Biomechanical Role of Epsin in Influenza A Virus Entry
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Reagents
2.2. Generation of Influenza A Virus Strains with Different Morphologies
2.3. Fluorescence Labeling of IAV
2.4. Inhibiting Clathrin-Mediated Endocytosis
2.5. TIRF Microscopy
2.6. D Structured Illumination Microscopy
2.7. Live-Cell Lattice Light Sheet Microscopy
2.8. Colocalization Analysis for Fixed Cells
2.9. IAV Particle Tracking for Lattice Light Sheet Microscopy
2.10. Flow Cytometry
2.11. Infectious Virus Measurement by Plaque Assay
2.12. Statistics and Reproducibility
3. Results
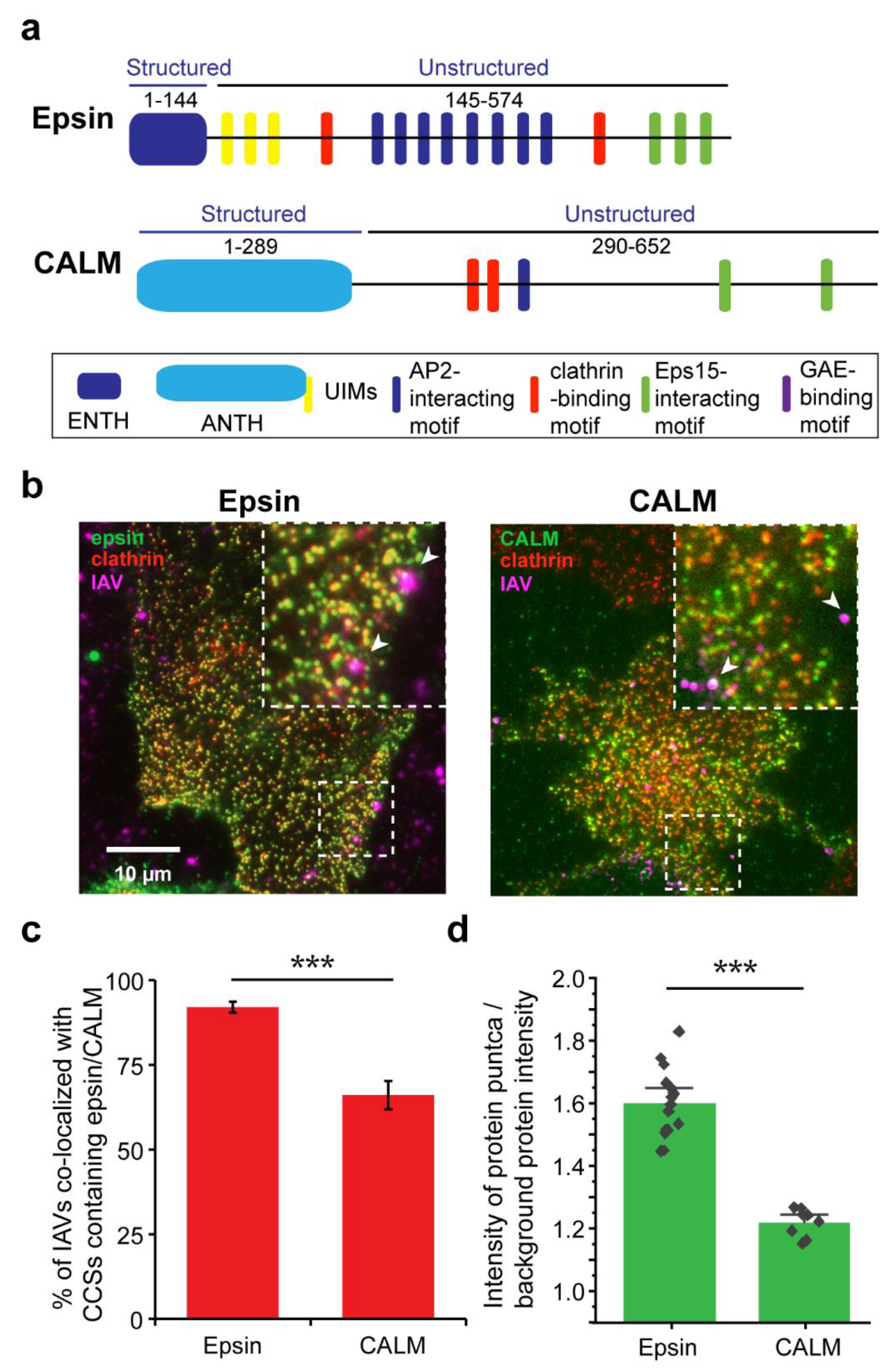
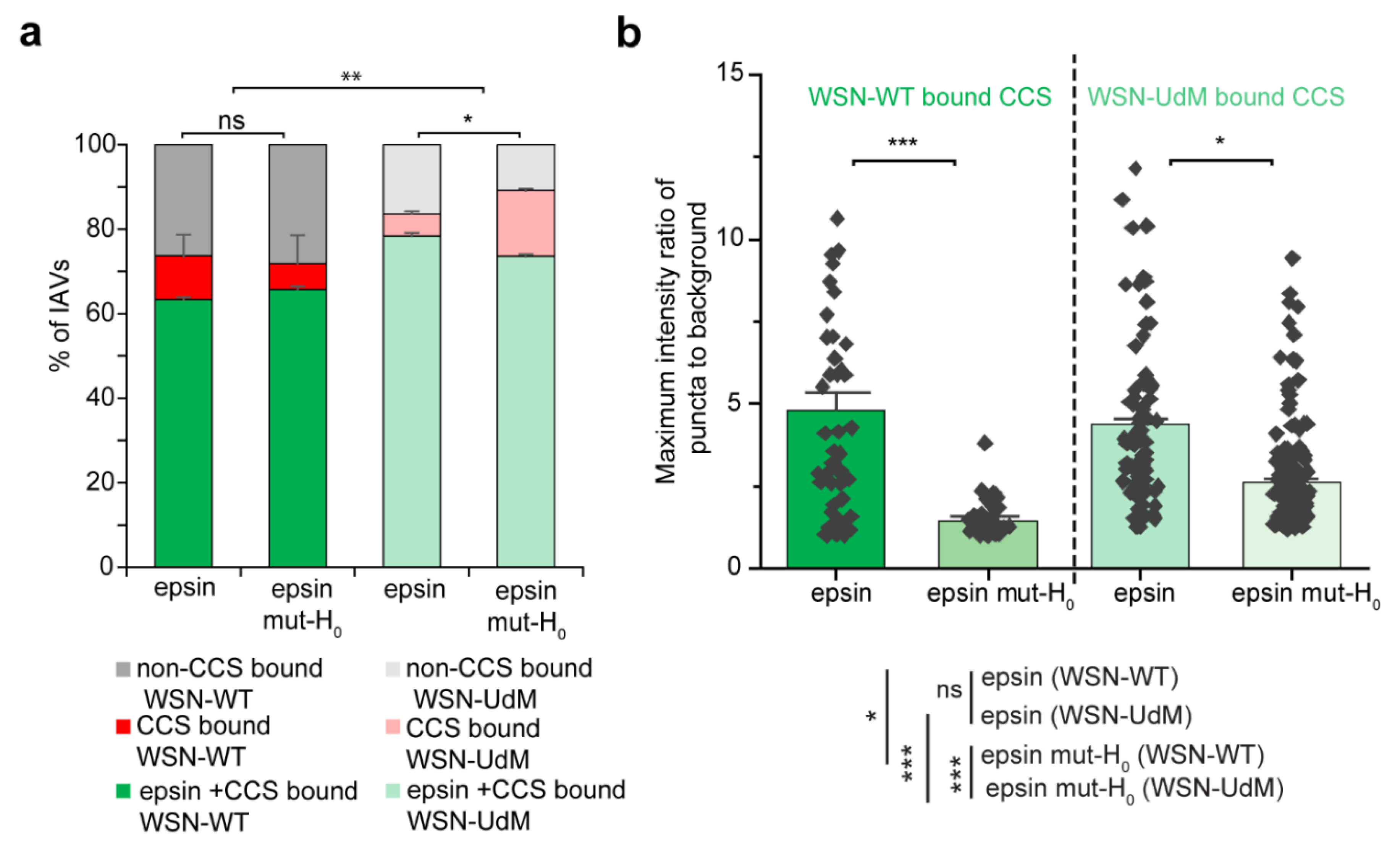
3.1. IAVs Co-Localize with Clathrin-Coated Structures (CCSs) Containing ENTH/ANTH Proteins
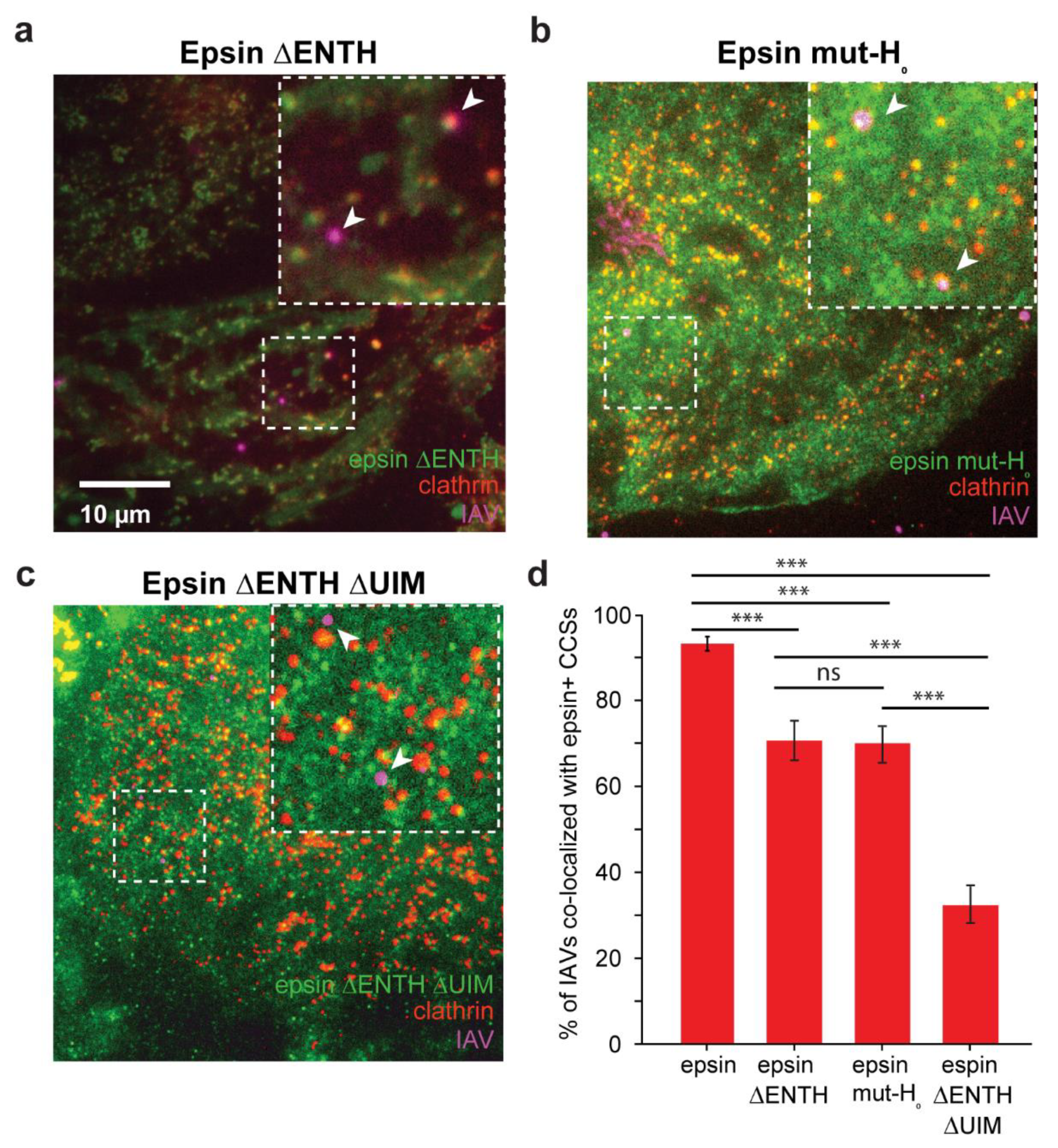
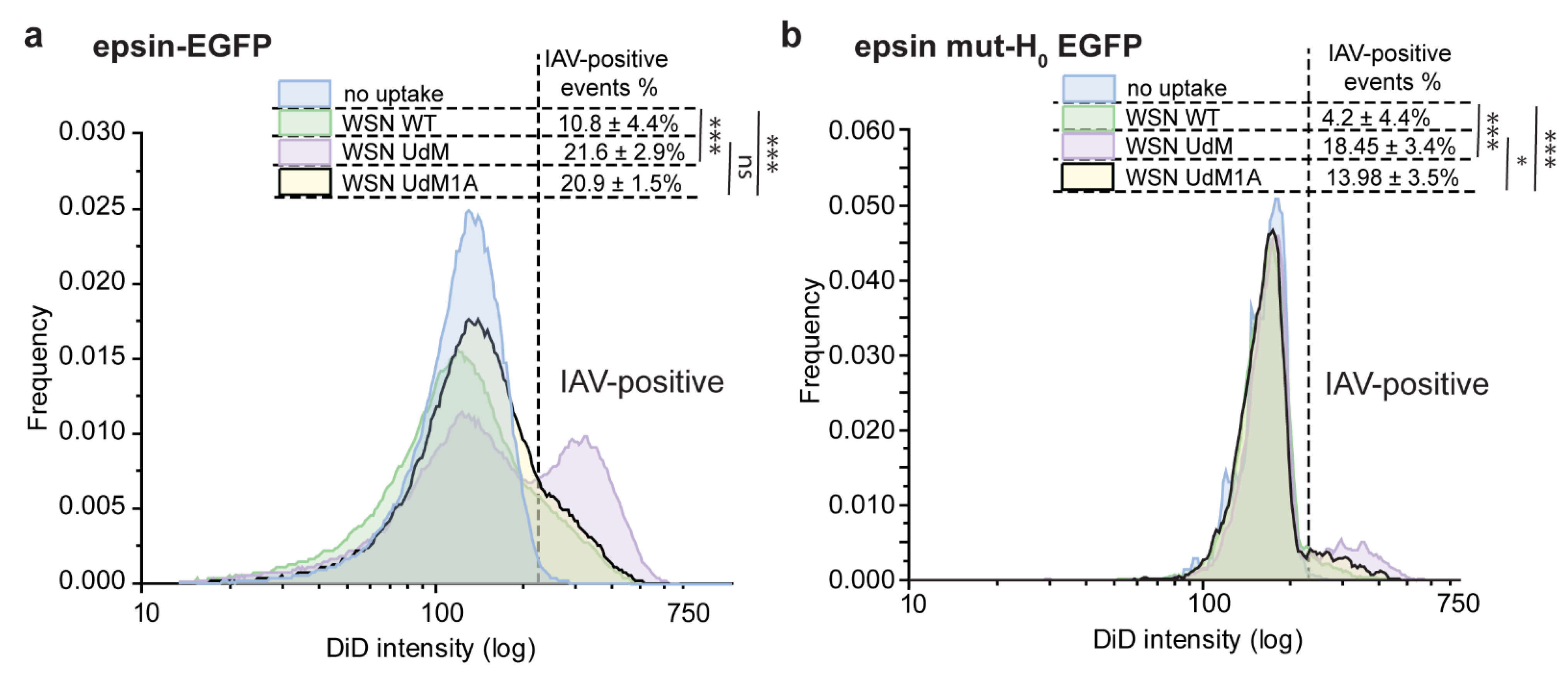
3.2. Colocalization of IAVs to CCSs and Internalization Are Disrupted by Overexpression of Epsin Mutants without Functional ENTH Domain
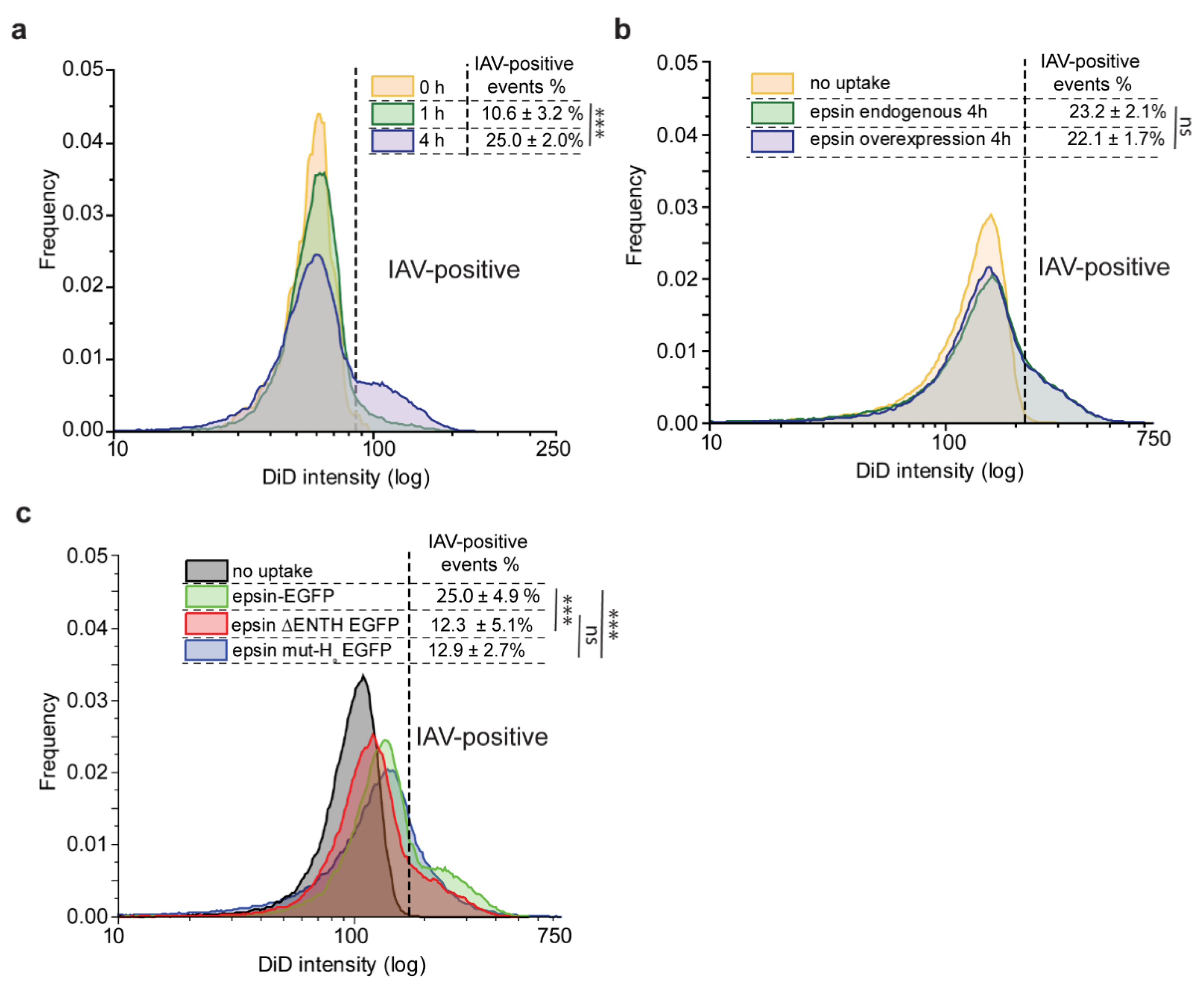
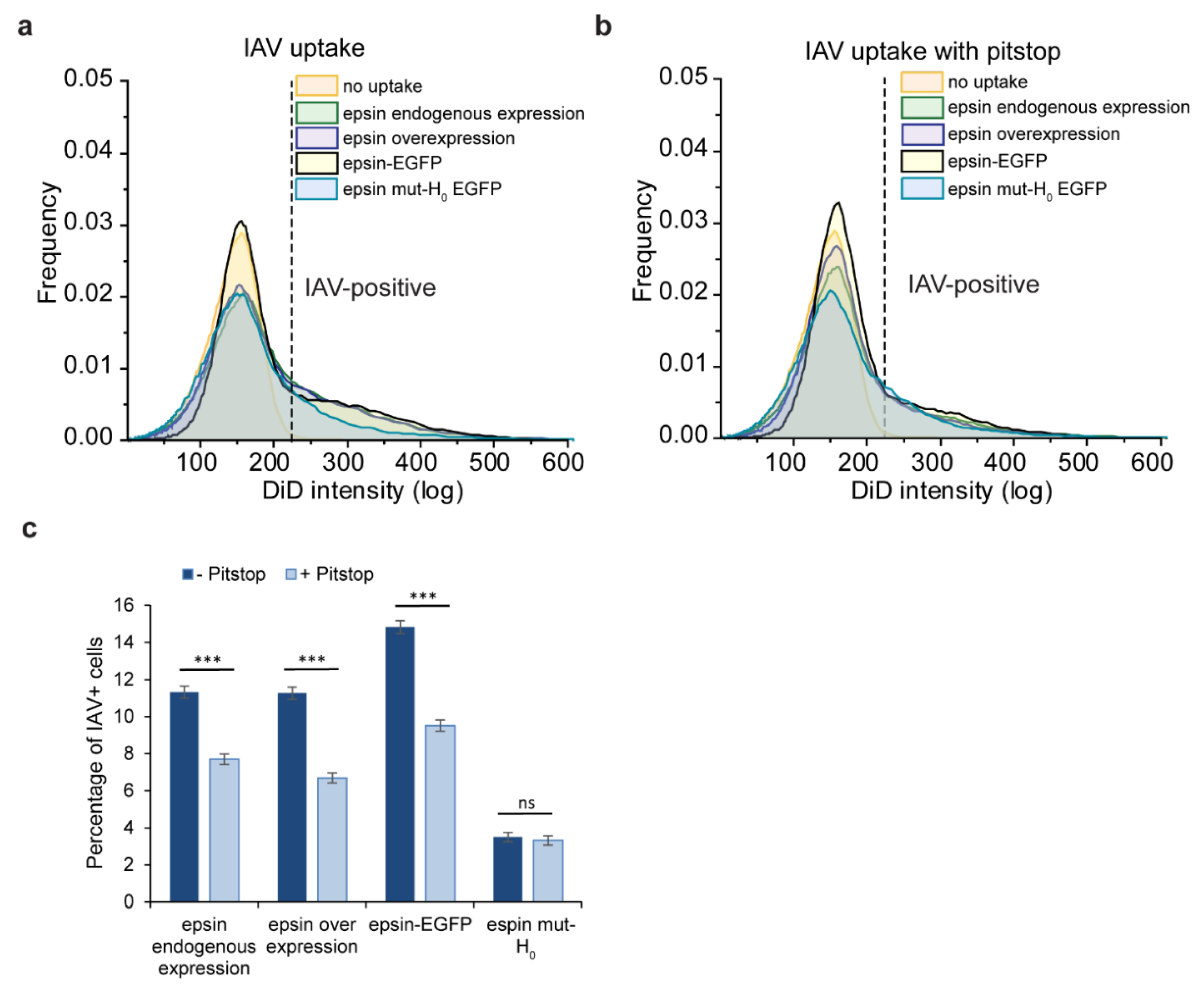
3.3. Bulk Uptake of IAVs Is Disrupted by the Overexpression of Epsin Derivatives Lacking Functional ENTH Domain in RPE Cells
3.4. Bulk Uptake of IAVs Is Disrupted by the Disruption of CME
