Ceramide Phosphoethanolamine as a Possible Marker of Periodontal Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacteria
2.3. Proteins
2.4. Lipid Extraction from Bacteria
2.5. Lipidomic Analysis of a Total Extract of Lipids from Porphyromonas Gingivalis and Tannerella Forsythia
2.5.1. Nuclear Magnetic Resonance Analysis
2.5.2. HPLC-Electrospray Ionization-Mass Spectrometry Analysis
2.6. Clinical Periodontal Examination
2.7. Dental Plaque and Saliva Sampling
2.8. Total Lipid Extraction from Clinical Supragingival Plaque and Saliva Samples
2.9. Thin-Layer Chromatography
2.10. Dot Blot
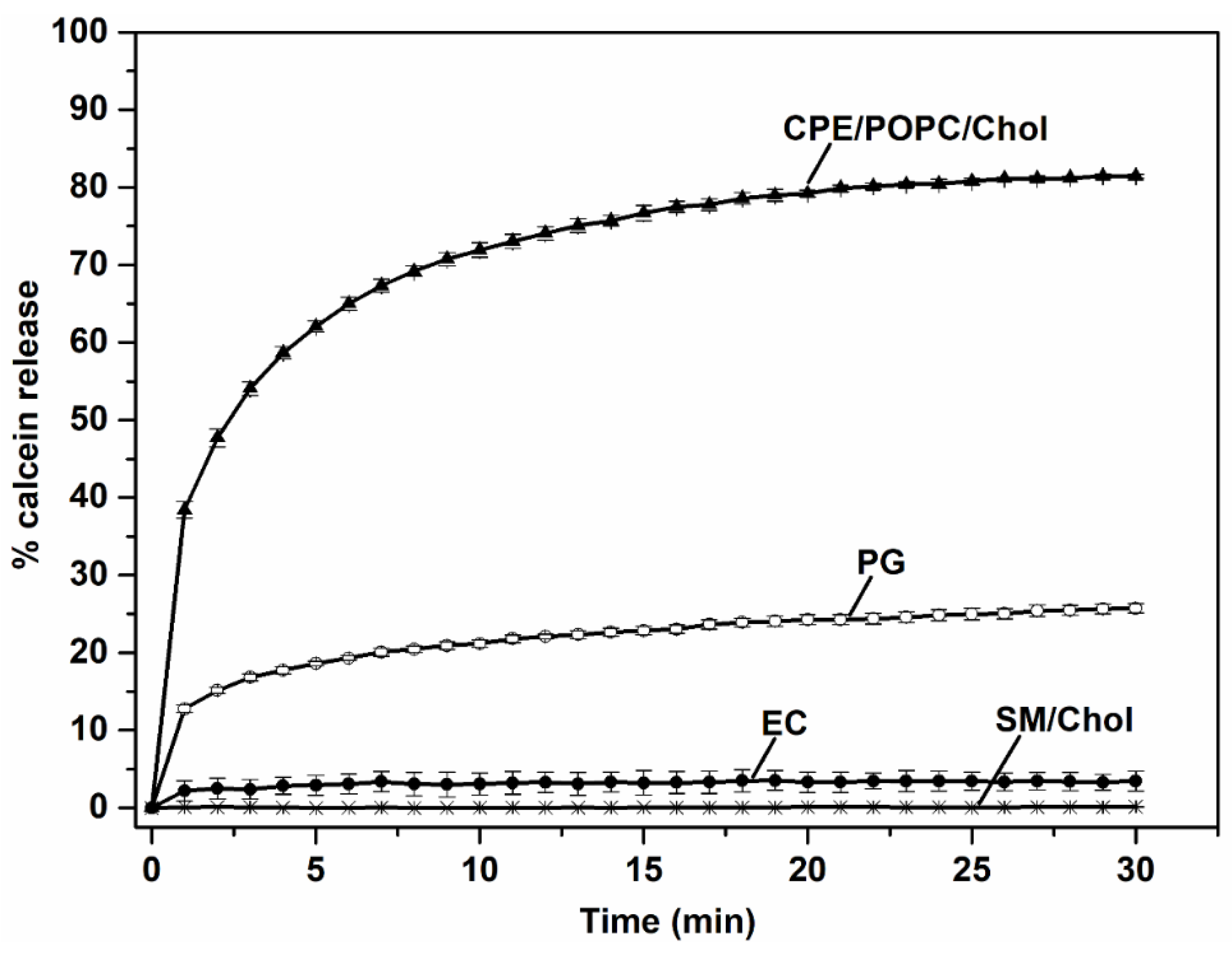
2.11. Permeabilization of Small Unilamellar Vesicles
2.12. Statistical Analysis
3. Results and Discussion
3.1. Lipidomic Analysis of the Total Extract of Lipids from Porphyromonas Gingivalis and Tannerella Forsythia Show the Presence of CPE Species
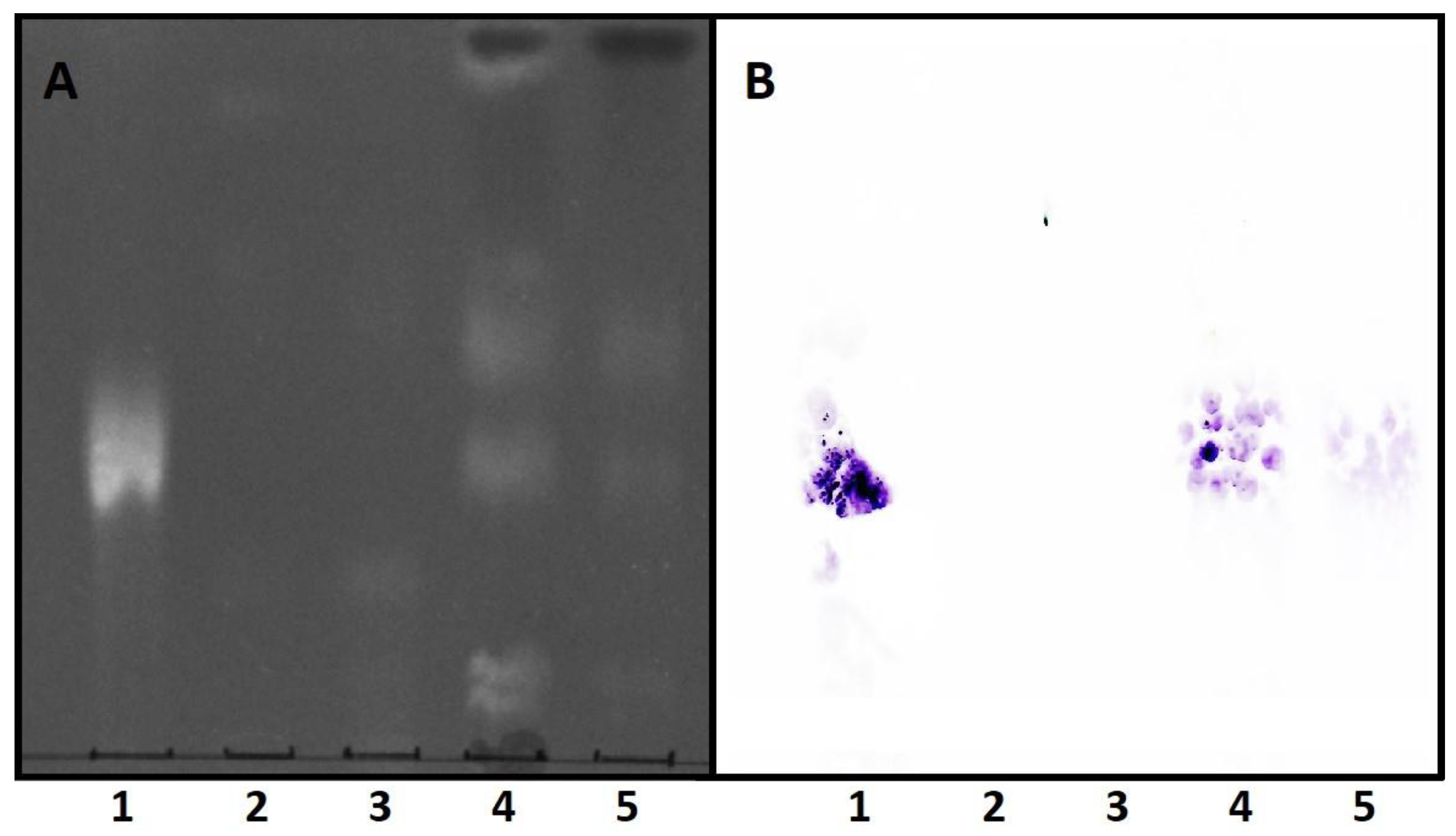
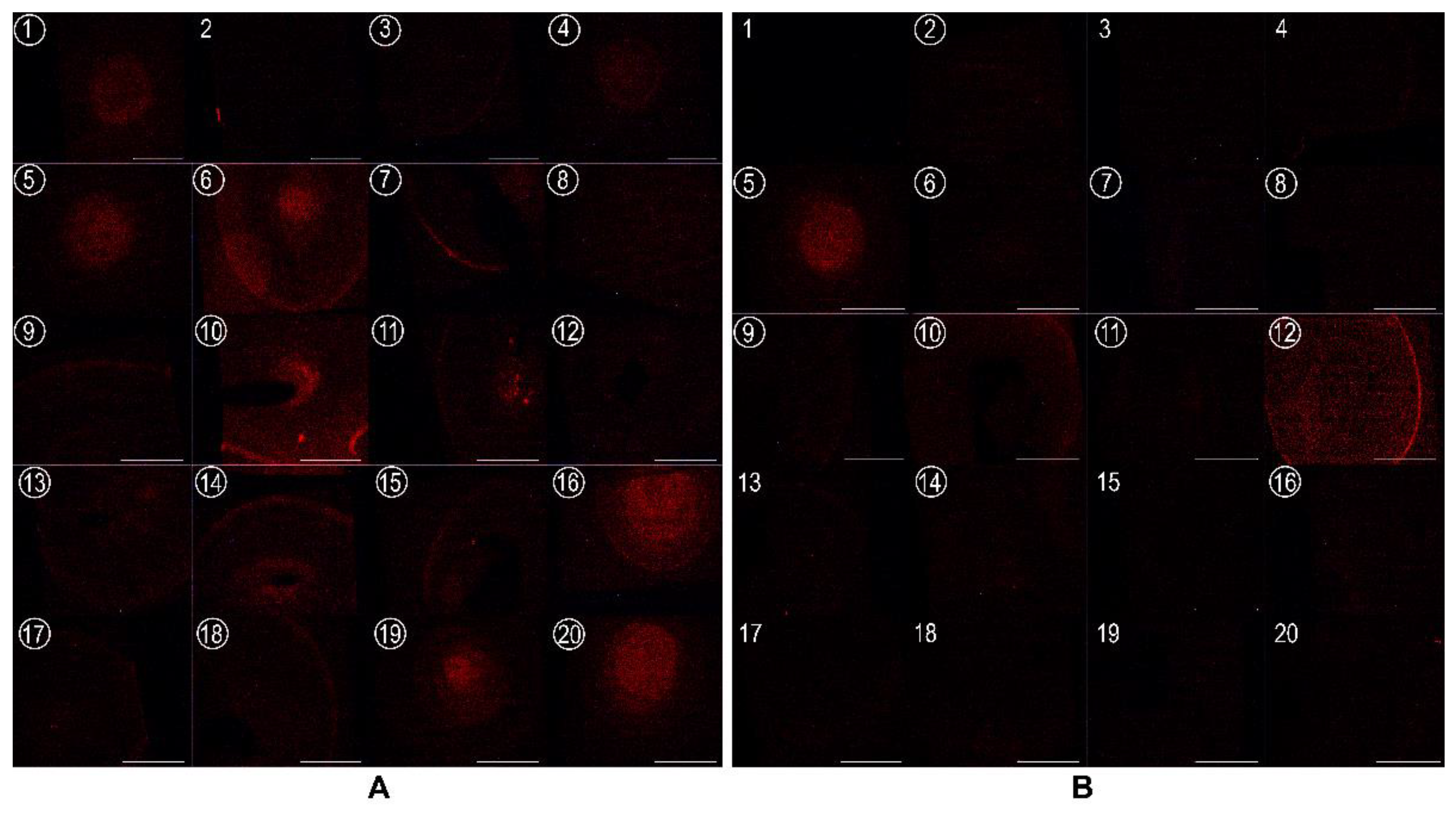
3.2. EryA Specifically Interacts with Porphyromonas Gingivalis-Produced CPE Species
3.3. EryA as a Sensor of CPE in Lipid Extracts from Plaque Samples from Periodontal Patients
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, L.; Lamster, I.; Greenspan, J.; Pitts, N.; Scully, C.; Warnakulasuriya, S. Global burden of oral diseases: Emerging concepts, management and interplay with systemic health. Oral Dis. 2016, 22, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.; Abajobir, A.A.; Abate, K.H.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abdulkader, R.S.; Abdulle, A.M.; Abebo, T.A.; Abera, S.F.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef] [Green Version]
- Kassebaum, N.J.; Smith, A.G.C.; Bernabé, E.; Fleming, T.D.; Reynolds, A.E.; Vos, T.; Murray, C.J.L.; Marcenes, W.; GBD 2015 Oral Health Collaborators. Global, Regional, and National Prevalence, Incidence, and Disability-Adjusted Life Years for Oral Conditions for 195 Countries, 1990–2015: A Systematic Analysis for the Global Burden of Diseases, Injuries, and Risk Factors. J. Dent. Res. 2017, 96, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya and Bhattacharyya Periodontal Infections in Children. Available online: https://lupinepublishers.com/pediatric-dentistry-journal/fulltext/periodontal-infections-in-children.ID.000197.php (accessed on 1 March 2022).
