1. Introduction
Nitrate (NO
3−) is the main source of nitrogen, the important biogenic element for terrestrial plants [
1,
2]. Nitrate concentrations in soils vary widely and are often in the micromolar range, limiting plant growth [
3]. Both low-affinity (LATS) and high-affinity (HATS) nitrate transport systems operate in plants, participating in NO
3− uptake by roots, translocation to shoots and allocation of N among tissues [
1,
3,
4,
5,
6,
7,
8,
9,
10].
The
AtNPF6.3 (
CHL1/
NRT1.1) gene from
Arabidopsis thaliana was the first to be cloned among the genes of the NO
3− transporting proteins [
11]. The study of Tsay and coworkers [
11] initiated identification of multiple homologous proteins, now grouped into a large family of low-affinity nitrate transporters: the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY (NPF/NRT1/PTR). There are 53 members of this family in
A. thaliana [
10]. NPF proteins exhibit different substrate specificity in plants. They transport not only nitrate but also other substances, including nitrite, peptides, amino acids, glucosinolates, auxin, abscisic acid and gibberellins [
10,
12]. In spite of belonging to the family of low-affinity transporters, AtNPF6.3 is involved in NO
3− absorption by roots in both a low-affinity and a high-affinity mode, depending on the external nitrate concentration. AtNPF6.3 displays biphasic Michaelis–Menten saturation kinetics as a HATS (Km ≈ 50 µM) at external nitrate concentrations less than 0.2–0.5 mM and as a LATS (Km ≈ 4 mM) at higher nitrate concentrations [
7,
13]. Switching between low-affinity and high-affinity modes is regulated by the phosphorylation status of Thr101 residue at the N-terminus in AtNPF6.3 [
13,
14]. The investigation of AtNPF6.3 crystal structure gave rise to the hypothesis that the switching mechanism of the dual-affinity nitrate transporter is based on a coupling–decoupling of homo-dimers of this protein. AtNPF6.3 in an unphosphorylated, structurally coupled state (dimer), functions as a low-affinity transporter. Phosphorylation of Thr101 uncouples the dimers and shifts the protein to a high-affinity state [
15,
16]. Using the
Xenopus oocyte expression system, AtNPF6.3 was shown to be an electrogenic proton-coupled nitrate transporter, suggesting symport stoichiometry of H
+/NO
3− > 1 [
7,
17].
Apart from transport functions, AtNPF6.3 serves as a nitrate sensor involved in the control of nitrate assimilation and developmental processes, including the regulation of expression of nitrate-related genes, root system development and breaking seed dormancy [
10,
12,
18,
19,
20,
21,
22].
A. thaliana NPF6.3 homologs have been identified in different plant species including
Brassica napus (
BnNPF6.3/NRT1.1B/NRT1.2) [
23],
Oryza sativa (
OsNPF6.3–6.5/NRT1.1A-C) [
12,
24,
25],
Zea mays (
ZmNPF6.4/NRT1.1A, ZmNPF6.6/NRT1.1B, ZmNRT1.1C-D) [
24,
25,
26],
Medicago truncatula (MtNPF6.8/NRT1.3) [
27] and
Sorghum bicolor (
SbNPF6.5/NRT1.1B) [
28]. Transport functions of the AtNPF6.3 homologs have been investigated using the
Xenopus oocyte expression system. MtNPF6.8/NRT1.3, like AtNPF6.3, demonstrated dual-affinity kinetics with high (Km = 41.6 µM) and low (Km = 7.2 mM) nitrate affinity [
27]. Some homologs demonstrated functional divergence from AtNPF6.3. For example, the Km for NO
3− uptake by BnNPF6.3/NRT1.2 was in the low-affinity range and grew from 4 to 14 mM as the membrane voltage was changed from −40 mV to −180 mV [
23]. By contrast,
OsNPF6.4/NRT1.1A displayed only low-affinity nitrate kinetics with a Km of 9 mM [
29].
Salinization reduces nitrate availability to plants through competition between NO
3− and Cl
− for anionic transporters [
30,
31,
32,
33]; this could be crucial, especially for those plants growing in soils with low (micromolar) nitrate concentrations. However, few studies have investigated the role of AtNPF6.3 homologs in chloride transport. Using
Xenopus oocytes, voltage clamp,
36Cl
−, and
15N uptake techniques, Wen with coworkers [
26] investigated anion transport by two AtNPF6.3 homologs from
Z. mays, ZmNPF6.4/NRT1.1A and ZmNPF6.6/NRT1.1B. In the absence of NaCl, ZmNPF6.4 functioned as a low-affinity nitrate transporter with efflux activity, while ZmNPF6.6 demonstrated nitrate transporting activity with high-affinity kinetics (Km = 210 µM). However, under added Cl
−, both proteins transported Cl
−, ZmNPF6.4/NRT1.1A with high-affinity saturation kinetics (Km = 390 µM) and ZmNPF6.6/NRT1.1B with linear kinetics, in a wide range of external Cl
− concentrations. Higher uptake at acidic pH, compared to weakly alkaline pH, indicated that both transporters operated as Cl
−/H
+ symporters. Competition between NO
3− and Cl
− was shown for both transporters, with ZmNPF6.4 exhibiting Cl
− selectivity, while ZmNPF6.6 was selective for NO
3−. The authors attributed the differences in the transport functions of the two
Z. mays proteins, NPF6.4/NRT1.1A and NPF6.6/NRT1.1B, to different key polar residues which are involved in anion binding in the anion conducting tunnel [
15,
16]. The residue responsible for nitrate binding is presented by histidine (His 356) in ZmNPF6.6, while some other residue or domain, which remains to be identified, is involved in binding chloride in ZmNPF6.4 [
26].
In other experiments,
A. thaliana,
O. sativa and
Nicotiana benthamiana plants grown in a nutrient solution with NH
4+ as a nitrogen source were found to be hypersensitive to NaCl stress [
33]. For
AtNPF6.3 mutant plants, the authors showed that the salt hypersensitivity was a result of Cl
− overaccumulation due to AtNPF6.3 Cl
− transport activity, which was up-regulated by NH
4+; salt stress hypersensitivity was alleviated in the presence of competing NO
3− anion.
Under saline conditions, halophytes, plants inhabiting saline soils, are suggested to regulate the absorption of nutrients more efficiently than glycophytes, plants that are not salt-tolerant [
32,
34,
35,
36]. However, available information on anion-transporting proteins of halophytes is scarce. Gene cloning and elucidation of functional features of anion transporters from halophytes are important for unraveling the mechanisms of plant salinity tolerance and improving crop salt tolerance by genetic engineering (reviewed by [
37,
38,
39,
40]).
Here, we describe cloning
SaNPF6.3, a putative ortholog of
A. thaliana NPF6.3/
NRT1.1, from the euhalophyte
Suaeda altissima. The genus
Suaeda belongs to the Amaranthaceae (Chenopodiaceae), many members of which inhabit highly saline soils and are characterized by extreme salt tolerance [
41,
42].