3.5. Visualization of Cellular Entry of Filament-Forming IAVs Using Lattice Light Sheet Microscopy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doherty, G.J.; McMahon, H.T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P.; Helenius, A. Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 2014, 6, a016972. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endo-cytosis. J. Biol. Chem. J. Biol. Chem. 2021, 296, 100306. [Google Scholar]
- Piccini, L.E.; Castilla, V.; Damonte, E.B. Dengue-3 Virus Entry into Vero Cells: Role of Clathrin-Mediated Endocytosis in the Outcome of Infection. PLoS ONE 2015, 10, e0140824. [Google Scholar] [CrossRef] [PubMed]
- van der Schaar, H.M.; Rust, M.J.; Chen, C.; van der Ende-Metselaar, H.; Wilschut, J.; Zhuang, X.; Smit, J.M. Dissecting the Cell Entry Pathway of Dengue Virus by Single-Particle Tracking in Living Cells. PLOS Pathog. 2008, 4, e1000244. [Google Scholar] [CrossRef]
- Hu, T.; Frieman, M.; Wolfram, J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotechnol. 2020, 15, 247–249. [Google Scholar] [CrossRef]
- Daecke, J.; Fackler, O.T.; Dittmar, M.T.; Kraäusslich, H.-G. Involvement of Clathrin-Mediated Endocytosis in Human Immu-nodeficiency Virus Type 1 Entry. J. Virol. 2005, 79, 1581–1594. [Google Scholar] [CrossRef]
