Amyloid Precursor Protein Changes Arrangement in a Membrane and Its Structure Depending on the Cholesterol Content
Abstract
1. Introduction
2. Materials and Methods
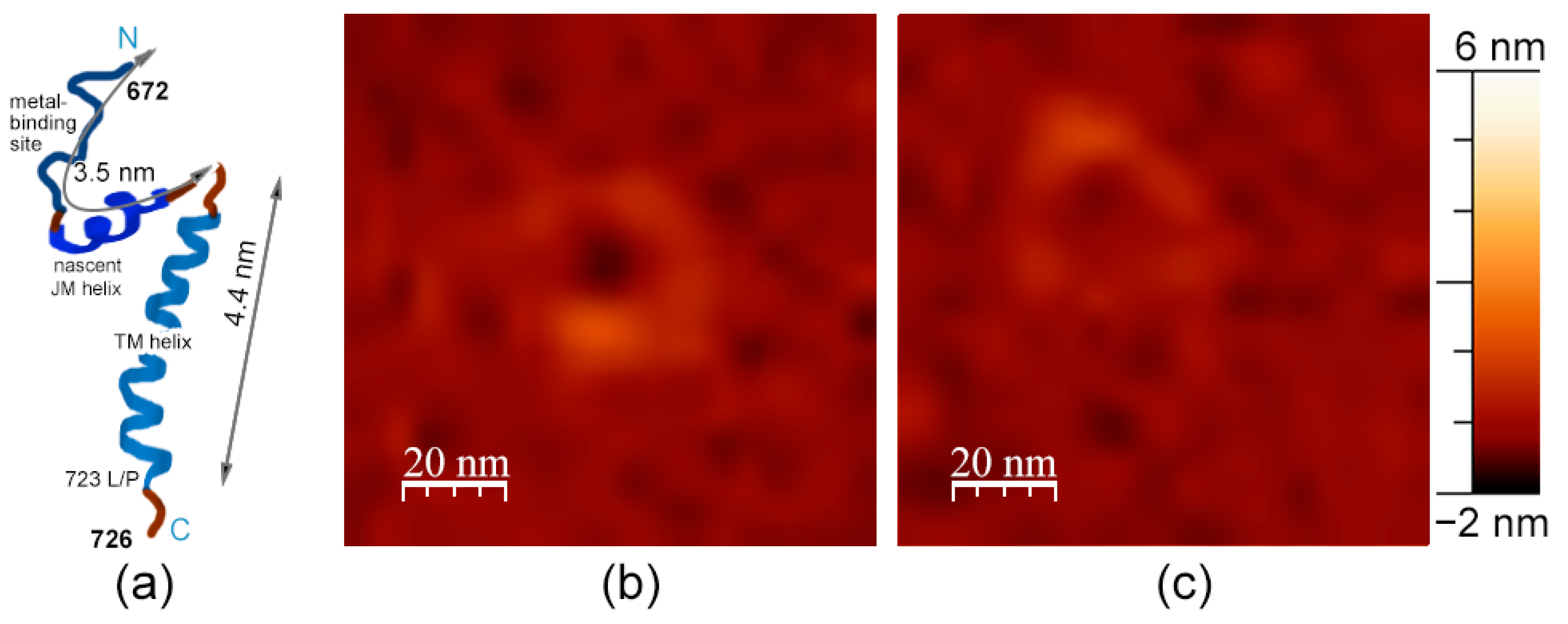
3. Results
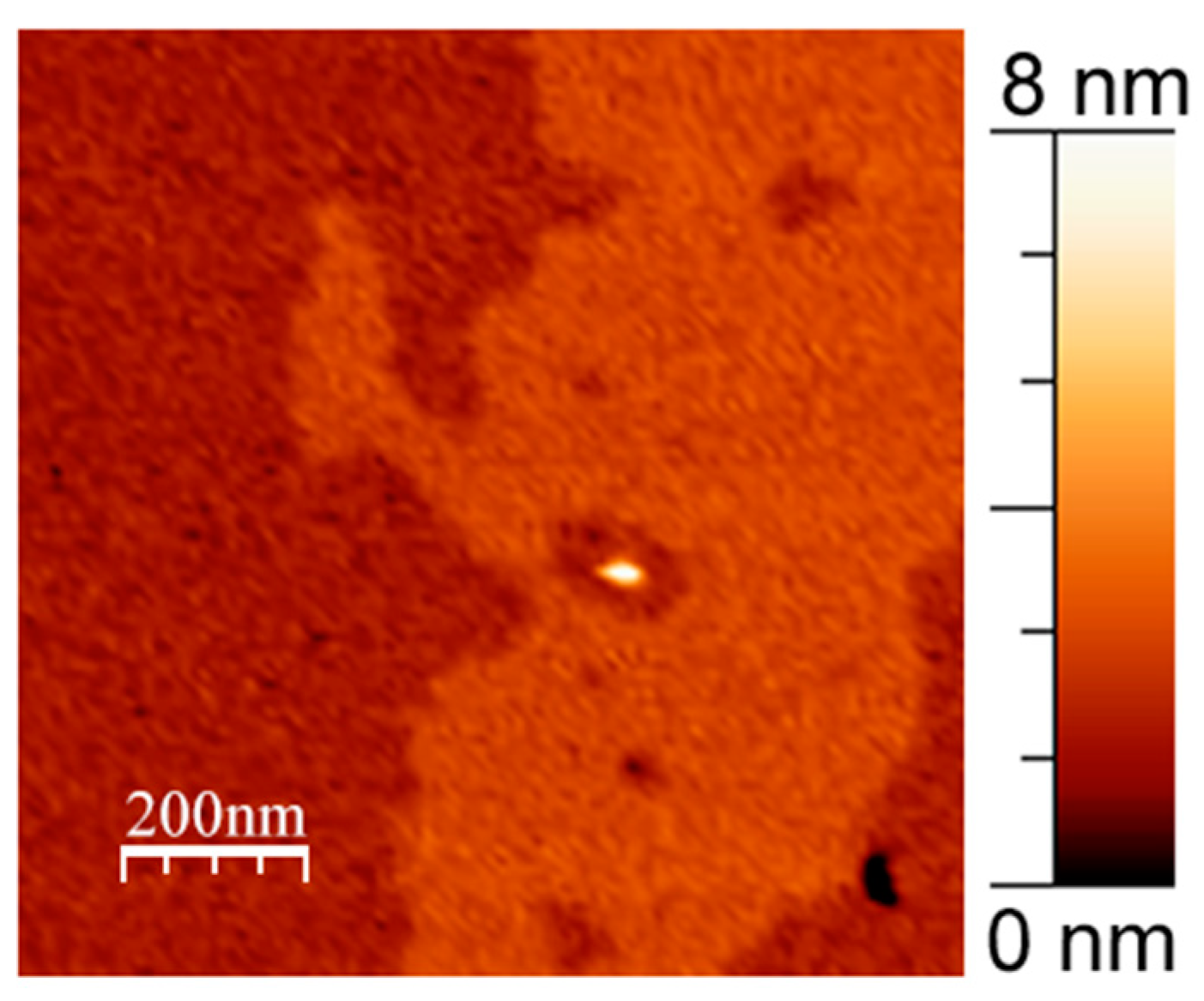
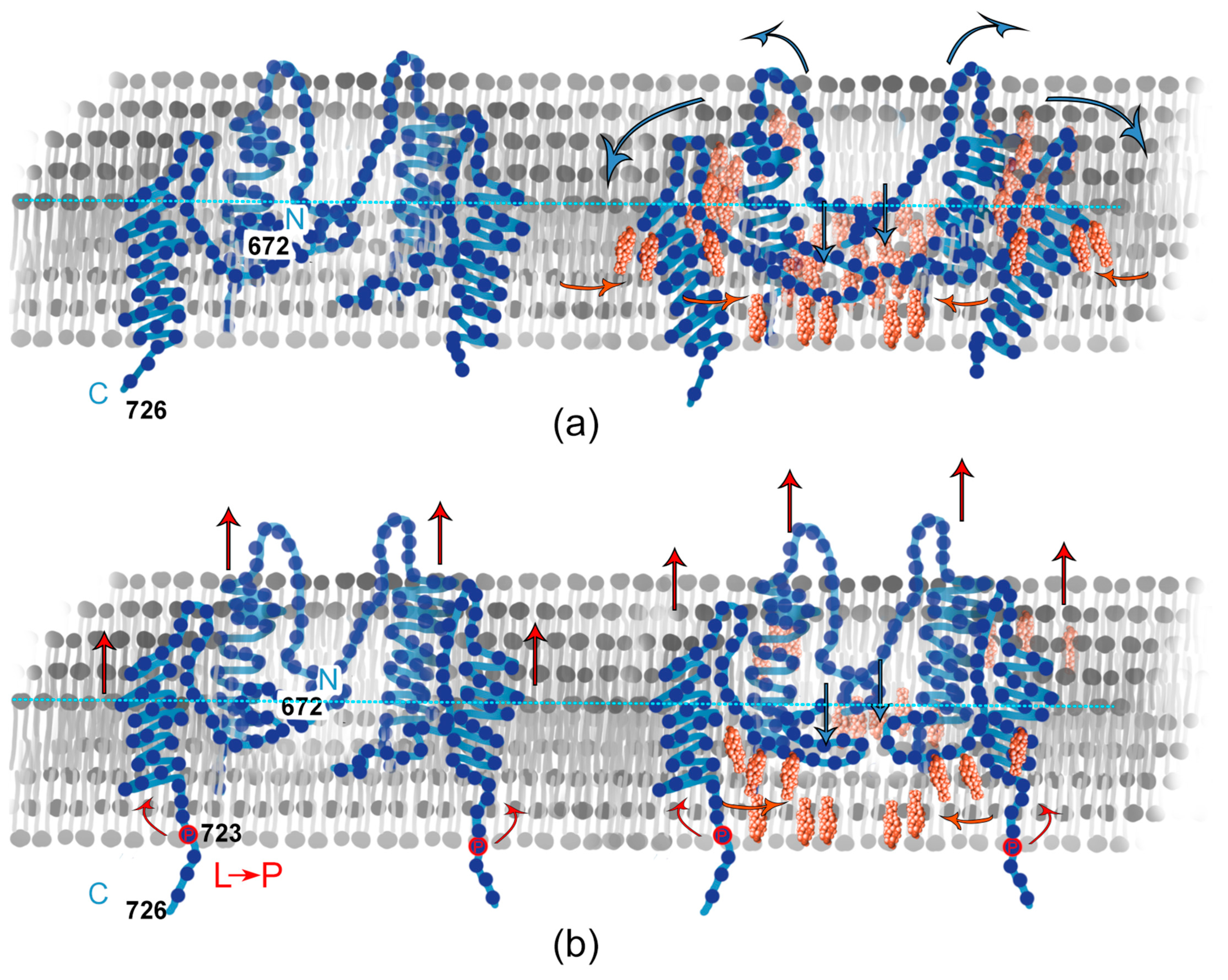
3.1. APP and Lipid Rafts
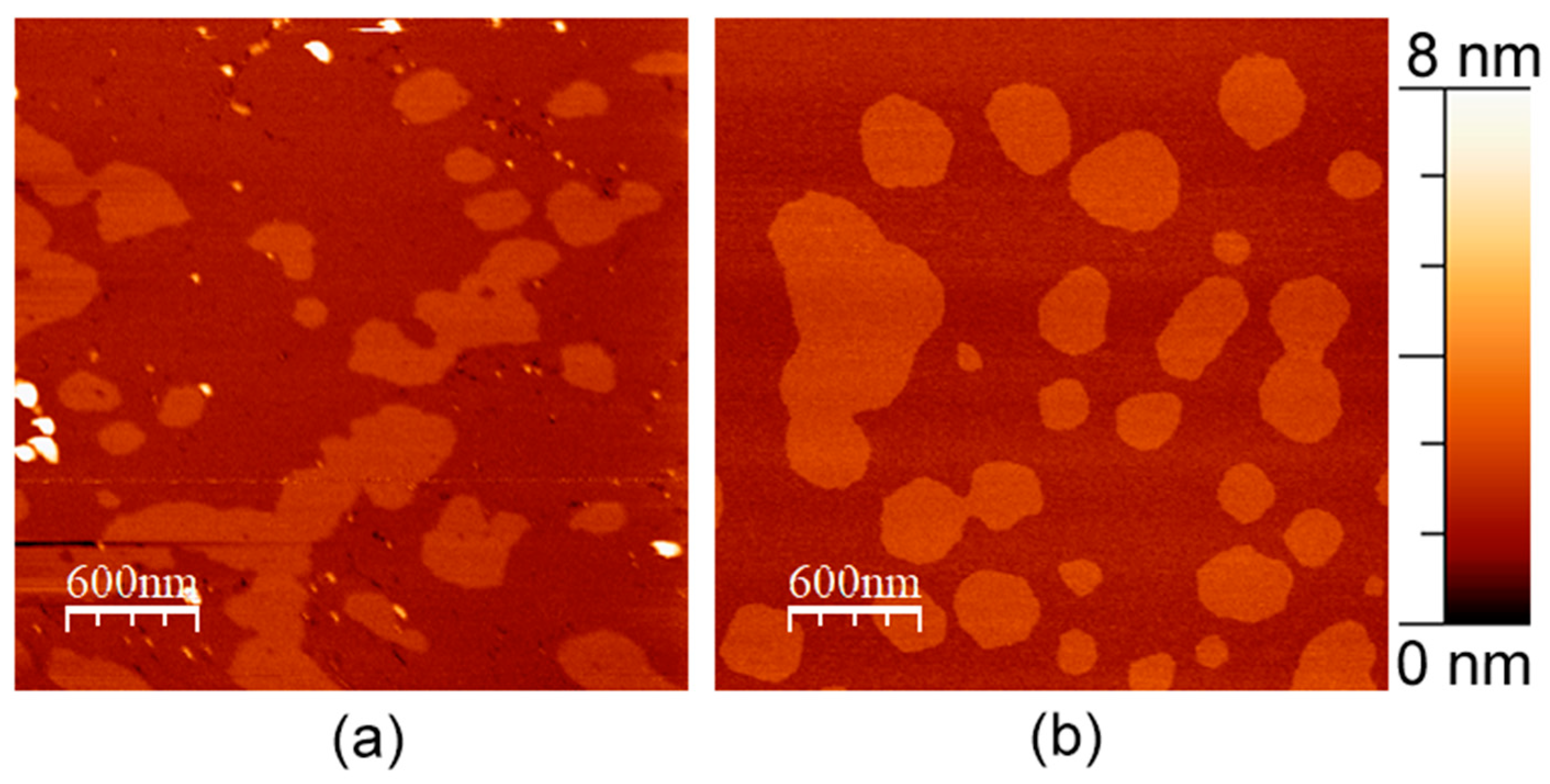
3.2. APP and Cholesterol Content
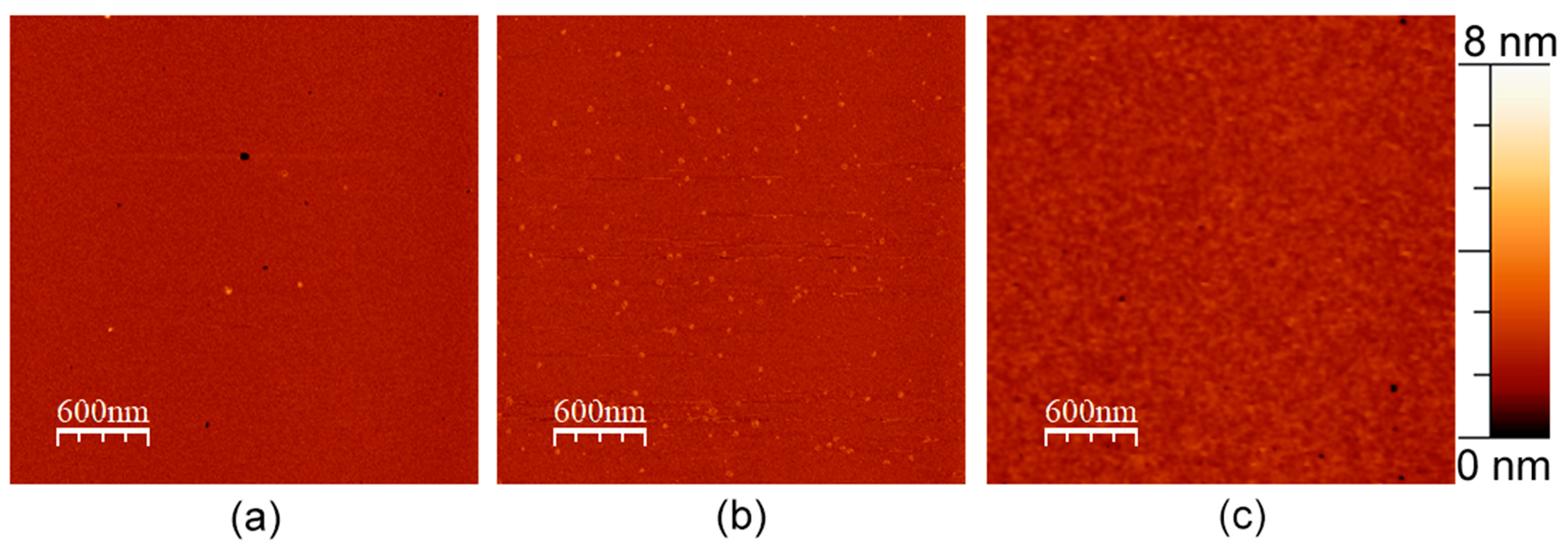
3.3. APP in a Membrane without Cholesterol
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s Disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.E. Diagnosis of Dementia. Psychosomatics 1979, 20, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Li, H.; Gaddam, S.S.K.; Justice, N.J.; Robertson, C.S.; Zheng, H. Amyloid Precursor Protein Revisited: Neuron-Specific Expression and Highly Stable Nature of Soluble Derivatives. J. Biol. Chem. 2012, 287, 2437–2445. [Google Scholar] [CrossRef]
- Lee, J.-H.; Yang, D.-S.; Goulbourne, C.N.; Im, E.; Stavrides, P.; Pensalfini, A.; Chan, H.; Bouchet-Marquis, C.; Bleiwas, C.; Berg, M.J.; et al. Faulty Autolysosome Acidification in Alzheimer’s Disease Mouse Models Induces Autophagic Build-up of Aβ in Neurons, Yielding Senile Plaques. Nat. Neurosci. 2022, 25, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsic Disorder in Proteins Associated with Neurodegenerative Diseases. Front. Biosci. Landmark Ed. 2009, 14, 5188–5238. [Google Scholar] [CrossRef]
- Lührs, T.; Ritter, C.; Adrian, M.; Riek-Loher, D.; Bohrmann, B.; Döbeli, H.; Schubert, D.; Riek, R. 3D Structure of Alzheimer’s Amyloid-Beta(1-42) Fibrils. Proc. Natl. Acad. Sci. USA 2005, 102, 17342–17347. [Google Scholar] [CrossRef]