- Ghallab, N.A. Diagnostic potential and future directions of biomarkers in gingival crevicular fluid and saliva of periodontal diseases: Review of the current evidence. Arch. Oral Biol. 2018, 87, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Dubey, P.; Mittal, N. Periodontal diseases—A brief review. Int. J. Oral Health Dent. 2020, 6, 177–187. [Google Scholar] [CrossRef]
- Harvey, J.D. Periodontal Microbiology. Dent. Clin. North Am. 2017, 61, 253–269. [Google Scholar] [CrossRef]
- Nelson, R.G.; Shlossman, M.; Budding, L.M.; Pettitt, D.J.; Saad, M.F.; Genco, R.J.; Knowler, W.C. Periodontal Disease and NIDDM in Pima Indians. Diabetes Care 1990, 13, 836–840. [Google Scholar] [CrossRef]
- Lovegrove, J.M. Dental plaque revisited: Bacteria associated with periodontal disease. J. N. Z. Soc. Periodontol. 2004, 7–21. [Google Scholar]
- Wang, J.; Qi, J.; Zhao, H.; He, S.; Zhang, Y.; Wei, S.; Zhao, F. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci. Rep. 2013, 3, 1843. [Google Scholar] [CrossRef]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primer 2017, 3, 7038. [Google Scholar] [CrossRef]
- Armitage, G.C. Analysis of gingival crevice fluid and risk of progression of periodontitis. Periodontol. 2000 2004, 34, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Barros, S.P.; Williams, R.; Offenbacher, S.; Morelli, T. Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontol. 2000 2016, 70, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Wassall, R.R.; Preshaw, P.M. Clinical and technical considerations in the analysis of gingival crevicular fluid. Periodontol. 2000 2016, 70, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Henson, B.S.; Camargo, P.M.; Wong, D.T. The clinical value of salivary biomarkers for periodontal disease. Periodontol. 2000 2009, 51, 25–37. [Google Scholar] [CrossRef]
- Kinney, J.S.; Morelli, T.; Braun, T.; Ramseier, C.A.; Herr, A.E.; Sugai, J.V.; Shelburne, C.E.; Rayburn, L.A.; Singh, A.K.; Giannobile, W.V. Saliva/Pathogen Biomarker Signatures and Periodontal Disease Progression. J. Dent. Res. 2011, 90, 752–758. [Google Scholar] [CrossRef] [Green Version]
- Korte, D.L.; Kinney, J. Personalized medicine: An update of salivary biomarkers for periodontal diseases. Periodontol. 2000 2016, 70, 26–37. [Google Scholar] [CrossRef]
- Jaedicke, K.M.; Preshaw, P.M.; Taylor, J.J. Salivary cytokines as biomarkers of periodontal diseases. Periodontol. 2000 2016, 70, 164–183. [Google Scholar] [CrossRef]
- Loos, B.G.; Tjoa, S. Host-derived diagnostic markers for periodontitis: Do they exist in gingival crevice fluid? Periodontol. 2000 2005, 39, 53–72. [Google Scholar] [CrossRef]
- Buduneli, N.; Kinane, D.F. Host-derived diagnostic markers related to soft tissue destruction and bone degradation in periodontitis. J. Clin. Periodontol. 2011, 38, 85–105. [Google Scholar] [CrossRef]
- Mäntylä, P.; Stenman, M.; Kinane, D.F.; Tikanoja, S.; Luoto, H.; Salo, T.; Sorsa, T. Gingival crevicular fluid collagenase-2 (MMP-8) test stick for chair-side monitoring of periodontitis. J. Periodontal Res. 2003, 38, 436–439. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Yan, H.; Huang, L. Salivary matrix metalloproteinase (MMP)-8 as a biomarker for periodontitis. Medicine 2018, 97, e9642. [Google Scholar] [CrossRef] [PubMed]
