CrossRef]
- Wang, B.; Guo, C. Concentration-dependent effects of cholesterol on the dimerization of Amyloid-β peptides in lipid bilayers. ACS Chem. Neurosci. 2022, 13, 2709–2718. [Google Scholar] [CrossRef]
- Isu, U.H.; Polasa, A.; Moradi, M. Differential Behavior of Conformational Dynamics in Active and Inactive States of Cannabinoid Receptor 1. J. Phys. Chem. B 2024, 128, 8437–8447. [Google Scholar] [CrossRef]

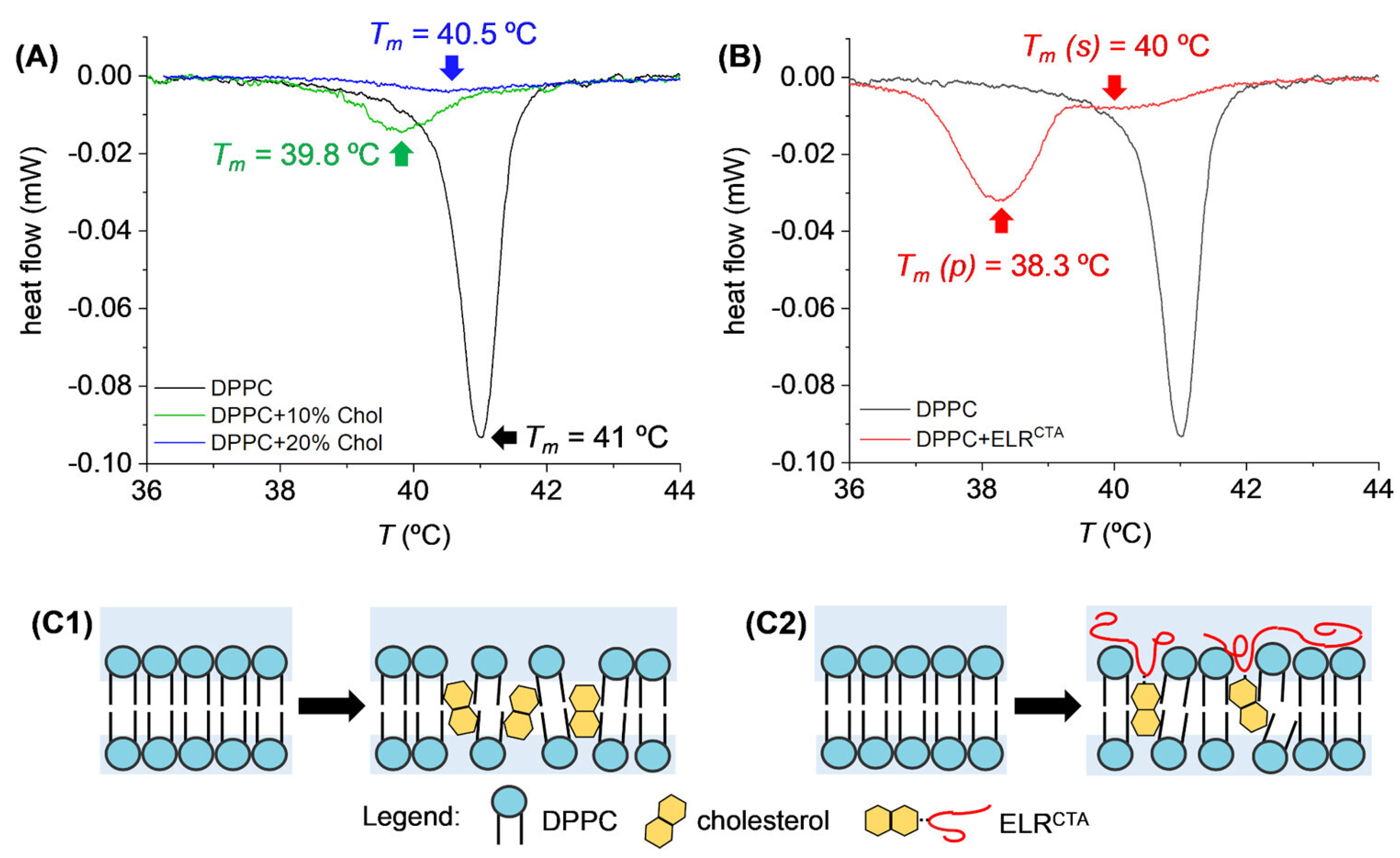

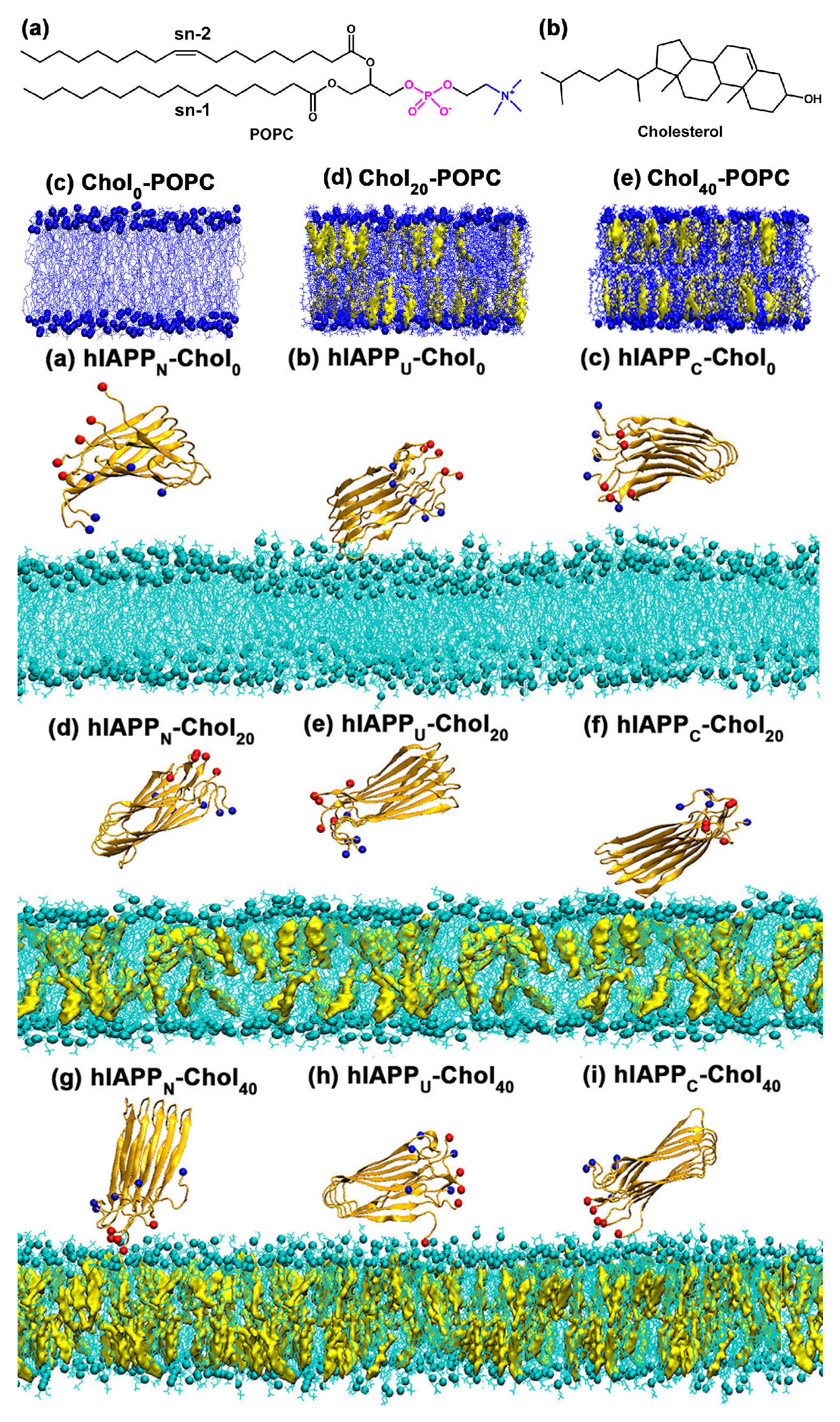
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khodadadi, E.; Khodadadi, E.; Chaturvedi, P.; Moradi, M. Comprehensive Insights into the Cholesterol-Mediated Modulation of Membrane Function Through Molecular Dynamics Simulations. Membranes 2025, 15, 173. https://doi.org/10.3390/membranes15060173
Khodadadi E, Khodadadi E, Chaturvedi P, Moradi M. Comprehensive Insights into the Cholesterol-Mediated Modulation of Membrane Function Through Molecular Dynamics Simulations. Membranes. 2025; 15(6):173. https://doi.org/10.3390/membranes15060173
Chicago/Turabian StyleKhodadadi, Ehsaneh, Ehsan Khodadadi, Parth Chaturvedi, and Mahmoud Moradi. 2025. "Comprehensive Insights into the Cholesterol-Mediated Modulation of Membrane Function Through Molecular Dynamics Simulations" Membranes 15, no. 6: 173. https://doi.org/10.3390/membranes15060173
APA StyleKhodadadi, E., Khodadadi, E., Chaturvedi, P., & Moradi, M. (2025). Comprehensive Insights into the Cholesterol-Mediated Modulation of Membrane Function Through Molecular Dynamics Simulations. Membranes, 15(6), 173. https://doi.org/10.3390/membranes15060173