- Urban, A.S.; Pavlov, K.V.; Kamynina, A.V.; Okhrimenko, I.S.; Arseniev, A.S.; Bocharov, E.V. Structural Studies Providing Insights into Production and Conformational Behavior of Amyloid-β Peptide Associated with Alzheimer’s Disease Development. Mol. Basel Switz. 2021, 26, 2897. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Q.; Zhang, Y.; Xu, H. Proteolytic Processing of Alzheimer’s β-Amyloid Precursor Protein. J. Neurochem. 2012, 120, 9–21. [Google Scholar] [CrossRef]
- Gouras, G.K.; Olsson, T.T.; Hansson, O. β-Amyloid Peptides and Amyloid Plaques in Alzheimer’s Disease. Neurother. J. Am. Soc. Exp. Neurother. 2015, 12, 3–11. [Google Scholar] [CrossRef]
- Takahashi, R.H.; Nagao, T.; Gouras, G.K. Plaque Formation and the Intraneuronal Accumulation of β-Amyloid in Alzheimer’s Disease. Pathol. Int. 2017, 67, 185–193. [Google Scholar] [CrossRef]
- Piller, C. Blots on a Field? Science 2022, 377, 358–363. [Google Scholar] [CrossRef]
- Morris, G.P.; Clark, I.A.; Vissel, B. Inconsistencies and Controversies Surrounding the Amyloid Hypothesis of Alzheimer’s Disease. Acta Neuropathol. Commun. 2014, 2, 135. [Google Scholar] [CrossRef]
- Rosenblum, W.I. Why Alzheimer Trials Fail: Removing Soluble Oligomeric Beta Amyloid Is Essential, Inconsistent, and Difficult. Neurobiol. Aging 2014, 35, 969–974. [Google Scholar] [CrossRef]
- Levy, E.; Carman, M.D.; Fernandez-Madrid, I.J.; Power, M.D.; Lieberburg, I.; van Duinen, S.G.; Bots, G.T.A.M.; Luyendijk, W.; Frangione, B. Mutation of the Alzheimer’s Disease Amyloid Gene in Hereditary Cerebral Hemorrhage, Dutch Type. Science 1990, 248, 1124–1126. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N. Chapter 11-Alzheimer’s Disease. In Brain Lipids in Synaptic Function and Neurological Disease; Fantini, J., Yahi, N., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 253–277. ISBN 978-0-12-800111-0. [Google Scholar]