S. altissima is one of the most salt tolerant plants and is able to perform its life cycle at NaCl concentrations up to 1M [
43]. The ability of SaNPF6.3 to transport nitrate was examined by functional complementation analysis in the mutant strain Δ
ynt1 of the yeast
Hansenula (
Ogatae)
polymorpha.
H. polymorpha is a suitable model organism to study nitrate assimilation pathways in plants since it is able to take up and metabolize nitrate as the only nitrogen source [
44,
45,
46,
47,
48]. Gene
YNT1 (yeast nitrate transporter 1) encodes the only high-affinity nitrate transporter in
H. polymorpha. In the mutant strain Δ
ynt1, the gene
YNT1 has been deleted. The nitrate transporting activity of SaNPF6.3 was also validated by checking the ability of Δ
ynt1 expressing
SaNPF6.3 to absorb NO
3− from nitrate-containing media. The relative
SaNPF6.3 transcript abundance in
S. altissima organs was measured for plants grown at various nitrate and chloride concentrations in nutrient solutions. The changes in
SaNPF6.3 expression which were induced by changing the nitrate availability and salinity in the medium were also studied.
2. Material and Methods
2.1. Plant Material
Seeds of
Suaeda altissima (L.) Pall. were collected from plants growing in the wild on the shores of the salt lake Elton, located in the Volgograd region in Russia. Seeds were stratified at +4 °C over 3 days and then germinated in wet sand. Seed germination and further plant growth were carried out in a growth chamber under controlled conditions at 24 °C and relative humidity of 60–70%. The plants were illuminated with high pressure sodium lamps DNaZ_400 (Reflux, Novocherkassk, Russia) at a light flux of 300 µmol photons m
−2 s
−1 and photoperiod of 16 h, with an 8 h dark period. Fourteen days after germination, the seedlings were transplanted to an aerated Robinson and Downton nutrient solution (NS) [
49] in 3-L opaque glass containers (5 plants per container) with low (0.5 mM) or high (15 mM) nitrate concentrations. Plants were grown hydroponically under the same environmental conditions until they were 45 days old, when they were used in most experiments. For total RNA extraction, organs of
S. altissima plants (roots, leaves, stems, flowers) were sampled (approximately 1 g fresh weight of each sample) and frozen in liquid nitrogen for further use.
To study the long-term salinity effect on the expression of SaNPF6.3 in Suaeda organs, NaCl was applied to the NS after 7 d. To avoid salt shock, NaCl was added gradually in increments of 50 or 100 mM per day, up to the final concentrations of 250 or 750 mM; no NaCl was added to the NS for control plants. To study the effect of salt shock on the expression of SaNPF6.3, NaCl was raised to 250 mM in a single addition. SaNPF6.3 transcript levels were determined after 31 d for both the long-term NaCl treatment and for the NaCl shock, when the plants had reached 45 d of age. In order to investigate the effects of changes in nitrite and ammonium ions on the expression of SaNPF6.3, KNO2 and (NH4)2SO4 were added to the NS at a final concentration of 5 mM after 31 d of plant growth in NS, with both a low (0.5 mM) or a high (15 mM) nitrate concentration. The effects of an increase in nitrate concentration in the NS from 0.5 mM up to 5 mM and the transfer of plants grown at 15 mM nitrate to the non-nitrate medium were also studied using plants of the same age. A comparative assessment of SaNPF6.3 expression levels in different organs of S. altissima was performed for 21- and 45-day-old plants (7 and 31 days of growth in hydroponics, respectively) in roots, stems and leaves and for 60-day-old plants (at 46 day of growth in hydroponics) in flowers.
To obtain xylem exudates, the shoots of 55-day-old S. altissima plants grown under standard conditions in the NS medium but supplemented with nitrate and chloride at various concentrations (the ion concentrations are indicated in the corresponding figure), were cut off 2 cm above the root collar. Silicone tubes were placed on the stem stumps, and the exudates were collected in the tubes for 48 h at 24 °C. In the experiments where the dependence of NO3− concentration in the xylem exudate on NO3− concentration in the medium was determined, the plants were grown in the presence of 100 mM NaCl in NS. Two days before the exudate collection, the plants were transferred to modified NS, in which 1 mM KH2PO4 and 4 mM Ca(NO3)2 were replaced by 1 mM NaH2PO4 and 4 mM CaCl2, respectively, while KNO3 was added in various concentrations. In order to investigate the dependence of Cl− concentration in the xylem exudate on Cl− concentration in the medium, NaCl was applied to the NS gradually after 7 days of plant growth in hydroponics without NaCl, as indicated above.
2.2. Yeast Strain and Vectors Used in the Study
Methylotrophic yeast (
Hansenula polymorpha) double auxotrophic strains DL-1 (
leu2 ura3 genotype) (wild-type strain, WT strain) and yeast integrative vectors pCCUR2 and pCHLX were used in this study. The strain DL-1 (
leu2 ura3) was transformed with plasmids pCCUR2 and pCHLX carrying the URA and LEU genes, respectively, to ensure the growth of the yeast strains without additional nitrogen sources, leucine and uracil, when performing complementation tests. Plasmids pCCUR2 and pCHLX were kindly provided by Dr. Michael Agafonov (Federal Research Center “Fundamentals of Biotechnology”, Russian Academy of Sciences, Moscow, Russia). Yeast cells were transformed by the lithium method [
50] or by electroporation [
51] using an Eppendorf device (Eppendorf, Framingham, MA, USA).
2.3. Extraction of Total RNA from Plant Material and the First-Strand cDNA Synthesis
Total RNA from
S.altissima plant organs was isolated by the hot phenolic method [
52] and used as a template for the total first-strand cDNA synthesis. For amplification of the 3′- and 5′- ends of the
SaNPF6.3 transcript by the Step-Out RACE method, the first-strand cDNA was synthesized on the total RNA template, isolated from
Suaeda roots, using MINT revertase (Evrogen, Moscow, Russia). Full-length cDNA of
SaNPF6.3 gene was also amplified on the total RNA template, isolated from
Suaeda roots. To obtain full-length cDNA of
SaNPF6.3 and quantify the representation of the gene transcripts in
S. altissima organs, first-strand cDNA synthesis was performed on total RNA templates using (dT)15 primer and MMLV revertase (Evrogen, Moscow, Russia).