- Wiegand, T.; Fratini, M.; Frey, F.; Yserentant, K.; Liu, Y.; Weber, E.; Galior, K.; Ohmes, J.; Braun, F.; Herten, D.-P.; et al. Forces during cellular uptake of viruses and nanoparticles at the ventral side. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Stachowiak, J.C.; Brodsky, F.; Miller, E. A cost–benefit analysis of the physical mechanisms of membrane curvature. Nat. Cell Biol. 2013, 15, 1019–1027. [Google Scholar] [CrossRef]
- Sun, S.X.; Wirtz, D. Mechanics of Enveloped Virus Entry into Host Cells. Biophys. J. 2006, 90, L10–L12. [Google Scholar] [CrossRef]
- Joseph, J.G.; Liu, A.P. Mechanical Regulation of Endocytosis: New Insights and Recent Advances. Adv. Biosyst. 2020, 4, 1900278. [Google Scholar] [CrossRef]
- Gefen, A. Effects of Virus Size and Cell Stiffness on Forces, Work, and Pressures Driving Membrane Invagination in a Recep-tor-Mediated Endocytosis. J. Biomech. Eng. 2010, 132, 084501. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, Y.; Yuan, H.; Gao, H.; Zhang, S. Role of Nanoparticle Geometry in Endocytosis: Laying Down to Stand Up. Nano Lett. 2013, 13, 4546–4550. [Google Scholar] [CrossRef]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nano-particle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef]
- Dudek, A.M.; Zabaleta, N.; Zinn, E.; Pillay, S.; Zengel, J.; Porter, C.; Franceschini, J.S.; Estelien, R.; Carette, J.E.; Zhou, G.L.; et al. GPR108 Is a Highly Conserved AAV Entry Factor. Mol. Ther. 2019, 28, 367–381. [Google Scholar] [CrossRef]