- Mattson, M.P. Cellular Actions of Beta-Amyloid Precursor Protein and Its Soluble and Fibrillogenic Derivatives. Physiol. Rev. 1997, 77, 1081–1132. [Google Scholar] [CrossRef]
- Nilsberth, C.; Westlind-Danielsson, A.; Eckman, C.B.; Condron, M.M.; Axelman, K.; Forsell, C.; Stenh, C.; Luthman, J.; Teplow, D.B.; Younkin, S.G.; et al. The “Arctic” APP Mutation (E693G) Causes Alzheimer’s Disease by Enhanced Aβ Protofibril Formation. Nat. Neurosci. 2001, 4, 887–893. [Google Scholar] [CrossRef]
- Yoshioka, K.; Miki, T.; Katsuya, T.; Ogihara, T.; Sakaki, Y. The 717Val→Ile Substitution in Amyloid Precursor Protein Is Associated with Familial Alzheimer’s Disease Regardless of Ethnic Groups. Biochem. Biophys. Res. Commun. 1991, 178, 1141–1146. [Google Scholar] [CrossRef]
- Murrell, J.R.; Hake, A.M.; Quaid, K.A.; Farlow, M.R.; Ghetti, B. Early-Onset Alzheimer Disease Caused by a New Mutation (V717L) in the Amyloid Precursor Protein Gene. Arch. Neurol. 2000, 57, 885. [Google Scholar] [CrossRef]
- Kwok, J.B.J.; Li, Q.-X.; Hallupp, M.; Whyte, S.; Ames, D.; Beyreuther, K.; Masters, C.L.; Schofield, P.R. Novel Leu723Pro Amyloid Precursor Protein Mutation Increases Amyloid Β42(43) Peptide Levels and Induces Apoptosis. Ann. Neurol. 2000, 47, 249–253. [Google Scholar] [CrossRef]
- Dimitrov, M.; Alattia, J.-R.; Lemmin, T.; Lehal, R.; Fligier, A.; Houacine, J.; Hussain, I.; Radtke, F.; Dal Peraro, M.; Beher, D.; et al. Alzheimer’s Disease Mutations in APP but Not γ-Secretase Modulators Affect Epsilon-Cleavage-Dependent AICD Production. Nat. Commun. 2013, 4, 2246. [Google Scholar] [CrossRef]