2.5. Identification of the Full Length SaNPF6.3 Coding Sequence
Partial coding sequence (the middle fragment) of the
SaNPF6.3 gene was obtained by us previously (GenBank ID: MK580125.1) [
53]. Here, based on this sequence, the forward and reverse primer sets were designed for amplification of the 5′- and 3′-end sequences of
SaNPF6.3 cDNA. The 5′- and 3′-end sequences of
SaNPF6.3 cDNA were determined by the Step-Out RACE technology (kit #SKS03, Evrogen, Moscow, Russia). The cDNA fragments were amplified on the total cDNA template using Encyclo DNA polymerase (#PK002, Evrogen, Moscow, Russia). The 5′-sequence of
SaNPF6.3 cDNA (1152 bp) was amplified with primers SaNPF6.3_r (round 1) and SaNPF6.3_5′RACE_R1 (round 2). The 3′-sequence of
SaNPF6.3 cDNA (722 bp) was amplified with SaNPF6.3_f (round 1) and SaNPF6.3_F1 (round 2). All amplicons obtained, including also the middle cDNA fragment amplified from the cDNA template with a primer pair SaNPF6.3_f and SaNPF6.3_r, were cloned into pAL2-T vector (Evrogen, Moscow, Russia) for replication in
E. coli cells and the following sequencing. Subsequently, the partial sequences (middle fragment, 5′- and 3′- ends of
SaNPF6.3 cDNA) were assembled in silico using SnapGene software 5.0.8 (
https://www.snapgene.com/snapgene-viewer (accessed on 29 July 2023)). The resulting coding sequence of 1788 bp contained open reading frame (ORF) for the protein of 596 aa. This sequence was used for the design of primers for the amplification of full-length
SaNPF6.3 cDNA on the total first-strand cDNA template.
2.6. Cloning of the Full-Length SaNPF6.3 cDNA in the Yeast Vector pCHLX
The full-length coding sequence of
SaNPF6.3 was amplified from the total first-strand cDNA using primer pairs SaNPF6.3_Gib_F and SaNPF6.3_Gib_R. The full-length
SaNPF6.3 cDNA was cloned into the yeast integrative vector pCHLX [
54] under the control of the inducible nitrate reductase promoter
pYNR1 and terminator
tYNR1 of
H. polymorpha. Promoter
pYNR1 and terminator
tYNR1 sequences were amplified from the
H. polymorpha genomic DNA template using primer pairs pYNR1_F and pYNR1_R, and tYNR1_F and tYNR1_R. The first 10 cycles of amplification of the promoter, terminator and gene coding sequences were performed using Encyclo polymerase (No. PK002, Evrogen, Moscow, Russia); the next 25 cycles were performed using CloneAmp HiFi PCR Premix kit (No. 639298, Clontech, Mountain View, CA, USA). The pCHLX vector was linearized in the Hind III and EcoRI restriction sites and ligated with the synthesized
pYNR1,
tYNR1 and
SaNPF6.3 sequences using a Gibson assembly kit (No. E5510, SkyGen, NEB, Ipswich, MA, USA) to produce the pCHLX-
pYNR1-
SaNPF6.3-
tYNR1 construct (further denoted as pCHLX-
SaNPF6.3). The cloned sequences were verified by sequencing. The
SaNPF6.3 cDNA sequence (1788 bp) was deposited in GenBank (GenBank ID: OQ330855).
The pCHLX-
GFP-SaNPF6.3 construct was also obtained for the expression of SaNPF6.3 fused at the N-terminus with green fluorescent protein (GFP) in yeast cells. To achieve this, three fragments were ligated in one reaction: (1) GFP coding sequence amplified from pTR vector [
55] with pYNR1_GFP_F and TEV_GFP_R primers (2)
SaNPF6.3 coding sequence amplified from the first cDNA strand with TEV_SaNPF6.3_F and SaNPF6.3_Gib_R primers; (3) plasmid pCHLX linearized by inverted PCR from the pCHLX-
SaNPF6.3 construct with the pYNR1_R3 and tYNR1_F primer pair. Ligation was carried out using Gibson assembly kit (NEB, Ipswich, MA, USA).
2.7. Cloning of AtNPF6.3 Coding Sequence in the Yeast Vector pCHLX
To control the functional complementation of the
H. polymorpha mutant Δ
ynt1, the mutant was transformed by the construct carrying the known nitrate transporter gene
AtNPF6.3 (GenBank ID: NP_563899.1) from
Arabidopsis. The coding sequence of
AtNPF6.3 was amplified from the
Arabidopsis total first-strand cDNA, using primer pairs
AtNPF6.3_Gib_F and
AtNPF6.3_Gib_R. The total first-strand cDNA was synthesized on the total RNA template, isolated from roots of adult
Arabidopsis plants. Like
SaNPF6.3 cDNA, full-length
AtNPF6.3 coding sequence was cloned into the vector pCHLX [
54] under the control of the inducible nitrate reductase promoter
pYNR1 and terminator
tYNR1 of
H. polymorpha. For ligation with
AtNPF6.3, nitrate reductase promoter and terminator sequences were amplified from the
H. polymorpha genomic DNA template using primer pairs pYNR1_F and pYNR1_R2, and tYNR1_F2 and tYNR1_R. All stages of the amplification and ligation of the sequences were similar to how it was carried out during the amplification and ligation of the sequences in the case of
SaNPF6.3 cloning. The construct pCHLX-
pYNR1-AtNPF6.3-tYNR1 (pCHLX-
AtNPF6.3) was obtained for the transformation of mutant Δ
ynt1 cells.
2.8. Implementation of Amino Acid Substitutions in the Amino Acid Sequence of the SaNPF6.3 Protein
To obtain point amino acid substitutions of Tyr358His, Thr106Asp and Thr106Ala in the SaNPF6.3 protein sequence, the pCHLX-pYNR1-SaNPF6.3-tYNR1 construct was linearized by inverse PCR with primer pairs SaNPF6.3_1074_F and SaNPF6.3_Tyr358His_R, SaNPF6.3_318_F and SaNPF6.3_Thr106Asp_R, and SaNPF6.3_318_F and SaNPF6.3_Thr106Ala_R, respectively, using a ready-to-use CloneAmp HiFi PCR Premix kit (TakaraBio, San Jose, CA, USA). The resulting linear forms of PCR products with corresponding nucleotide substitutions were converted into circular forms. For this purpose, the 5′-ends of the linearized plasmid DNA were phosphorylated using T4-polynucleotide kinase. The reaction mixture for the phosphorylation, containing T4 DNA-ligase buffer, 250 ng linearized DNA and 10 U T4-polynucleotid kinase (SibEnzyme, Novosibirsk, Russia), was incubated for 15 min at 37 °C. The phosphorylated product was ligated by adding 2.5 U T4 ligase (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA) to the reaction mixture and incubating the mixture for 16–18 h at room temperature. The ligase was inactivated by incubation of the mixture at 65 °C for 15 min. Plasmid pCHLX-SaNPF6.3, which served as a template for amplification, was removed from the mixture by treatment with methyl-dependent DNA endonuclease Mal I (SibEnzyme, Novosibirsk, Russia). The mixture was purified from proteins by treatment with phenol-chloroform, and plasmid DNA was resuspended in mQH2O for subsequent transformation of competent E. coli cells.