- Franco, C.; Patricia, H.-R.; Timo, S.; Claudia, B.; Marcela, H. Matrix Metalloproteinases as Regulators of Periodontal Inflammation. Int. J. Mol. Sci. 2017, 18, 440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathnayake, N.; Åkerman, S.; Klinge, B.; Lundegren, N.; Jansson, H.; Tryselius, Y.; Sorsa, T.; Gustafsson, A. Salivary Biomarkers for Detection of Systemic Diseases. PLoS ONE 2013, 8, e61356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangbulla, V.; Nirola, A.; Gupta, M.; Batra, P.; Gupta, M. Salivary IgA, Interleukin-1β and MMP-8 as Salivary Biomarkers in Chronic Periodontitis Patients. Chin. J. Dent. Res. Off. J. Sci. Sect. Chin. Stomatol. Assoc. CSA 2017, 20, 43–51. [Google Scholar] [CrossRef]
- Noack, B.; Kipping, T.; Tervahartiala, T.; Sorsa, T.; Hoffmann, T.; Lorenz, K. Association between serum and oral matrix metalloproteinase-8 levels and periodontal health status. J. Periodontal Res. 2017, 52, 824–831. [Google Scholar] [CrossRef]
- Kushlinskii, N.E.; Solovykh, E.A.; Karaoglanova, T.B.; Bayar, U.; Gershtein, E.S.; Troshin, A.A.; Kostyleva, O.I.; Grinin, V.M.; Maksimovskaya, L.N.; Yanushevitch, O.O. Content of Matrix Metalloproteinase-8 and Matrix Metalloproteinase-9 in Oral Fluid of Patients with Chronic Generalized Periodontitis. Bull. Exp. Biol. Med. 2011, 152, 240–244. [Google Scholar] [CrossRef]
- Gursoy, U.K.; Könönen, E.; Pradhan-Palikhe, P.; Tervahartiala, T.; Pussinen, P.J.; Suominen-Taipale, L.; Sorsa, T. Salivary MMP-8, TIMP-1, and ICTP as markers of advanced periodontitis. J. Clin. Periodontol. 2010, 37, 487–493. [Google Scholar] [CrossRef]
- Salazar, M.G.; Jehmlich, N.; Murr, A.; Dhople, V.M.; Holtfreter, B.; Hammer, E.; Völker, U.; Kocher, T. Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. J. Clin. Periodontol. 2013, 40, 825–832. [Google Scholar] [CrossRef]
- Taba, M.; Kinney, J.; Kim, A.S.; Giannobile, W.V. Diagnostic Biomarkers for Oral and Periodontal Diseases. Dent. Clin. N. Am. 2005, 49, 551–571. [Google Scholar] [CrossRef] [Green Version]
- Crone, H.D.; Bridges, R.G. The phospholipids of the housefly, Musca domestica. Biochem. J. 1963, 89, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Masood, M.A.; Yuan, C.; Acharya, J.K.; Veenstra, T.D.; Blonder, J. Quantitation of ceramide phosphorylethanolamines containing saturated and unsaturated sphingoid base cores. Anal. Biochem. 2010, 400, 259–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraut, R. Roles of sphingolipids in Drosophila development and disease. J. Neurochem. 2011, 116, 764–778. [Google Scholar] [CrossRef]
- Kellermann, M.Y.; Schubotz, F.; Elvert, M.; Lipp, J.S.; Birgel, D.; Prieto-Mollar, X.; Dubilier, N.; Hinrichs, K.-U. Symbiont–host relationships in chemosynthetic mussels: A comprehensive lipid biomarker study. Org. Geochem. 2012, 43, 112–124. [Google Scholar] [CrossRef]
- Hori, T.; Arakawa, I.; Sugita, M. Distribution of Ceramide 2-Aminoethylphosphonate and Ceramide Aminoethylphosphate (Sphingoethanolamine) in Some Aquatic Animals. J. Biochem. 1967, 62, 67–70. [Google Scholar] [CrossRef]
- Yang, T.K.; Mc Means, E.; Anderson, L.E.; Jenkin, H.M. Sphingophospholipids of species ofAedes andCulex mosquito cells cultivated in suspension culture from logarithmic and stationary phases of growth. Lipids 1974, 9, 1009–1013. [Google Scholar] [CrossRef]
- O’Connor, J.D.; Polito, A.J.; Monroe, R.E.; Sweeley, C.C.; Bleber, L.L. Characterization of invertebrate sphingolipid bases: Occurrence of eicosasphinga-4, 11-dienine and eicosasphing-11-enine in scorpion. Biochim. Biophys. Acta BBA-Lipids Lipid Metab. 1970, 202, 195–197. [Google Scholar] [CrossRef]