- Bocharov, E.V.; Nadezhdin, K.D.; Urban, A.S.; Volynsky, P.E.; Pavlov, K.V.; Efremov, R.G.; Arseniev, A.S.; Bocharova, O.V. Familial L723P Mutation Can Shift the Distribution between the Alternative APP Transmembrane Domain Cleavage Cascades by Local Unfolding of the Ε-Cleavage Site Suggesting a Straightforward Mechanism of Alzheimer’s Disease Pathogenesis. ACS Chem. Biol. 2019, 14, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.L.; Vassar, R. The Alzheimer’s Disease Beta-Secretase Enzyme, BACE1. Mol. Neurodegener. 2007, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Kumar-Singh, S.; Dewachter, I.; Moechars, D.; Lübke, U.; De Jonghe, C.; Ceuterick, C.; Checler, F.; Naidu, A.; Cordell, B.; Cras, P.; et al. Behavioral Disturbances without Amyloid Deposits in Mice Overexpressing Human Amyloid Precursor Protein with Flemish (A692G) or Dutch (E693Q) Mutation. Neurobiol. Dis. 2000, 7, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Ariga, T.; Kobayashi, K.; Hasegawa, A.; Kiso, M.; Ishida, H.; Miyatake, T. Characterization of High-Affinity Binding between Gangliosides and Amyloid Beta-Protein. Arch. Biochem. Biophys. 2001, 388, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Di Scala, C.; Yahi, N.; Lelièvre, C.; Garmy, N.; Chahinian, H.; Fantini, J. Biochemical Identification of a Linear Cholesterol-Binding Domain within Alzheimer’s β Amyloid Peptide. ACS Chem. Neurosci. 2013, 4, 509–517. [Google Scholar] [CrossRef]
- Beel, A.J.; Mobley, C.K.; Kim, H.J.; Tian, F.; Hadziselimovic, A.; Jap, B.; Prestegard, J.H.; Sanders, C.R. Structural Studies of the Transmembrane C-Terminal Domain of the Amyloid Precursor Protein (APP): Does APP Function as a Cholesterol Sensor? Biochemistry 2008, 47, 9428–9446. [Google Scholar] [CrossRef]
- Barrett, P.J.; Song, Y.; Van Horn, W.D.; Hustedt, E.J.; Schafer, J.M.; Hadziselimovic, A.; Beel, A.J.; Sanders, C.R. The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol. Science 2012, 336, 1168–1171. [Google Scholar] [CrossRef]