2.9. Production of a Deletion Mutant Δynt1 of H. polymorpha
A mutant with a deleted
YNT1 gene (GenBank ID: CP080316.1),
ynt1::BleoR(ZeoR), was derived from the wild-type strain DL-1 (
leu2,
ura3) of
H. polymorpha through homologous recombination by an “one-step gene disruption” method [
56]. To carry out the knockout of the gene
YNT1, the zeocin resistance gene (
ZeoR) was incorporated into the
YNT1 locus through homologous recombination at the
YNT1 gene sites [
44]. For this purpose, PCR fragments corresponding to “left” and “right” flanking parts of the
YNT1 gene and the zeocin resistance gene
ZeoR amplified from the pVR2 vector [
57] were cloned into the bacterial vector pBlueScript KS(II)+ using Gibson assembly kit (NEB, Ipswich, MA, USA). “Left” and “Right” fragments of the
YNT1 gene and the gene
ZeoR were amplified using primer pairs ZeoYNTRfl_F and pBlueYNTRfl_R, pBlueYNTLfl_F and YNTLflZeo_R, and ZeoCas1 and ZeoCas2, respectively. From the resulting construct, pBlueScript KS(II)+
YNT1_L-ZeoR- YNT1_R, a fragment of
YNT1_L-ZeoR- YNT1_R, was excised by Hind III and Pst I restriction endonucleases (SibEnzym, Novosibirsk, Russia) and transferred into DL1 cells. The presence of the insert in the zeocin-resistant yeast clones was validated by PCR screening using the primer pair pBlueYNTLfl_F and pBlueYNTRfl_R. Growth of the selected colonies was tested on the agarized media containing nitrate and nitrite (in the concentration range of 0.2–5 mM) to exclude the occurrence of other spontaneous mutations in the genome which may occur when yeast is transformed by the lithium method.
The mutant strain Δynt1 (ynt1: BleoR(ZeoR), leu2, ura3) was additionally transformed with pCCUR2 and pCHLX integrative plasmids carrying the URA and LEU genes, respectively, to ensure the growth of the yeast strains without additional nitrogen sources in the selective media, namely, leucine and uracil, when performing complementation tests.
2.10. Cultivation of H. polymorpha and Transformants of Δynt1 Strain
Cells of
H. polymotpha WT strain, mutant strain Δ
ynt1, and transformants of the Δ
ynt1 strain were grown on a rich YPD medium (1% yeast extract, 2% peptone, 2% glucose) or a minimal synthetic SD medium (0.67% yeast nitrogen base without amino acids, 2% glucose) with (NH
4)
2SO
4 addition (medium SD + AS) or KNO
3 addition (medium SD + KNO
3) as nitrogen sources. The KNO
3 concentrations ranged from 0.2 to 5 mM. The media contained or did not include auxotrophic additions (0.02 mg/mg leucine, 0.04 mg/mL uracil). The yeast growth occurred at 37 °C for 2–3 days. All manipulations with
H. polymorpha were performed according to the protocols generally accepted for the yeast [
51]. Yeast transformants were selected on minimal selective media in the absence of leucine and/or uracil. Transformants that contained the insertion in the genome were validated by PCR with DL-1_Chr2_HpURA3_F or Hp_DL-1_Chr1_R primers for genomic DNA and standard M13_R or M13_F primers for pCCUR2 and pCHLX vectors.
2.11. Study of nitrate uptake by H. polymorpha cells expressing SaNPF6.3 and AtNPF6.3 Genes
Cells of H. polymorpha knockout mutant strains Δynt1 expressing SaNPF6.3 and AtNPF6.3, as well as WT cells, were grown to approximately 20 mg fresh weight/mL in a minimal SD medium (2% dextrose and 0.67% YNB containing sulphate ammonium) at 37 °C for 2 days. To induce the expression of nitrate transporter genes, cells were precipitated, washed with water, and cultured for 24 h (37 °C) in a minimal SD–AS medium (2% dextrose, 0.67% YNB without ammonium sulphate) supplemented with 20 mM NaNO3 as the only nitrogen source. After induction of the nitrate transporter genes, yeast cells were repeatedly washed with water and resuspended in a minimal SD–AS medium (2% dextrose; 0.176% YNB without sulphate ammonium) supplemented with 0.5 mM or 2 mM NaNO3. The yeast cultures were left to grow in these media for 18 h (37 °C); the absorption of nitrate by the cells was determined by measuring nitrate concentration decline in incubation media using an Elite-021 NO3− selective electrode (Niko-Analit, Moscow, Russia).
2.12. Immunoblotting
For SDS-PAAG protein electrophoresis and the following immunoblot analysis,
H. polymorpha WT cells and cells transformed with pCHLX-
GFP-SaNPF6.3 were grown in 10 mL of SD + AS medium (0.67% yeast nitrogen base without amino acids, 2% glucose; ammonium sulphate as a nitrogen source was a component of the yeast nitrogen base) for 24 hr. Then, the yeast cells were transferred to SD + KNO
3 medium (0.17% yeast nitrogen base without amino acids and ammonium sulphate, 2% glucose + 10 mM KNO
3 as a sole nitrogen source). After cultivation for 24 h in the NO
3−-containing medium, leading to the induction of nitrate transporter genes, the cells were precipitated and lysed in 300 µL of A buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1% SDS, 1 mM PMSF) with 300 µL glass beads using Vortex. The lysate was centrifuged (13,000×
g, 10 min, +4 °C) and the proteins from the supernatant were analyzed by SDS-PAAG electrophoresis and immunoblotting. The protein concentration in the supernatant was determined by the bicinchoninic acid method [
58]. Electrophoresis was performed in 8% separating gel, loading 50 µg of protein per lane. The semi-dry transfer of the proteins from PAAG matrix to the nitrocellulose membrane (NC) (pore diameter of 0.45 µm; Schleicher & Schuell, Dassel, Germany) was carried out at 100 mA for 1 h using transfer device (Helicon, Moscow, Russia). For immunodetection of the recombinant protein GFP-SaNPF6.3, polyclonal anti-GFP antibodies (Evrogen, Moscow, Russia) were used as primary antibodies and horseradish peroxidase-conjugated (HRP) antibodies (Imtek Ltd., Moscow, Russia) as secondary ones. After treatment with a blocking solution (5% Carnation non-fat dry milk (Nestle, Vevey, Switzerland) in TBS with 0.1% Tween-20), the membrane was incubated with the primary antibodies (overnight at +4 °C) and then with the secondary antibodies (2 h). Primary antibodies were used at a 1:2000 dilution in TBS–Tw–BSA solution (1×TBS, 0.2% Tween 20, 1% BSA, 0.01% NaN
3), and secondary antibodies were used at a 1:10,000 dilution in the TBS–Tw–DM solution (1×TBS, 0.2% Tween 20, 5% Carnation non-fat dry milk). After incubation with the antibodies, the membrane was washed with TBS–Tw solution (1×TBS, 0.1% Tween 20) and immunoreactive bands were visualized using an ECL kit (GE Healthcare, Piscataway, NJ, USA). A ChemiDoc XRS+ System gel documentation system (BioRad, Hercules, CA, USA) was used for visualization.