- Büning, H. AAV Entry: Filling in the Blanks. Mol. Ther. 2020, 28, 346–347. [Google Scholar] [CrossRef]
- Barro, R.; Ursúa, J.; Weng, J. The Coronavirus and the Great Influenza Pandemic: Lessons from the “Spanish Flu” for the Coronavirus’s Potential Effects on Mortality and Economic Activity. Natl. Bur. Econ. Res. 2020, 26866. [Google Scholar] [CrossRef]
- Monto, A.S.; Fukuda, K. Lessons from Influenza Pandemics of the Last 100 Years. Clin. Infect. Dis. 2019, 5, 951–957. [Google Scholar] [CrossRef]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Move-ment. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef]
- Böttcher-Friebertshäuser, E.; Garten, W.; Matrosovich, M.; Klenk, H.D. The Hemagglutinin: A Determinant of Pathogenicity. Influenza Pathogenesis and Control—Volume I. In Current Topics in Microbiology and Immunology; Compans, R., Oldstone, M., Eds.; Springer: Cham, Switzerland, 2014; Volume 385, pp. 3–34. [Google Scholar] [CrossRef]
- Chen, C.; Zhuang, X. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc. Natl. Acad. Sci. USA 2008, 105, 11790–11795. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.G.; Osorio, C.; Yee, V.; Agrawal, A.; Liu, A.P. Complimentary action of structured and unstructured domains of epsin supports clathrin-mediated endocytosis at high tension. Commun. Biol. 