- Nadezhdin, K.D.; Bocharova, O.V.; Bocharov, E.V.; Arseniev, A.S. Structural and Dynamic Study of the Transmembrane Domain of the Amyloid Precursor Protein. Acta Naturae 2011, 3, 69–76. [Google Scholar] [CrossRef]
- Hanbouch, L.; Schaack, B.; Kasri, A.; Fontaine, G.; Gkanatsiou, E.; Brinkmalm, G.; Camporesi, E.; Portelius, E.; Blennow, K.; Mourier, G.; et al. Specific Mutations in the Cholesterol-Binding Site of APP Alter Its Processing and Favor the Production of Shorter, Less Toxic Aβ Peptides. Mol. Neurobiol. 2022, 59, 7056–7073. [Google Scholar] [CrossRef]
- Pinigin, K.V.; Kondrashov, O.V.; Jiménez-Munguía, I.; Alexandrova, V.V.; Batishchev, O.V.; Galimzyanov, T.R.; Akimov, S.A. Elastic Deformations Mediate Interaction of the Raft Boundary with Membrane Inclusions Leading to Their Effective Lateral Sorting. Sci. Rep. 2020, 10, 4087. [Google Scholar] [CrossRef]
- Galimzyanov, T.R.; Lyushnyak, A.S.; Aleksandrova, V.V.; Shilova, L.A.; Mikhalyov, I.I.; Molotkovskaya, I.M.; Akimov, S.A.; Batishchev, O.V. Line Activity of Ganglioside GM1 Regulates the Raft Size Distribution in a Cholesterol-Dependent Manner. Langmuir 2017, 33, 3517–3524. [Google Scholar] [CrossRef]
- Krasnobaev, V.D.; Galimzyanov, T.R.; Akimov, S.A.; Batishchev, O.V. Lysolipids Regulate Raft Size Distribution. Front. Mol. Biosci. 2022, 9, 1021321. [Google Scholar] [CrossRef]
- Trabelsi, S.; Zhang, S.; Lee, T.R.; Schwartz, D.K. Linactants: Surfactant Analogues in Two Dimensions. Phys. Rev. Lett. 2008, 100, 037802. [Google Scholar] [CrossRef]
- Krasnobaev, V.D.; Batishchev, O.V. The Role of Lipid Domains and Physical Properties of Membranes in the Development of Age-Related Neurodegenerative Diseases. Biochem. Mosc. Suppl. Ser. Membr. Cell Biol. 2022, 16, 268–281. [Google Scholar] [CrossRef]