- Abeytunga, D.T.U.; Glick, J.J.; Gibson, N.J.; Oland, L.A.; Somogyi, A.; Wysocki, V.H.; Polt, R. Presence of unsaturated sphingomyelins and changes in their composition during the life cycle of the moth Manduca sexta. J. Lipid Res. 2004, 45, 1221–1231. [Google Scholar] [CrossRef] [Green Version]
- Broad, T.E.; Dawson, R.M.C. Formation of ceramide phosphorylethanolamine from phosphatidylethanolamine in the rumen protozoon Entodinium caudatum (Short Communication). Biochem. J. 1973, 134, 659–662. [Google Scholar] [CrossRef] [Green Version]
- Kaneshiro, E.S.; Jayasimhulu, K.; Sul, D.; Erwin, J.A. Identification and initial characterizations of free, glycosylated, and phosphorylated ceramides of Paramecium. J. Lipid Res. 1997, 38, 2399–2410. [Google Scholar] [CrossRef]
- Moreau, R.A.; Young, D.H.; Danis, P.O.; Powell, M.J.; Quinn, C.J.; Beshah, K.; Slawecki, R.A.; Dilliplane, R.L. Identification of ceramide-phosphorylethanolamine in Oomycete plant pathogens: Pythium ultimum, phytophthora infestans, and Phytophthora capsici. Lipids 1998, 33, 307–317. [Google Scholar] [CrossRef]
- Murakami, C.; Sakane, F. Sphingomyelin synthase-related protein generates diacylglycerol via the hydrolysis of glycerophospholipids in the absence of ceramide. J. Biol. Chem. 2021, 296, 100454. [Google Scholar] [CrossRef] [PubMed]
- Bickert, A.; Ginkel, C.; Kol, M.; vom Dorp, K.; Jastrow, H.; Degen, J.; Jacobs, R.L.; Vance, D.E.; Winterhager, E.; Jiang, X.-C.; et al. Functional characterization of enzymes catalyzing ceramide phosphoethanolamine biosynthesis in mice [S]. J. Lipid Res. 2015, 56, 821–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichols, F.C.; Yao, X.; Bajrami, B.; Downes, J.; Finegold, S.M.; Knee, E.; Gallagher, J.J.; Housley, W.J.; Clark, R.B. Phosphorylated Dihydroceramides from Common Human Bacteria Are Recovered in Human Tissues. PLoS ONE 2011, 6, e16771. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Muto, Y.; Tanaka-Bandoh, K.; Watanabe, K.; Ueno, K. Sphingolipid Composition in Bacteroides Species. Anaerobe 1995, 1, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Nichols, F.C.; Rojanasomsith, K. Porphyromonas gingivalis lipids and diseased dental tissues. Oral Microbiol. Immunol. 2006, 21, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Kraševec, N.; Skočaj, M.; Maček, P.; Anderluh, G.; Sepčić, K. Fungal aegerolysin-like proteins: Distribution, activities, and applications. Appl. Microbiol. Biotechnol. 2015, 99, 601–610. [Google Scholar] [CrossRef]
- Butala, M.; Novak, M.; Kraševec, N.; Skočaj, M.; Veranič, P.; Maček, P.; Sepčić, K. Aegerolysins: Lipid-binding proteins with versatile functions. Semin. Cell Dev. Biol. 2017, 72, 142–151. [Google Scholar] [CrossRef]
- Panevska, A.; Skočaj, M.; Modic, Š.; Razinger, J.; Sepčić, K. Aegerolysins from the fungal genus Pleurotus—Bioinsecticidal proteins with multiple potential applications. J. Invertebr. Pathol. 2021, 186, 107474. [Google Scholar] [CrossRef]
- Grundner, M.; Panevska, A.; Sepčić, K.; Skočaj, M. What Can Mushroom Proteins Teach Us about Lipid Rafts? Membranes 2021, 11, 264. [Google Scholar] [CrossRef]
