Is Lipid Specificity Key to the Potential Antiviral Activity of Mouthwash Reagent Chlorhexidine against SARS-CoV-2?
Abstract
1. Introduction
2. Materials and Methods
2.1. Chlorhexidine Structure and Parameter
2.2. Molecular Dynamics Simulations
2.2.1. Simulation of Plasma and Viral Membranes
2.2.2. Conventional CHX-Membrane Simulations
2.3. Potential of Mean Force (PMF) Calculations
2.3.1. Free-Energy of CHX Binding to Membrane
2.3.2. Free-Energy of Pore Formation
3. Results
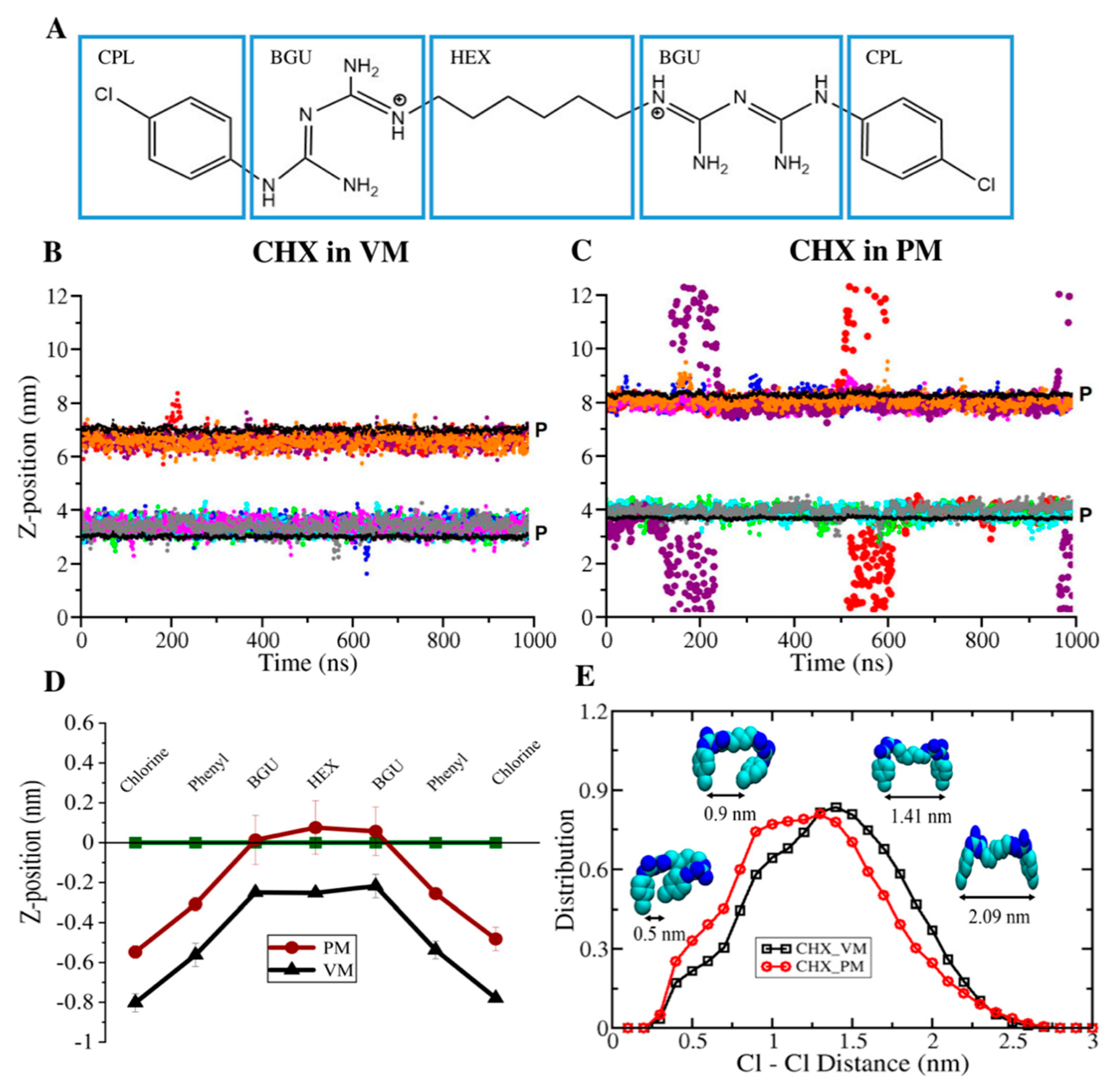
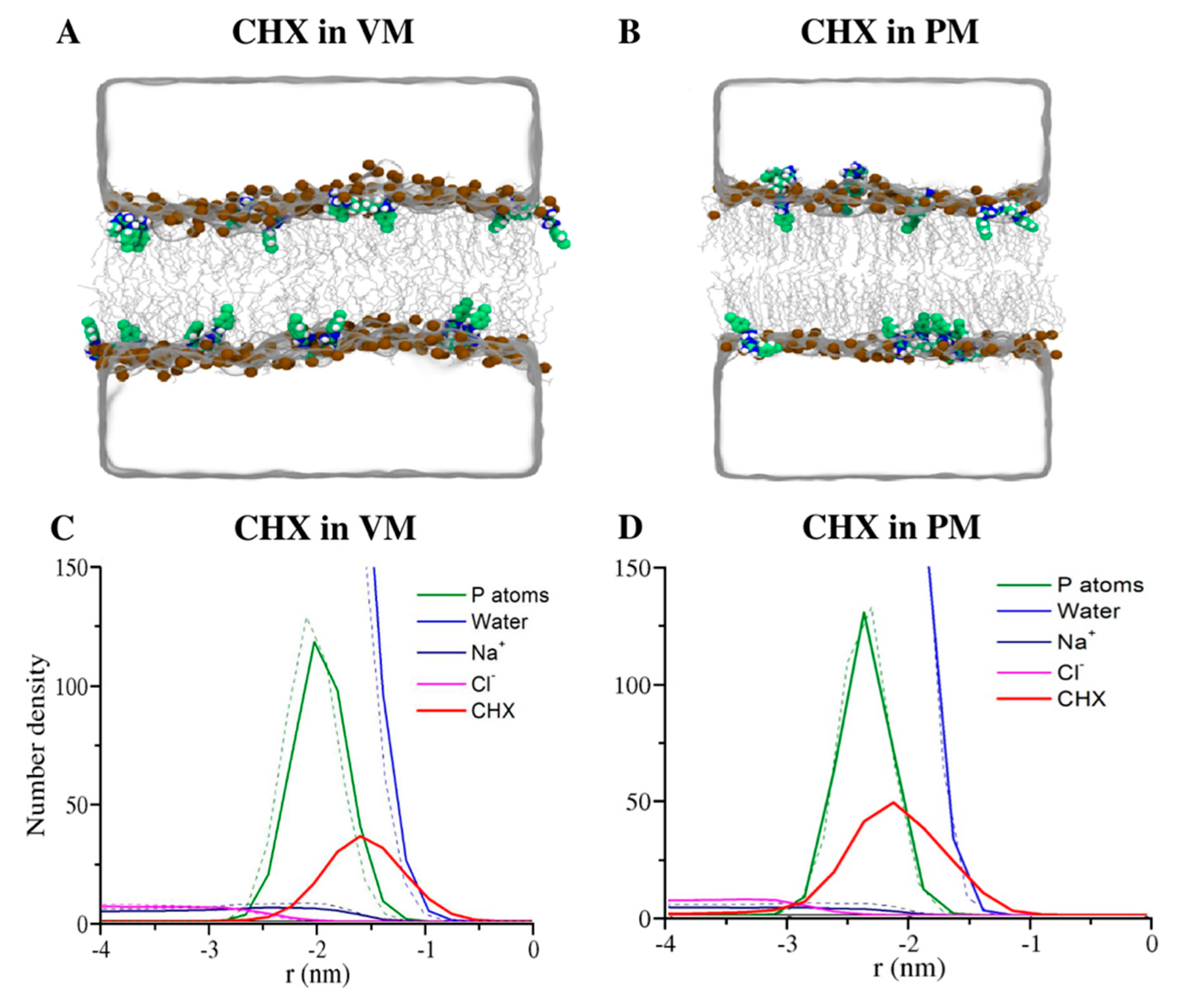
3.1. CHX is More Stable in the Viral Model Membrane Than in the Plasma Membrane
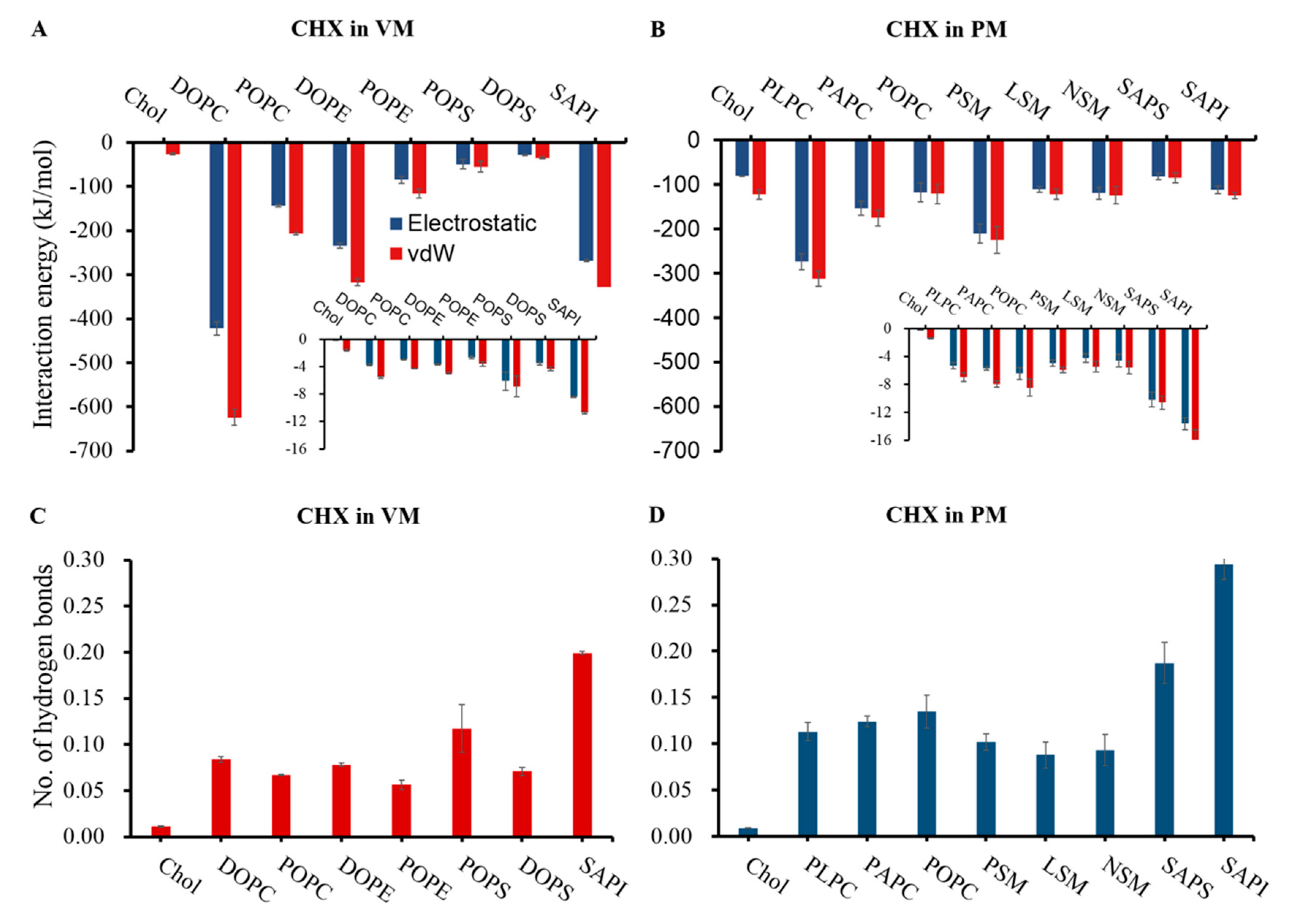