- Bocharova, O.V.; Urban, A.S.; Nadezhdin, K.D.; Bocharov, E.V.; Arseniev, A.S. Cell-Free Expression of the APP Transmembrane Fragments with Alzheimer’s Disease Mutations Using Algal Amino Acid Mixture for Structural NMR Studies. Protein Expr. Purif. 2016, 123, 105–111. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef]
- Grimm, M.O.W.; Grimm, H.S.; Pätzold, A.J.; Zinser, E.G.; Halonen, R.; Duering, M.; Tschäpe, J.A.; De Strooper, B.; Müller, U.; Shen, J.; et al. Regulation of Cholesterol and Sphingomyelin Metabolism by Amyloid-Beta and Presenilin. Nat. Cell Biol. 2005, 7, 1118–1123. [Google Scholar] [CrossRef]
- Beel, A.J.; Sakakura, M.; Barrett, P.J.; Sanders, C.R. Direct Binding of Cholesterol to the Amyloid Precursor Protein: An Important Interaction in Lipid–Alzheimer’s Disease Relationships? Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2010, 1801, 975–982. [Google Scholar] [CrossRef]
- Bocharov, E.V.; Mineev, K.S.; Pavlov, K.V.; Akimov, S.A.; Kuznetsov, A.S.; Efremov, R.G.; Arseniev, A.S. Helix-Helix Interactions in Membrane Domains of Bitopic Proteins: Specificity and Role of Lipid Environment. Biochim. Biophys. Acta Biomembr. 2017, 1859, 561–576. [Google Scholar] [CrossRef]
- Bocharov, E.V.; Gremer, L.; Urban, A.S.; Okhrimenko, I.S.; Volynsky, P.E.; Nadezhdin, K.D.; Bocharova, O.V.; Kornilov, D.A.; Zagryadskaya, Y.A.; Kamynina, A.V.; et al. All-d-Enantiomeric Peptide D3 Designed for Alzheimer’s Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-β Precursors. J. Med. Chem. 2021, 64, 16464–16479. [Google Scholar] [CrossRef]