2.13. Determination of SaNPF6.3 Localization in Yeast Cells
The images of yeast cells transformed with pCHLX-GFP-SaNPF6.3 were obtained with a laser scanning confocal microscope LSM-710-NLO (Carl Zeiss, Jena, Germany) equipped with a 63x oil immersion objective Plan-Apochromat, with numerical aperture of 1.4 and ZEN 2010 software 1.1.2.0 (Carl Zeiss, Jena, Germany). For analysis, the suspension of yeast cells was transferred to sterilized Petri dishes with a thin glass bottom. The fluorescence signals were detected in confocal channel mode with a confocal diaphragm of 46 μm in diameter, image size 1024 × 1024 pixels (132 nm/pixel), and a scanning rate of 1.27 μs/pixel (1.33 s/image). The GFP fluorescence was excited at λex = 488 nm and visualized in the 490–555 nm range. The transmitted laser light was recorded with a separate T-PMT detector.
2.14. Quantitative Analysis of SaNPF6.3 Transcripts in S. altissima Organs
Quantitative analysis of SaNPF6.3 transcripts was performed by qRT-PCR using a LightCycler® 96 System (Roche Diagnostics Corporation, Indianapolis, IN, USA). The cDNA templates for the amplification of SaNPF6.3 fragments were synthesized on total RNAs templates, isolated from different organs of S. altissima plants grown in the NS with various nitrate and NaCl concentrations or the plants subjected to NaCl shock. A ready-to-use reaction mixture with intercalating dye SYBR Green I (Evrogen, Moscow, Russia) was used. The S. altissima gene of elongation factor 1 alpha SaeEF1alpha (GenBank ID: MN076325.1) and protein phosphatase gene SaPP2A (GenBank ID:OP752355) were used as the reference genes. To amplify the SaeEF1alpha and SaPP2A fragments, SaeEF1alfa_F1 and SaeEF1alfa_R1, and SaPP2A_F1 and SaPP2A_R1 primer pairs, respectively, were used. The results are based on three replicates. The results obtained were processed by the LightCycler 96SW 1.1 software. Similar results were obtained with SaeEF1alpha and SaPP2A, hence, the data are presented only for the former gene. For the amplification of SaNPF6.3 fragment, the primer pair SaNPF6.3_F1 and SaNPF6.3_R1 was used.
2.15. Determination of NO3− and Cl− Concentrations in Xylem Exudates
Xylem exudates were mineralized at 400 °C, the solid residues obtained were diluted with mQ water and the nitrate concentrations in the solutions were determined using an Elite-021 NO3−-selective electrode (Niko-Analit, Moscow, Russia). Cl− was assayed by titration with Hg2+ using a Top Buret H digital burette (Eppendorf, Wesseling-Berzdorf, Germany).
2.16. Bioinformatic Analysis of Amino Acid Sequences
Multiple sequence alignment of amino acid sequences was performed by MAFFT software, version 7 (
https://www.ebi.ac.uk/Tools/msa/mafft/, accessed on 4 July 2023) and visualized by Jalview software, version 2.11.2.7 (
https://www.jalview.org/ (accessed on 4 July 2023)). A phylogenetic analysis of plant NPF family proteins was carried out by Molecular Evolutionary Genetic Analysis (MEGA) 11 software (version 11,
https://www.megasoftware.net/, accessed on 4 July 2023), using the maximum likelihood method based on the Jones–Taylor–Thornton model [
59] (1000 bootstrap replications were performed). Protein topology was predicted by DeepTMHMM software (version 1.0.24,
https://dtu.biolib.com/DeepTMHMM, accessed on 26 September 2023). The 3D protein structure was predicted by SWISS-MODEL (
https://swissmodel.expasy.org/interactive, accessed on 26 September 2023).
2.17. Statistics
Data processing (mean, standard errors) and the production of graphs were performed using Sigma Plot software (version 14.0). Statistical analysis of the data was made by one-way analysis of variance (ANOVA). Statistical calculations were carried out in the Microsoft Excel program (version 2019). Standard errors are given. Different letters indicate significant difference (p-value < 0.05).
4. Discussion
From the euhalophyte
Suaeda altissima, we cloned the coding sequence of the
SaNPF6.3 gene, homologous to the
AtNPF6.3/
AtNRT1.1 encoding a dual-affinity nitrate transporter/transceptor in the model plant
A. thaliana. The protein SaNPF6.3 belongs to the large NPF/NRT1/PTR family of transporters, including proteins involved in the transport of nitrate, nitrite, peptides, amino acids, glucosinolates, auxin, ABA and gibberellins [
10,
13,
65]. Phylogenetic analysis showed evolutionary relations of SaNPF6.3 to NPF6.3 family proteins (
Figure 1) and displayed a larger similarity of SaNPF6.3 to NPF6.3 members from other families of dicotyledonous plants rather than families of monocotyledons. A SaNPF6.3 topological model matched those of the NPF6.3 proteins from other plants, in particular AtNPF6.3 (
Figure 2b). In silico analysis based on SaNPF6.3 multiple sequence alignment with homologous proteins from other plants (
Figure 2) revealed that SaNPF6.3 has features inherent in the NRT1/NPF/PTR family of anion transporters.
(1) A conserved threonine residue, Thr106, in the motif RxxT (
Figure 2), a putative phosphorylation site, is found at the N-terminus of SaNPF6.3 in TMH3 (
Figure 1c). Similar threonine residues are present at equivalent positions in other NPF6.3 transporters. In AtNPF6.3, an equivalent Thr101 was shown to be responsible for switching between a high- and a low-affinity AtNPF6.3 mode [
14,
60]. The switching was accomplished through alterations in the Thr101 phosphorylation status which was dependent on nitrate availability in the growth medium. At low nitrate concentrations (<1 mM), Thr101 is phosphorylated by CBL-INTERACTING PROTEIN KINASE 23 (CIPK23) and AtNPF6.3 operates as a high-affinity transporter. In the range of millimolar nitrate concentrations, Thr101 is dephosphorylated and AtNRT1.1 operates as a low-affinity nitrate transporter [
14,
60]. However, the anion-transporting activities of
Zea mays proteins, ZmNPF6.6 and ZmNPF6.4, unlike AtNPF6.3, appear unlikely to be regulated by the phosphorylation status of equivalent threonine residues [
26]. In ZmNPF6.6, the point mutations Thr104Ala disrupting phosphorylation and Thr104Asp mimicking a phosphorylation event retained the high-affinity nitrate transport activity, although with a lower Km for nitrate. Thr106Ala and Thr106Asp substitutions in chloride-transporting ZmNPF6.4 eliminated the saturable curve in chloride uptake, with influx activity becoming linear as external chloride concentrations increased [
26]. The authors suggested that the transport activity of ZmNPF6.6 and ZmNPF6.4 changed in a phosphorylation-independent but threonine-dependent manner. We made similar substitutions in the
S. altissima protein: Thr106Ala and Thr106Asp. Growth of the mutant yeast strain Δ
ynt1 transformed with the modified and unmodified versions of
SaNPF6.3 on the minimal SR medium containing nitrate did not differ markedly (data not presented), testifying against the role of a Thr106 phosphorylation status in switching SaNPF6.3 affinity to nitrate.