2020, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ford, M.G.J.; Mills, I.G.; Peter, B.J.; Vallis, Y.; Praefcke, G.; Evans, P.R.; McMahon, H.T. Curvature of clathrin-coated pits driven by epsin. Nature 2002, 419, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.E.; Mathiasen, S.; Bright, N.A.; Pierre, F.; Kelly, B.T.; Kladt, N.; Schauss, A.; Merrifield, C.J.; Stamou, D.; Höning, S.; et al. Regulates Clathrin-Coated Vesicle Size and Maturation by Directly Sensing and Driving Membrane Curvature. Dev. Cell 2015, 33, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Dadonaite, B.; Vijayakrishnan, S.; Fodor, E.; Bhella, D.; Hutchinson, E.C. Filamentous influenza viruses. J. Gen. Virol. 2016, 97, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Leser, G.P.; Lamb, R.A. Filamentous Influenza Virus Enters Cells via Macropinocytosis. J. Virol. 2012, 86, 10950–10960. [Google Scholar] [CrossRef]
- De Vries, E.; Tscherne, D.M.; Wienholts, M.J.; Cobos-Jiménez, V.; Scholte, F.; García-Sastre, A.; Rottier, P.J.M.; De Haan, C.A.M. Dissection of the Influenza A Virus Endocytic Routes Reveals Macropinocytosis as an Alternative Entry Pathway. PLOS Pathog. 2011, 7, e1001329. [Google Scholar] [CrossRef]
- Neumann, G.; Watanabe, T.; Ito, H.; Watanabe, S.; Goto, H.; Gao, P.; Hughes, M.; Perez, D.R.; Donis, R.; Hoffmann, E.; et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 1999, 96, 9345–9350. [Google Scholar] [CrossRef]