- Vetrivel, K.S.; Meckler, X.; Chen, Y.; Nguyen, P.D.; Seidah, N.G.; Vassar, R.; Wong, P.C.; Fukata, M.; Kounnas, M.Z.; Thinakaran, G. Alzheimer Disease Abeta Production in the Absence of S-Palmitoylation-Dependent Targeting of BACE1 to Lipid Rafts. J. Biol. Chem. 2009, 284, 3793–3803. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Vetrivel, K.S.; Drisdel, R.C.; Meckler, X.; Gong, P.; Leem, J.Y.; Li, T.; Carter, M.; Chen, Y.; Nguyen, P.; et al. S-Palmitoylation of Gamma-Secretase Subunits Nicastrin and APH-1. J. Biol. Chem. 2009, 284, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Marchant, R.E.; Zagorski, M.G. ABri Peptide Associated with Familial British Dementia Forms Annular and Ring-like Protofibrillar Structures. Amyloid 2004, 11, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Canale, C.; Oropesa-Nuñez, R.; Diaspro, A.; Dante, S. Amyloid and Membrane Complexity: The Toxic Interplay Revealed by AFM. Semin. Cell Dev. Biol. 2018, 73, 82–94. [Google Scholar] [CrossRef]
- Kayed, R.; Pensalfini, A.; Margol, L.; Sokolov, Y.; Sarsoza, F.; Head, E.; Hall, J.; Glabe, C. Annular Protofibrils Are a Structurally and Functionally Distinct Type of Amyloid Oligomer *. J. Biol. Chem. 2009, 284, 4230–4237. [Google Scholar] [CrossRef]
- Zhu, M.; Han, S.; Zhou, F.; Carter, S.A.; Fink, A.L. Annular Oligomeric Amyloid Intermediates Observed by in Situ Atomic Force Microscopy *. J. Biol. Chem. 2004, 279, 24452–24459. [Google Scholar] [CrossRef]
- Song, Y.; Hustedt, E.J.; Brandon, S.; Sanders, C.R. Competition between Homodimerization and Cholesterol Binding to the C99 Domain of the Amyloid Precursor Protein. Biochemistry 2013, 52, 5051–5064. [Google Scholar] [CrossRef]
- Abad-Rodriguez, J.; Ledesma, M.D.; Craessaerts, K.; Perga, S.; Medina, M.; Delacourte, A.; Dingwall, C.; De Strooper, B.; Dotti, C.G. Neuronal Membrane Cholesterol Loss Enhances Amyloid Peptide Generation. J. Cell Biol. 2004, 167, 953–960. [Google Scholar] [CrossRef]