(2) In TMH7 of SaNPF6.3, there is tyrosine residue Tyr358 (
Figure 1c and
Figure 2), the equivalent position of which is occupied by His in a number of NPF6.3 homologues including AtNPF6.3 [
11], ZmNPF6.6 [
26] and several others [
16]. The mutation of His356Ala abolished both high- and low-affinity nitrate transporter activities in AtNPF6.3, suggesting that His356 is required for nitrate binding [
15,
16]. SaNPF6.3 is not the only NPF6.3 transporter containing Tyr at the equivalent positions (
Figure 2,
Table S3). Tyr was also found in dual-affinity nitrate transporters MtNPF6.8 (Tyr350) [
27] and OsNPF6.3 (Tyr366) [
25], as well as in the high-affinity chloride and low-affinity nitrate transporter ZmNPF6.4 (Tyr370) [
26]. In a comparative study of anion transport by ZmNPF6.6 containing His362 and ZmNPF6.4 containing Tyr370, the authors found that in the absence of chloride, ZmNPF6.6 operates as a pH-dependent non-biphasic high-affinity nitrate-specific transporter, while ZmNPF6.4 acts as a low-affinity nitrate transporter. In the presence of chloride, ZmNPF6.6 switched to low-affinity, while ZmNPF6.4 to a high-affinity chloride transporter [
26]. The authors hypothesized that ZmNPF6.4
in planta is likely to be a component of the root chloride uptake system. SaNPF6.3 bearing Tyr358 should presumably share more features with ZmNPF6.4 than with ZmNPF6.6 and function more as a transporter of chloride than of nitrate. However, SaNPF6.3 transported nitrate in a test with functional complementation of the yeast mutant Δ
ynt1, lacking the only nitrate transporter (
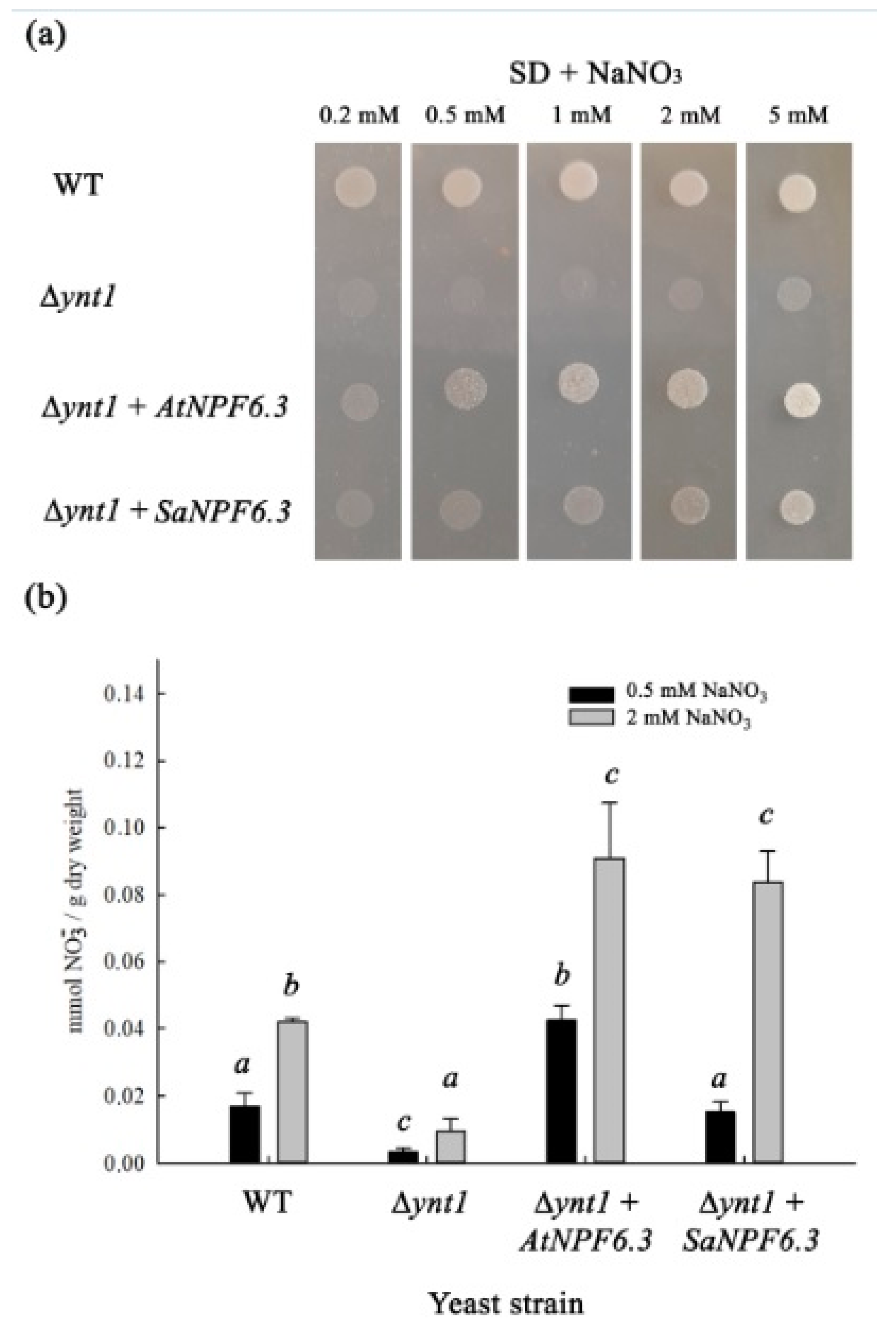
Figure 3a,b).
(3) Analogs of the conserved residues (Lys165 and Glu476), forming ionic bonds to maintain the AtNPF6.3 in a functional state [
15,
16], were found in SaNPF6.3 (Lys168 and Glu482) (
Figure 2).
(4) In TMH1 of SaNPF6.3, there is an ExxER motif (EACER) conserved among NPF6.3 family members (
Figure 2), an analog of which (EAVER) in AtNPF6.3 is involved in proton binding and the coupling of anion and proton transport [
15,
16].
(5) The presence of conserved proline residue between TMH10 and TMH11 was found to be a common property of all examined NPF6.3 homologs including AtNPF6.3 (Pro492) (
Figure 2) [
15,
16]. This proline residue is essential for signaling functions of AtNPF6.3 [
18]. We found an analog of this residue in SaNPF6.3 (Pro498) (
Figure 2).