- Bourmakina, S.V.; Garcia-Sastre, A. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 2003, 84, 517–527. [Google Scholar] [CrossRef]
- Bedi, S.; Noda, T.; Kawaoka, Y.; Ono, A. A Defect in Influenza A Virus Particle Assembly Specific to Primary Human Mac-rophages. mBio 2018, 9, e01916-18. [Google Scholar] [CrossRef]
- Aguet, F.; Antonescu, C.N.; Mettlen, M.; Schmid, S.L.; Danuser, G. Advances in Analysis of Low Signal-to-Noise Images Link Dynamin and AP2 to the Functions of an Endocytic Checkpoint. Dev. Cell 2013, 26, 279–291. [Google Scholar] [CrossRef]
- Tinevez, J.-Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Rust, M.J.; Zhuang, X. Endocytosis of influenza viruses. Microbes Infect. 2004, 6, 929–936. [Google Scholar] [CrossRef]
- Rust, M.J.; Lakadamyali, M.; Zhang, F.; Zhuang, X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 2004, 11, 567–573. [Google Scholar] [CrossRef]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Protein Amphipathic Helix Insertion: A Mechanism to Induce Membrane Fission. Front. Cell Dev. Biol. 2019, 7, 291. [Google Scholar] [CrossRef]
- Legendre-Guillemin, V.; Wasiak, S.; Hussain, N.K.; Angers, A.; McPherson, P.S. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 2004, 117, 9–18. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Decuzzi, P.; Ferrari, M. The Receptor-Mediated Endocytosis of Nonspherical Particles. Biophys. J. 2008, 94, 3790–3797. [Google Scholar] [CrossRef][Green Version]