3.2. CHX has a Stronger Binding Affinity to the Viral Membrane
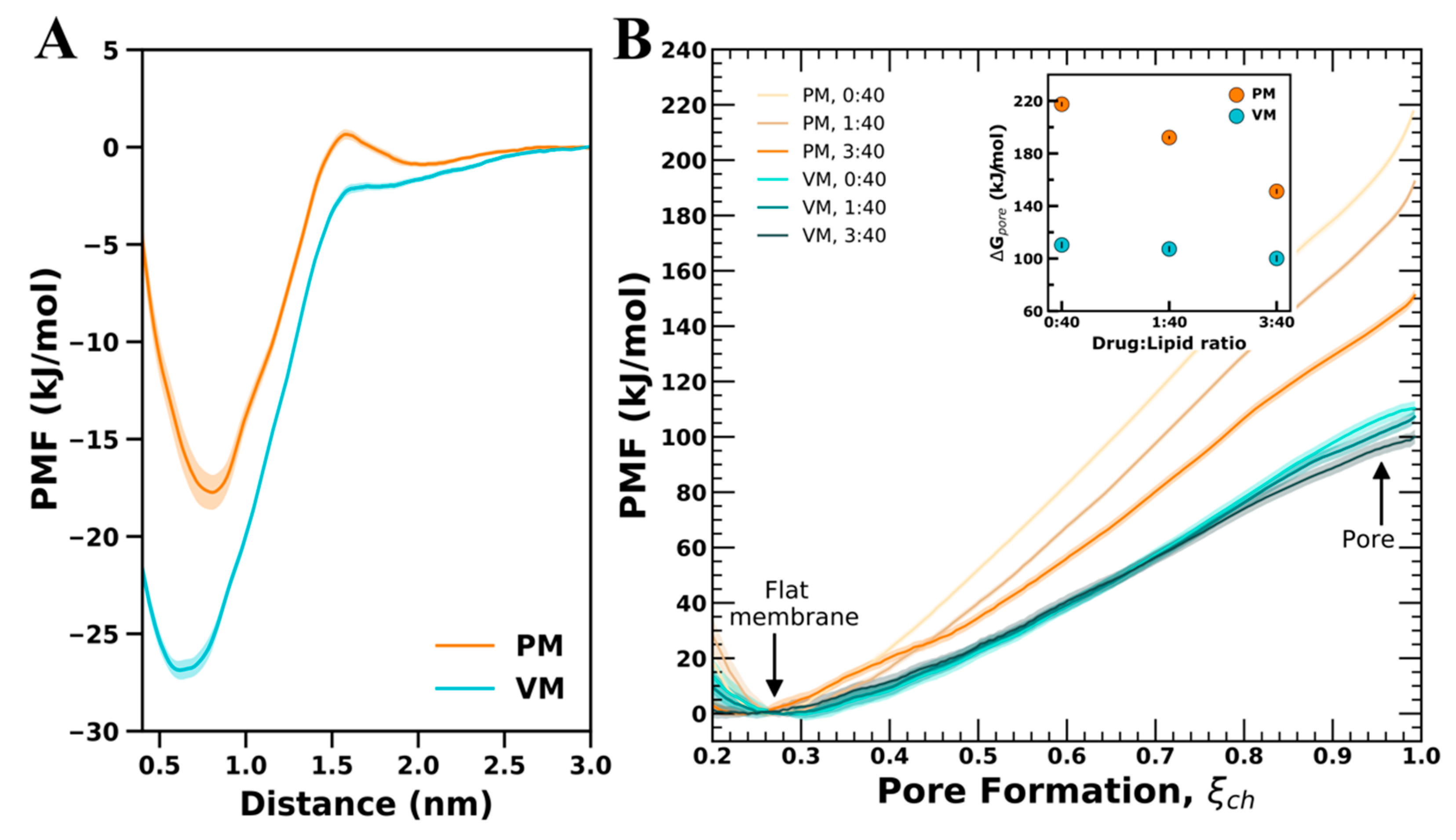
3.3. Free-Energy of CHX Binding to PM and VM
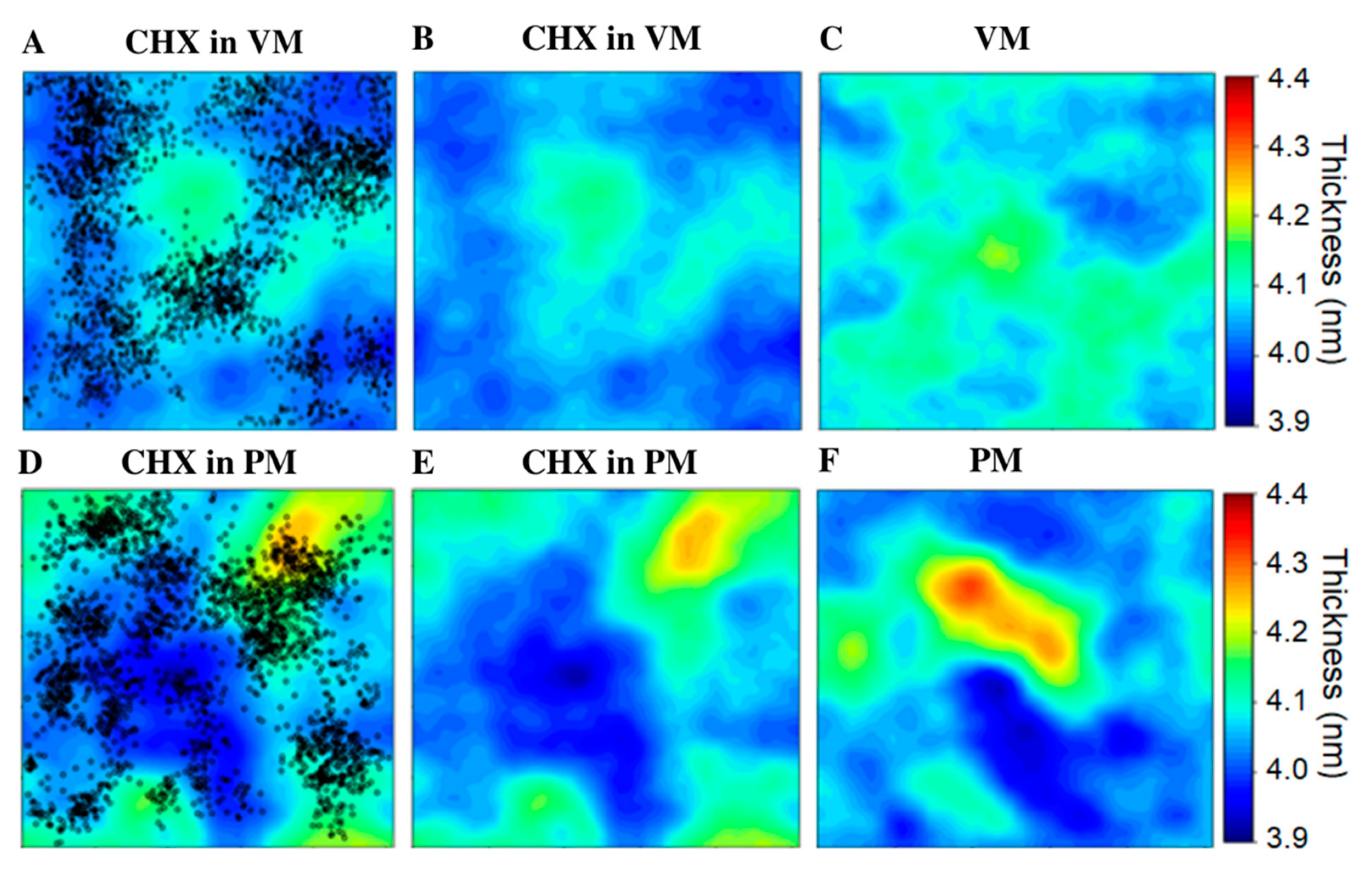
3.4. Membrane Perturbing Effects of CHX on Plasma vs. Viral Membrane
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- World Health Organization. Transmission of SARS-CoV-2: Implications for infection prevention precautions. Scientific Brief. 9 July 2020. Available online: https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions (accessed on 9 July 2020).
- World Health Organization. Modes of transmission of virus causing COVID-19: Implications for IPC precaution recommendations. Scientific Brief. 27 March 2020. Available online: https://reliefweb.int/report/world/modes-transmission-virus-causing-covid-19-implications-ipc-precaution-recommendations?gclid=CjwKCAjwi6WSBhA-EiwA6Niok21fzFfm-eehGL_aKng5xomluE6pW-mJFWqjiWe6UGSfEiE3slJwJBoCWpwQAvD_BwE (accessed on 27 March 2020).