SaNPF6.3 is likely an ortholog of AtNPF6.3/NRT1.1, a dual-affinity nitrate transporter/transceptor from
A. thaliana. The latter assumption is based on several points.
(I) The
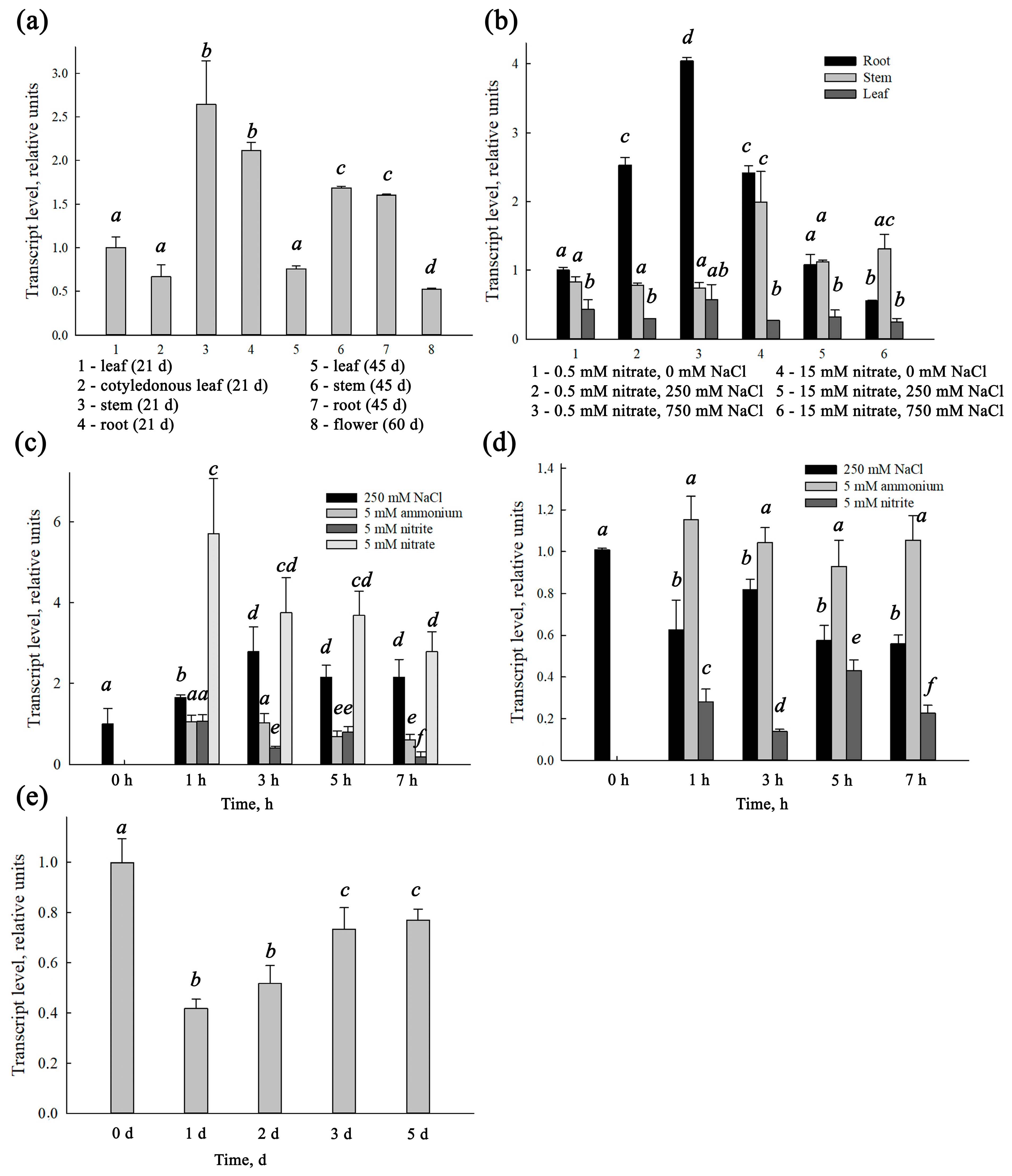
SaNPF6.3 expression occurred under a variety of environmental conditions similar to the expression of the
AtNPF6.3, the gene encoding dual-affinity nitrate transporter AtNPF6.3. The expression of
SaNPF6.3 was observed under external nitrate concentrations corresponding to both high- and low-affinity modes of the protein operation, as well as in the presence of the reduced nitrogen compounds, ammonium and nitrite ions, at salinization of the medium and without it (
Figure 5). The gene was transcribed in the root, stem, leaf and flower; the transcription occurred at different ontogenetic stages (
Figure 5). Expression levels were higher in the roots and stems than in the other organs, indicating a particularly important role of SaNPF6.3 in nitrate uptake and transfer.
(II) The recovery of
H. polymorpha mutant strain Δ
ynt1 growth on the minimal SD medium, in the presence of nitrate, when
SaNPF6.3 was expressed in the yeast cells, demonstrated clearly the nitrate transport properties of the
S. altissima protein (
Figure 3a). It should be noted that the nitrate concentrations applied, from 0.2 to 5 mM NO
3−, covered both high- and low-affinity ranges. The lack of full growth recovery of the Δ
ynt1 mutant transformed with
SaNPF6.3 may be due to a limited expression of this gene in the heterologous system used. Also, it is possible that only a small fraction of the synthesized SaNPF6.3 protein is delivered to the plasma membrane in the yeast cells, or a rapid degradation of the protein occurred. In line with this assumption, laser confocal microscopy demonstrated that in the mutant yeast cells expressing
GFP-SaNPF6.3, the recombinant protein was localized in the cytoplasm and vacuole (
Figure 4b).
(III) Nitrate uptake by the mutant Δ
ynt1 cells expressing
SaNPF6.3 demonstrated directly a nitrate-transporting function of SaNPF6.3 (
Figure 4b). The yeast transformants took up nitrate from the media containing this anion in concentrations of both 0.5 mM and 2.0 mM.
(IV) Like
AtNPF6.3 [
11,
66], the induction of
SaNPF6.3 expression in response to an increase in nitrate concentration in the NS, when plants are grown at a low nitrate availability (
Figure 5c), and the suppression of
SaNPF6.3 expression following the plants being transferred from sufficient nitrate supply to nitrate starvation (
Figure 5e) support also the participation of SaNPF6.3 in nitrate uptake. The decreased expression of
SaNPF6.3 in response to a nitrite addition at low nitrate availability (
Figure 4c) may be the result of an increased fraction of reduced nitrogen compounds in the cells during nitrate assimilation, which inhibits
SaNPF6.3 expression by the feedback.