- Willy, N.M.; Colombo, F.; Huber, S.; Smith, A.C.; Norton, E.G.; Kural, C.; Cocucci, E. CALM supports clathrin-coated vesicle completion upon membrane tension increase. Proc. Natl. Acad. Sci. USA 2021, 118, e2010438118. [Google Scholar] [CrossRef]
- Shaw, M.L.; Palese, P. Fields Virology, 6th ed.; LWW: Philadelphia, PA, USA, 2013. [Google Scholar]
- Herlo, R.; Lund, V.; Lycas, M.D.; Jansen, A.M.; Khelashvili, G.; Andersen, R.C.; Bhatia, V.; Pedersen, T.S.; Albornoz, P.B.; Johner, N.; et al. An Amphipathic Helix Directs Cellular Membrane Curvature Sensing and Function of the BAR Domain Protein PICK1. Cell Rep. 2018, 23, 2056–2069. [Google Scholar] [CrossRef]
- Zhang, S.; Gao, H.; Bao, G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, Z.; Deans, E.E.; Mittler, E.; Liu, M.; Chandran, K.; Ivanovic, T. The shape of pleomorphic virions determines resistance to cell-entry pressure. Nat. Microbiol. 2021, 6, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Cureton, D.K.; Massol, R.H.; Saffarian, S.; Kirchhausen, T.L.; Whelan, S.P.J. Vesicular Stomatitis Virus Enters Cells through Vesicles Incompletely Coated with Clathrin That Depend upon Actin for Internalization. PLOS Pathog. 2009, 5, e1000394. [Google Scholar] [CrossRef]