| APP Type | Type of Structure | Parameter | 10 mol. % Chol | 20 mol. % Chol | 33 mol. % Chol |
|---|---|---|---|---|---|
| WT | Clusters | quantity, units/μm2 | 2 ± 1 | 4 ± 1 | 2 ± 1 |
| average height, nm | 3 ± 2 | 4 ± 2 | 4 ± 2 | ||
| Concavities | quantity, units/μm2 | 12 ± 3 | 10 ± 3 | 14 ± 3 | |
| average depth, nm | 1.8 ± 0.4 | 2.0 ± 0.5 | 2.8 ± 0.5 | ||
| L723P | Clusters | quantity, units/μm2 | 15 ± 3 | 6 ± 2 | 7 ± 2 |
| average height, nm | 4 ± 2 | 3 ± 2 | 3 ± 2 | ||
| Concavities | quantity, units/μm2 | 4 ± 1 | 8 ± 1 | 15 ± 3 | |
| average depth, nm | 0.8 ± 0.2 | 0.8 ± 0.2 | 0.9 ± 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krasnobaev, V.D.; Bershatsky, Y.V.; Bocharova, O.V.; Bocharov, E.V.; Batishchev, O.V. Amyloid Precursor Protein Changes Arrangement in a Membrane and Its Structure Depending on the Cholesterol Content. Membranes 2023, 13, 706. https://doi.org/10.3390/membranes13080706
Krasnobaev VD, Bershatsky YV, Bocharova OV, Bocharov EV, Batishchev OV. Amyloid Precursor Protein Changes Arrangement in a Membrane and Its Structure Depending on the Cholesterol Content. Membranes. 2023; 13(8):706. https://doi.org/10.3390/membranes13080706
Chicago/Turabian StyleKrasnobaev, Vladimir D., Yaroslav V. Bershatsky, Olga V. Bocharova, Eduard V. Bocharov, and Oleg V. Batishchev. 2023. "Amyloid Precursor Protein Changes Arrangement in a Membrane and Its Structure Depending on the Cholesterol Content" Membranes 13, no. 8: 706. https://doi.org/10.3390/membranes13080706
APA StyleKrasnobaev, V. D., Bershatsky, Y. V., Bocharova, O. V., Bocharov, E. V., & Batishchev, O. V. (2023). Amyloid Precursor Protein Changes Arrangement in a Membrane and Its Structure Depending on the Cholesterol Content. Membranes, 13(8), 706. https://doi.org/10.3390/membranes13080706