It is possible that SaNPF6.3 mediates a low-affinity transport of chloride ions in addition to the dual-affinity transport of nitrate. The involvement of SaNPF6.3 in a low-affinity chloride transport is supported by increasing expression of the transporter gene in the response to NaCl addition to the NS. The stimulation of
SaNPF6.3 expression by NaCl in the presence of 0.5 mM nitrate observed at both the long-term salinity and under the salt shock conditions suggests the involvement of the transporter in the transport of chloride.
S. altissima, like other salt-accumulating halophytes, absorbs chloride in large quantities as a counter-ion for Na
+ passively entering the root cells. Further root-to-shoot translocation of Cl
− and Na
+, which play a role of ‘cheap’ osmotic compounds, contributes to maintaining the water potential gradient in the soil-root-shoot system [
43,
67,
68]. Another possible reason for the stimulation of
SaNPF6.3 expression in response to increasing NaCl concentration in the NS at low nitrate availability is a greater requirement for nitrate uptake under conditions of its competition with the chloride ion.
The decline in the expression of the transporter with increasing external NaCl concentration in the high external nitrate concentration (15 mM) may be a consequence of excessive amounts of the anions in the medium, both to meet the nitrogen requirements of plants and to maintain osmotic pressure in the cells. With the external NaCl concentration increasing, often accompanied by PM depolarization [
69], the transition point from active to passive chloride uptake can be attained [
70], decreasing the requirements of root cells in this transporter.
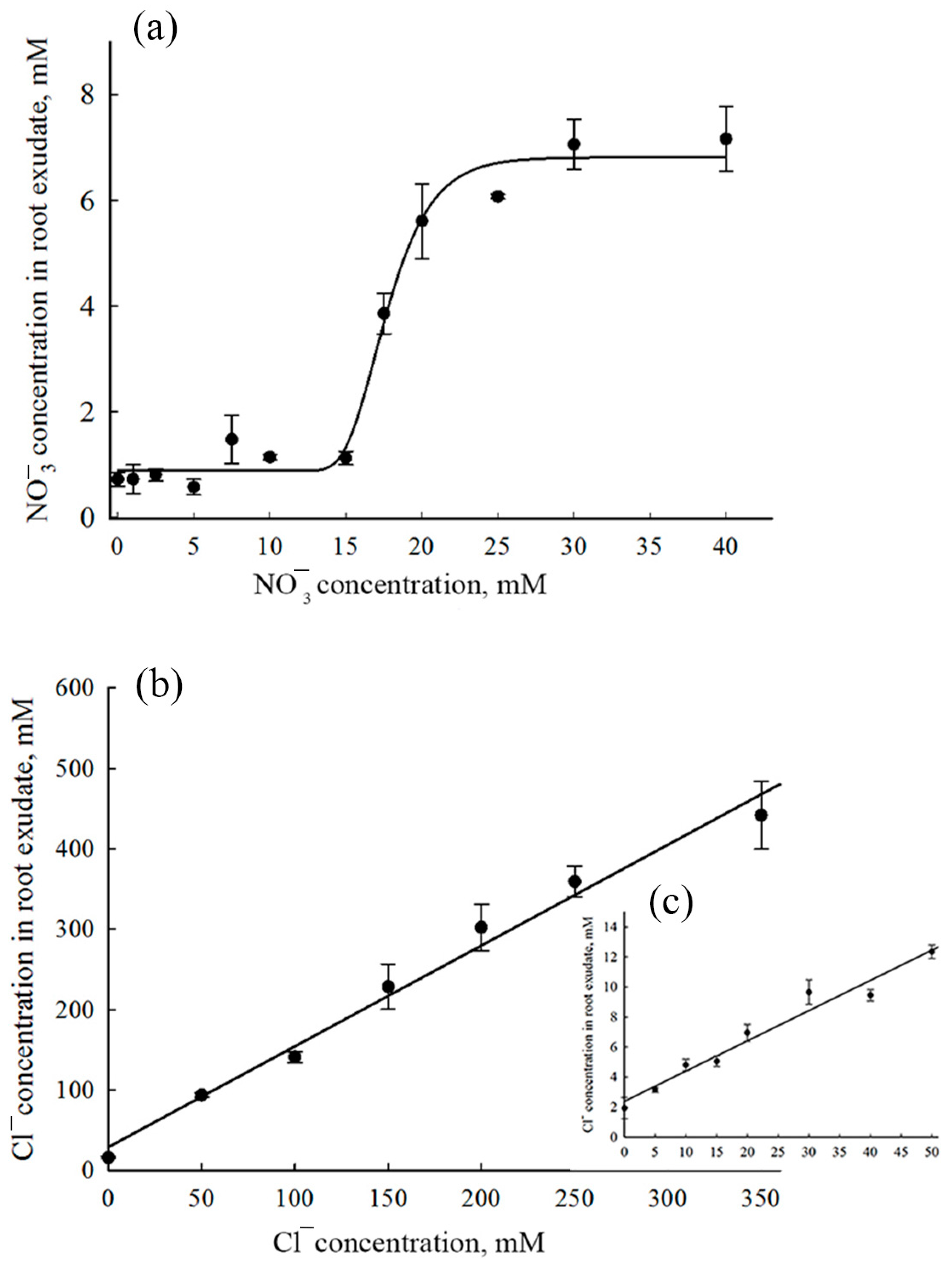
By collecting xylem exudate, we also examined the ability of
S. altissima roots to deliver nitrate and chloride to the above-ground organs (
Figure 6). The anion transfer to the shoots is based on the functions of the proteins responsible for absorption and root-to-shoot translocation of nitrate and chloride. The detached intact roots immersed in the nutrient solution represent a complex model system unable to demonstrate the function of individual transport proteins, such as SaNPF6.3. However, this system could be useful in ascertaining the physiological role of the protein operating in the whole organ, together with other proteins. As the ionic composition of the
S. altissima root exudate showed (
Figure 6), roots were able to ensure the delivery of both nitrate and chloride to the shoots. Chloride is required for the euhalophyte in a large quantity for the maintaining of cell turgor and the water potential gradient in the system of the whole plant [
43]. The nitrate transporting activity of SaNPF6.3 suggests the SaNPF6.3 involvement in nitrate absorption by
S. altissima roots. Two discrete levels observed for the NO
3− concentration in the exudate (
Figure 6a) may be attributed to high- and low-affinity modes of SaNPF6.3. However, another possibility is that another protein belonging to the NPF/NRT1 or NRT2 family is involved in nitrate absorption along with SaNPF6.3, or two different slow anion channels are activated for xylem loading as nitrate concentration increases. The ability to transfer chloride, along with nitrate, was demonstrated for two NPF homologues from
Z. mays, Zm-NPF6.6 and Zm-NPF6.4 [
26]. Given that in each of the dicotyledons studied, unlike in monocotyledons, only one AtNPF6.3 homolog has been found [
24], one would expect SaNPF6.3 to have properties that provide an ability for high-affinity nitrate transport in combination with low-affinity chloride transport.