- Cureton, D.K.; Massol, R.H.; Whelan, S.P.J.; Kirchhausen, T. The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis. PLOS Pathog. 2010, 6, e1001127. [Google Scholar] [CrossRef] [PubMed]
- Boulant, S.; Kural, C.; Zeeh, J.-C.; Ubelmann, F.; Kirchhausen, T. Actin dynamics counteract membrane tension during clath-rin-mediated endocytosis. Nature 2011, 13, 1124–1131. [Google Scholar] [CrossRef]
- Veiga, E.; Cossart, P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat. Cell Biol. 2005, 7, 894–900. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joseph, J.G.; Mudgal, R.; Lin, S.-S.; Ono, A.; Liu, A.P. Biomechanical Role of Epsin in Influenza A Virus Entry. Membranes 2022, 12, 859. https://doi.org/10.3390/membranes12090859
Joseph JG, Mudgal R, Lin S-S, Ono A, Liu AP. Biomechanical Role of Epsin in Influenza A Virus Entry. Membranes. 2022; 12(9):859. https://doi.org/10.3390/membranes12090859
Chicago/Turabian StyleJoseph, Jophin G., Rajat Mudgal, Shan-Shan Lin, Akira Ono, and Allen P. Liu. 2022. "Biomechanical Role of Epsin in Influenza A Virus Entry" Membranes 12, no. 9: 859. https://doi.org/10.3390/membranes12090859
APA StyleJoseph, J. G., Mudgal, R., Lin, S.-S., Ono, A., & Liu, A. P. (2022). Biomechanical Role of Epsin in Influenza A Virus Entry. Membranes, 12(9), 859. https://doi.org/10.3390/membranes12090859