- Bhat, H.B.; Ishitsuka, R.; Inaba, T.; Murate, M.; Abe, M.; Makino, A.; Kohyama-Koganeya, A.; Nagao, K.; Kurahashi, A.; Kishimoto, T.; et al. Evaluation of aegerolysins as novel tools to detect and visualize ceramide phosphoethanolamine, a major sphingolipid in invertebrates. FASEB J. 2015, 29, 3920–3934. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, T.; Ishitsuka, R.; Kobayashi, T. Detectors for evaluating the cellular landscape of sphingomyelin- and cholesterol-rich membrane domains. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2016, 1861, 812–829. [Google Scholar] [CrossRef] [PubMed]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-forming protein complexes from Pleurotus mushrooms kill western corn rootworm and Colorado potato beetle through targeting membrane ceramide phosphoethanolamine. Sci. Rep. 2019, 9, 5073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novak, M.; Krpan, T.; Panevska, A.; Shewell, L.K.; Day, C.J.; Jennings, M.P.; Guella, G.; Sepčić, K. Binding specificity of ostreolysin A6 towards Sf9 insect cell lipids. Biochim. Biophys. Acta BBA-Biomembr. 2020, 1862, 183307. [Google Scholar] [CrossRef] [PubMed]
- Tomita, T.; Noguchi, K.; Mimuro, H.; Ukaji, F.; Ito, K.; Sugawara-Tomita, N.; Hashimoto, Y. Pleurotolysin, a Novel Sphingomyelin-specific Two-component Cytolysin from the Edible Mushroom Pleurotus ostreatus, Assembles into a Transmembrane Pore Complex. J. Biol. Chem. 2004, 279, 26975–26982. [Google Scholar] [CrossRef] [Green Version]
- Ota, K.; Leonardi, A.; Mikelj, M.; Skočaj, M.; Wohlschlager, T.; Künzler, M.; Aebi, M.; Narat, M.; Križaj, I.; Anderluh, G.; et al. Membrane cholesterol and sphingomyelin, and ostreolysin A are obligatory for pore-formation by a MACPF/CDC-like pore-forming protein, pleurotolysin B. Biochimie 2013, 95, 1855–1864. [Google Scholar] [CrossRef]
- Lukoyanova, N.; Kondos, S.C.; Farabella, I.; Law, R.H.P.; Reboul, C.F.; Caradoc-Davies, T.T.; Spicer, B.A.; Kleifeld, O.; Traore, D.A.K.; Ekkel, S.M.; et al. Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin. PLoS Biol. 2015, 13, e1002049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milijaš Jotić, M.; Panevska, A.; Iacovache, I.; Kostanjšek, R.; Mravinec, M.; Skočaj, M.; Zuber, B.; Pavšič, A.; Razinger, J.; Modic, Š.; et al. Dissecting Out the Molecular Mechanism of Insecticidal Activity of Ostreolysin A6/Pleurotolysin B Complexes on Western Corn Rootworm. Toxins 2021, 13, 455. [Google Scholar] [CrossRef]
- Bligh and Dyer A Rapid Method of Total Lipid Extraction and Purification. Available online: https://cdnsciencepub.com/doi/abs/10.1139/o59-099 (accessed on 25 February 2022).
- Garbus, J.; DeLuca, H.F.; Loomans, M.E.; Strong, F.M. The Rapid Incorporation of Phosphate into Mitochondrial Lipids. J. Biol. Chem. 1963, 238, 59–63. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89, S159–S172. [Google Scholar] [CrossRef] [Green Version]
- Sepčić, K.; Berne, S.; Potrich, C.; Turk, T.; Maček, P.; Menestrina, G. Interaction of ostreolysin, a cytolytic protein from the edible mushroom Pleurotus ostreatus, with lipid membranes and modulation by lysophospholipids. JBIC J. Biol. Inorg. Chem. 2003, 270, 1199–1210. [Google Scholar] [CrossRef] [Green Version]
- Guschina, I.A.; Harris, K.M.; Maskrey, B.; Goldberg, B.; Lloyd, D.; Harwood, J.L. The microaerophilic flagellate, Trichomonas vaginalis, contains unusual acyl lipids but no detectable cardiolipin. J. Eukaryot. Microbiol. 2009, 56, 52–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaBach, J.P.; White, D.C. Identification of ceramide phosphorylethanolamine and ceramide phosphorylglycerol in the lipids of an anaerobic bacterium. J. Lipid Res. 1969, 10, 528–534. [Google Scholar] [CrossRef]
- Sevova, E.S.; Goren, M.A.; Schwartz, K.J.; Hsu, F.-F.; Turk, J.; Fox, B.G.; Bangs, J.D. Cell-free synthesis and functional characterization of sphingolipid synthases from parasitic trypanosomatid protozoa. J. Biol. Chem. 2010, 285, 20580–20587. [Google Scholar] [CrossRef] [Green Version]