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, G.; Filipowicz, N.A.; Randall, G.; Belov, G.A.; Kopek, B.G.; Wang, X. Host lipids in positive-strand RNA virus genome replication. Front. Microbiol. 2019, 10, 286. [Google Scholar] [CrossRef] [PubMed]
- Sicari, D.; Chatziioannou, A.; Koutsandreas, T.; Sitia, R.; Chevet, E. Role of the early secretory pathway in SARS-CoV-2 infection. J. Cell Biol. 2020, 219, e202006005. [Google Scholar] [CrossRef]
- Yan, B.; Chu, H.; Yang, D.; Sze, K.-H.; Lai, P.-M.; Yuan, S.; Shuai, H.; Wang, Y.; Kao, R.Y.-T.; Chan, J.F.-W.; et al. Characterization of the lipidomic profile of human coronavirus-infected cells: Implications for lipid metabolism remodeling upon coronavirus replication. Viruses 2019, 11, 73. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef]
- Kratzel, A.; Todt, D.; V’Kovski, P.; Steiner, S.; Gultom, M.; Thao, T.T.N.; Ebert, N.; Holwerda, M.; Steinmann, J.; Niemeyer, D.; et al. Inactivation of severe acute respiratory syndrome coronavirus 2 by WHO-recommended hand rub formulations and alcohols. Emerg. Infect. Dis. 2020, 26, 1592–1595. [Google Scholar] [CrossRef]
- O’Donnell, V.B.; Thomas, D.; Stanton, R.; Maillard, J.Y.; Murphy, R.C.; Jones, S.A.; Humphreys, I.; Wakelam, M.J.O.; Fegan, C.; Wise, M.P.; et al. Potential role of oral rinses targeting the viral lipid envelope in SARS-CoV-2 infection. Function 2020, 1, zqaa002. [Google Scholar] [CrossRef]
- World Health Organization. Advice for the Public: Coronavirus Disease (COVID-19). Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public (accessed on 10 May 2022).
- World Health Organization. Guidelines on Hand Hygiene in Health Care: First Global Patient Safety Challenge Clean Care Is Safer Care. Available online: https://pubmed.ncbi.nlm.nih.gov/23805438/ (accessed on 8 June 2022).
- Huang, N.; Perez, P.; Kato, T.; Mikami, Y.; Okuda, K.; Gilmore, R.C.; Conde, C.D.; Gasmi, B.; Stein, S.; Beach, M.; et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021, 27, 892–903. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Gan, F.; Du, Y.; Yao, Y. Salivary Glands: Potential reservoirs for COVID-19 asymptomatic infection. J. Dent. Res. 2020, 99, 989. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- To, K.K.; Tsang, O.T.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.; Cai, J.P.; Chan, J.M.; Chik, T.S.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Ather, A.; Parolia, A.; Ruparel, N.B. Efficacy of mouth rinses against SARS-CoV-2: A scoping review. Front. Dent. Med. 2021, 2, 648547. [Google Scholar] [CrossRef]
- Carrouel, F.; Goncalves, L.S.; Conte, M.P.; Campus, G.; Fisher, J.; Fraticelli, L.; Gadea-Deschamps, E.; Ottolenghi, L.; Bourgeois, D. Antiviral activity of reagents in mouth rinses against SARS-CoV-2. J. Dent. Res. 2021, 100, 124–132. [Google Scholar] [CrossRef]
- Fine, D.H.; Furgang, D.; Korik, I.; Olshan, A.; Barnett, M.L.; Vincent, J.W. Reduction of viable bacteria in dental aerosols by preprocedural rinsing with an antiseptic mouthrinse. Am. J. Dent. 1993, 6, 219–221. [Google Scholar]
- Koletsi, D.; Belibasakis, G.N.; Eliades, T. Interventions to reduce aerosolized microbes in dental practice: A systematic review with network meta-analysis of randomized controlled trials. J. Dent. Res. 2020, 99, 1228–1238. [Google Scholar] [CrossRef]