- Vacaru, A.M.; van den Dikkenberg, J.; Ternes, P.; Holthuis, J.C.M. Ceramide phosphoethanolamine biosynthesis in Drosophila is mediated by a unique ethanolamine phosphotransferase in the Golgi lumen. J. Biol. Chem. 2013, 288, 11520–11530. [Google Scholar] [CrossRef] [Green Version]
- Welti, R.; Mui, E.; Sparks, A.; Wernimont, S.; Isaac, G.; Kirisits, M.; Roth, M.; Roberts, C.W.; Botté, C.; Maréchal, E.; et al. Lipidomic analysis of Toxoplasma gondii reveals unusual polar lipids. Biochemistry 2007, 46, 13882–13890. [Google Scholar] [CrossRef] [Green Version]
- Batrakov, S.G.; Mosezhnyi, A.E.; Ruzhitsky, A.O.; Sheichenko, V.I.; Nikitin, D.I. The polar-lipid composition of the sphingolipid-producing bacterium Flectobacillus major. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2000, 1484, 225–240. [Google Scholar] [CrossRef]
- Nichols, F.C.; Riep, B.; Mun, J.; Morton, M.D.; Bojarski, M.T.; Dewhirst, F.E.; Smith, M.B. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis. J. Lipid Res. 2004, 45, 2317–2330. [Google Scholar] [CrossRef] [Green Version]
- Vieira, C.R.; Munoz-Olaya, J.M.; Sot, J.; Jiménez-Baranda, S.; Izquierdo-Useros, N.; Abad, J.L.; Apellániz, B.; Delgado, R.; Martinez-Picado, J.; Alonso, A.; et al. Dihydrosphingomyelin Impairs HIV-1 Infection by Rigidifying Liquid-Ordered Membrane Domains. Chem. Biol. 2010, 17, 766–775. [Google Scholar] [CrossRef]
- Hernández-Tiedra, S.; Fabriàs, G.; Dávila, D.; Salanueva, Í.J.; Casas, J.; Montes, L.R.; Antón, Z.; García-Taboada, E.; Salazar-Roa, M.; Lorente, M.; et al. Dihydroceramide accumulation mediates cytotoxic autophagy of cancer cells via autolysosome destabilization. Autophagy 2016, 12, 2213–2229. [Google Scholar] [CrossRef] [Green Version]
- Contreras, F.-X.; Basañez, G.; Alonso, A.; Herrmann, A.; Goñi, F.M. Asymmetric Addition of Ceramides but not Dihydroceramides Promotes Transbilayer (Flip-Flop) Lipid Motion in Membranes. Biophys. J. 2005, 88, 348–359. [Google Scholar] [CrossRef] [Green Version]
- Uemura, S.; Shishido, F.; Tani, M.; Mochizuki, T.; Abe, F.; Inokuchi, J. Loss of hydroxyl groups from the ceramide moiety can modify the lateral diffusion of membrane proteins in S. cerevisiae. J. Lipid Res. 2014, 55, 1343–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maula, T.; Al Sazzad, A.; Slotte, J.P. Influence of Hydroxylation, Chain Length, and Chain Unsaturation on Bilayer Properties of Ceramides. Biophys. J. 2015, 109, 1639–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, A.; Goñi, F.M. The Physical Properties of Ceramides in Membranes. Annu. Rev. Biophys. 2018, 47, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.R.; Listgarten, M.A.; Ebersole, J.L.; Strzempko, M.N.Y. Bacteroides forsythus sp. nov., a Slow-Growing, Fusiform Bacteroides sp. from the Human Oral Cavity. Int. J. Syst. Bacteriol. 1986, 36, 213–221. [Google Scholar] [CrossRef]
- Geiger, O.; Padilla-Gómez, J.; López-Lara, I.M. Bacterial Sphingolipids and Sulfonolipids. In Biogenesis of Fatty Acids, Lipids and Membranes; Geiger, O., Ed.; Handbook of Hydrocarbon and Lipid Microbiology; Springer International Publishing: Cham, Switzerland, 2019; pp. 123–137. ISBN 978-3-319-50430-8. [Google Scholar]
- Tomšič, K.; Rodič, K.; Sotošek, A.; Videmšek, P.; Seme, K.; Herrera, D.; Sanz, M.; Gašperšič, R. Do Differences in Cultivable Subgingival Species Exist between Different Periodontitis Stages and Grades? Oral Health Prev. Dent. 2021, 19, 15–24. [Google Scholar] [CrossRef]
- Fine, D.H.; Markowitz, K.; Fairlie, K.; Tischio-Bereski, D.; Ferrendiz, J.; Furgang, D.; Paster, B.J.; Dewhirst, F.E. A Consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis Is Present in Sites Prior to Bone Loss in a Longitudinal Study of Localized Aggressive Periodontitis. J. Clin. Microbiol. 2013, 51, 2850–2861. [Google Scholar] [CrossRef] [Green Version]
- Duran-Pinedo, A.E.; Chen, T.; Teles, R.; Starr, J.R.; Wang, X.; Krishnan, K.; Frias-Lopez, J. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014, 8, 1659–1672. [Google Scholar] [CrossRef]
- Popova, C.; Dosseva-Panova, V.; Panov, V. Microbiology of Periodontal Diseases. A Review. Biotechnol. Biotechnol. Equip. 2013, 27, 3754–3759. [Google Scholar] [CrossRef]
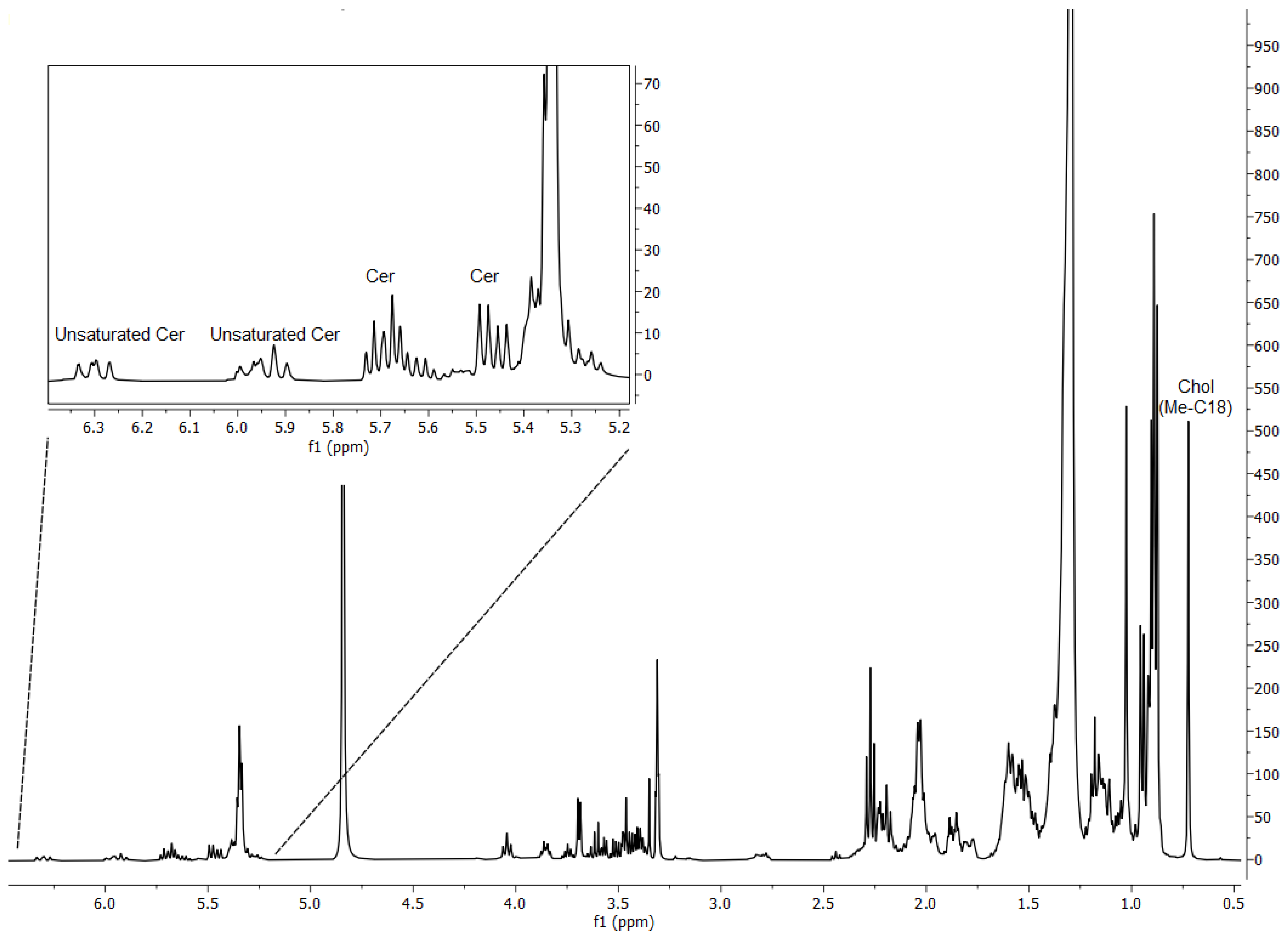
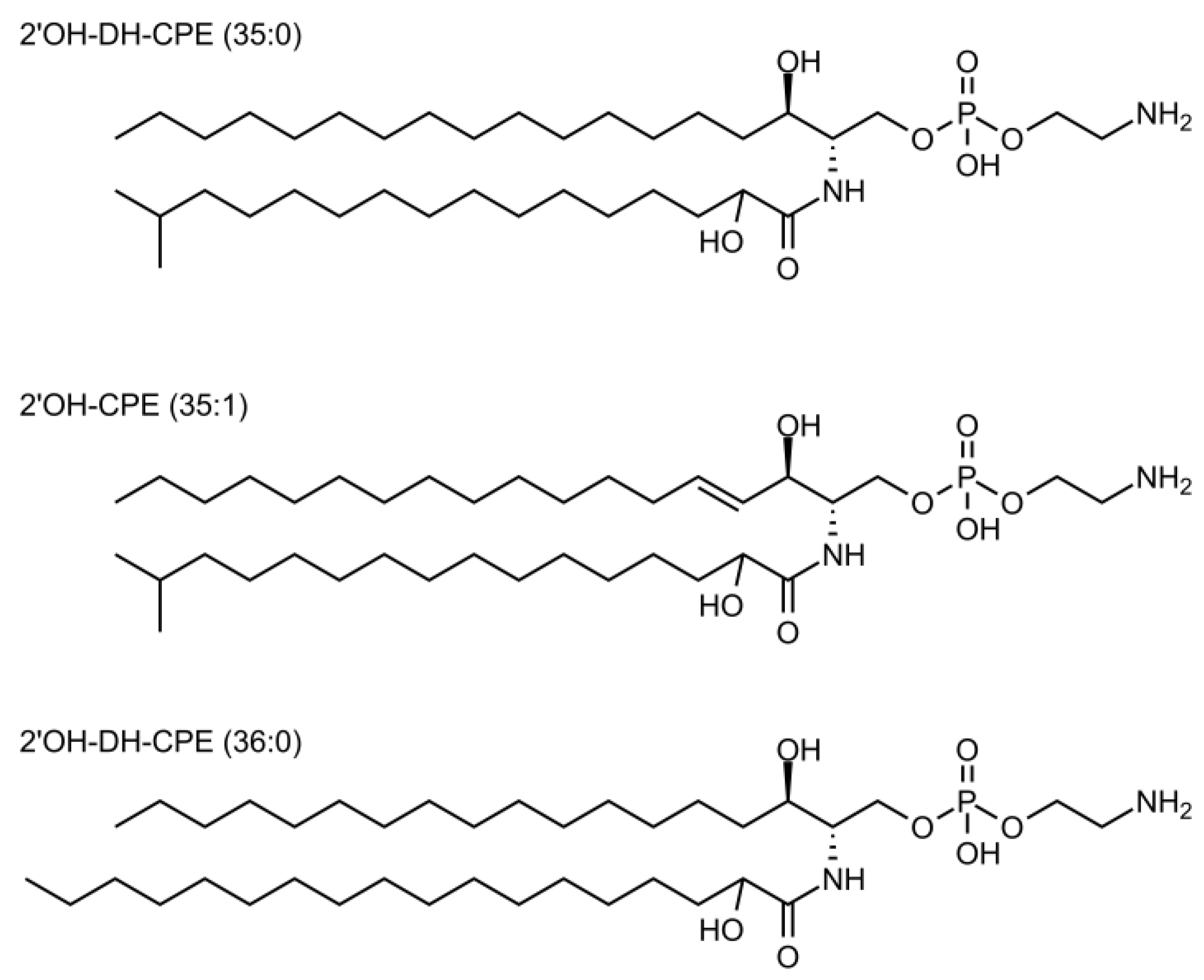
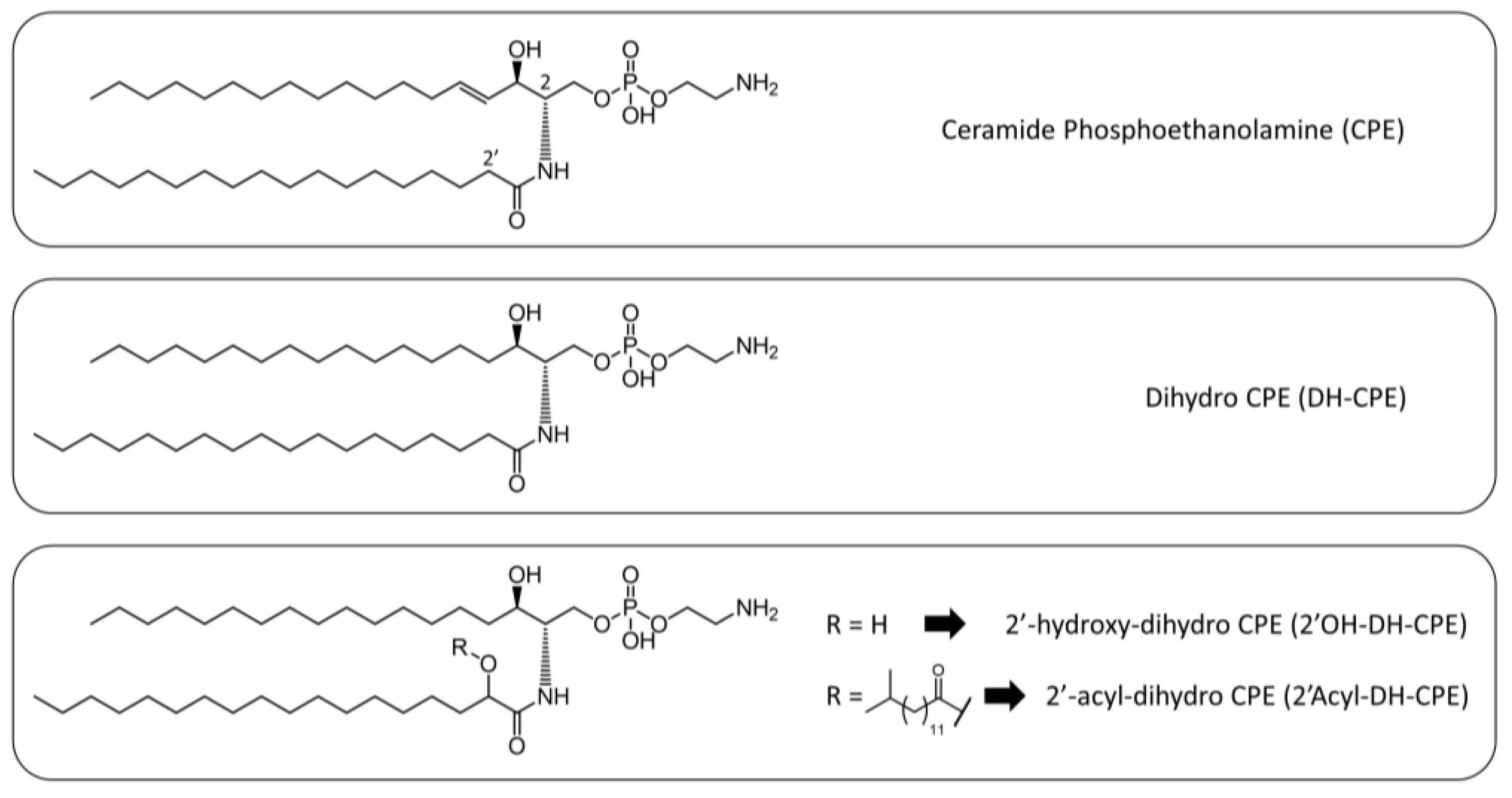

| Class | Relative Inter-Class Distribution (% w/w) |
|---|---|
| CPE | 0.02 |
| DH-CPE | 0.02 |
| 2′OH-DH-CPE | 26.6 |
| 2′-acyl-DH-CPE | N/A |
| Species | Relative Intra-Class Distribution (% Molar Fraction) |
|---|---|
| CPE (32:1) | 8.3 |
| CPE (33:1) | 11.2 |
| CPE (34:1) | 80.4 |
| DH-CPE (34:0) | 8.9 |
| DH-CPE (36:0) | 91.1 |
| 2′OH-DH-CPE (33:0) | 8.6 |
| 2′OH-DH-CPE (34:0) | 37.0 |
| 2′OH-DH-CPE (35:0) | 43.6 |
| 2′OH-DH-CPE (36:0) | 9.0 |
| 2′-acyl-DH-CPE (48:0) | 9.9 |
| 2′-acyl-DH-CPE (49:0) | 43.6 |
| 2′-acyl-DH-CPE (50:0) | 41.7 |
| 2′-acyl-DH-CPE (51:0) | 4.3 |
| Statistic | Value | 95% Cl |
|---|---|---|
| Sensitivity (True positive/ True positive + False positive) | 95.00% | 75.13% to 99.87% |
| Specificity (True negative/True negative + False positive) | 45.00% | 23.06% to 68.47% |
| Positive likelihood ratio (Sensitivity/1 − Specificity) | 1.73 | 1.15 to 2.6 |
| Negative likelihood ratio (1 − Sensitivity/Specificity) | 0.11 | 0.02 to 0.8 |
| Positive predictive value (True positive/ True positive + False positive) | 63.33% | 53.43% to 72.22% |
| Negative predictive value (True negative/ True negative + False negtive) | 90.00% | 55.64% to 98.48% |
| Accuracy (True positive + True negative/ True positive + True negative + False positive + False negative) | 70.00% | 53.47% to 83.44% |
| Disease prevalence | 50.00% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grundner, M.; Munjaković, H.; Tori, T.; Sepčić, K.; Gašperšič, R.; Oblak, Č.; Seme, K.; Guella, G.; Trenti, F.; Skočaj, M. Ceramide Phosphoethanolamine as a Possible Marker of Periodontal Disease. Membranes 2022, 12, 655. https://doi.org/10.3390/membranes12070655
Grundner M, Munjaković H, Tori T, Sepčić K, Gašperšič R, Oblak Č, Seme K, Guella G, Trenti F, Skočaj M. Ceramide Phosphoethanolamine as a Possible Marker of Periodontal Disease. Membranes. 2022; 12(7):655. https://doi.org/10.3390/membranes12070655
Chicago/Turabian StyleGrundner, Maja, Haris Munjaković, Tilen Tori, Kristina Sepčić, Rok Gašperšič, Čedomir Oblak, Katja Seme, Graziano Guella, Francesco Trenti, and Matej Skočaj. 2022. "Ceramide Phosphoethanolamine as a Possible Marker of Periodontal Disease" Membranes 12, no. 7: 655. https://doi.org/10.3390/membranes12070655
APA StyleGrundner, M., Munjaković, H., Tori, T., Sepčić, K., Gašperšič, R., Oblak, Č., Seme, K., Guella, G., Trenti, F., & Skočaj, M. (2022). Ceramide Phosphoethanolamine as a Possible Marker of Periodontal Disease. Membranes, 12(7), 655. https://doi.org/10.3390/membranes12070655