- Astani, A.; Reichling, J.; Schnitzler, P. Comparative study on the antiviral activity of selected monoterpenes derived from essential oils. Phytother. Res. 2010, 24, 673–679. [Google Scholar] [CrossRef]
- Bernstein, D.; Schiff, G.; Echler, G.; Prince, A.; Feller, M.; Briner, W. In vitro virucidal effectiveness of a 0.12%-chlorhexidine gluconate mouthrinse. J. Dent. Res. 1990, 69, 874–876. [Google Scholar] [CrossRef]
- Eggers, M.; Koburger-Janssen, T.; Eickmann, M.; Zorn, J. In vitro bactericidal and virucidal efficacy of povidone-iodine gargle/mouthwash against respiratory and oral tract pathogens. Infect. Dis. Ther. 2018, 7, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Omidbakhsh, N.; Sattar, S.A. Broad-spectrum microbicidal activity, toxicologic assessment, and materials compatibility of a new generation of accelerated hydrogen peroxide-based environmental surface disinfectant. Am. J. Infect. Control 2006, 34, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, A.; Hoskin, E.R.; Cugini, C.; Markowitz, K.; Chang, T.L.; Fine, D.H. Differential effects of antiseptic mouth rinses on SARS-CoV-2 infectivity in vitro. Pathogens 2021, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Komine, A.; Yamaguchi, E.; Okamoto, N.; Yamamoto, K. Virucidal activity of oral care products against SARS-CoV-2 in vitro. J. Oral Maxillofac. Surg. Med. Pathol. 2021, 33, 475–477. [Google Scholar] [CrossRef]
- Cavalcante-Leão, B.L.; de Araujo, C.-M.; Basso, I.-B.; Schroder, A.-G.-D.; Guariza-Filho, O.; Ravazzi, G.-C.; Gonçalves, F.-M.; Zeigelboim, B.-S.; Santos, R.-S.; Stechman-Neto, J. Is there scientific evidence of the mouthwashes effectiveness in reducing viral load in Covid-19? A systematic review. J. Clin. Exp. Dent. 2021, 13, e179. [Google Scholar] [CrossRef]
- Elzein, R.; Abdel-Sater, F.; Fakhreddine, S.; Hanna, P.A.; Feghali, R.; Hamad, H.; Ayoub, F. In vivo evaluation of the virucidal efficacy of chlorhexidine and povidone-iodine mouthwashes against salivary SARS-CoV-2. A randomized-controlled clinical trial. J. Evid. Based Dent. Pract. 2021, 21, 101584. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, J.T. Use of chlorhexidine to eradicate oropharyngeal SARS-CoV-2 in COVID-19 patients. J. Med. Virol. 2021, 93, 4370–4373. [Google Scholar] [CrossRef]
- Jain, A.; Grover, V.; Singh, C.; Sharma, A.; Das, D.K.; Singh, P.; Thakur, K.G.; Ringe, R.P. Chlorhexidine: An effective anticovid mouth rinse. J. Indian Soc. Periodontol. 2021, 25, 86–88. [Google Scholar]
- Meister, T.L.; Bruggemann, Y.; Todt, D.; Conzelmann, C.; Muller, J.A.; Gross, R.; Munch, J.; Krawczyk, A.; Steinmann, J.; Steinmann, J.; et al. Virucidal efficacy of different oral rinses against severe acute respiratory syndrome coronavirus 2. J. Infect. Dis. 2020, 222, 1289–1292. [Google Scholar] [CrossRef]
- Yoon, J.G.; Yoon, J.; Song, J.Y.; Yoon, S.Y.; Lim, C.S.; Seong, H.; Noh, J.Y.; Cheong, H.J.; Kim, W.J. Clinical significance of a high SARS-CoV-2 viral load in the saliva. J. Korean Med. Sci. 2020, 35, e195. [Google Scholar] [CrossRef]
- Balagopal, S.; Arjunkumar, R. Chlorhexidine: The gold standard antiplaque agent. J. Pharm. Sci. Res. 2013, 5, 270. [Google Scholar]
- Van Oosten, B.; Marquardt, D.; Komljenovic, I.; Bradshaw, J.P.; Sternin, E.; Harroun, T.A. Small molecule interaction with lipid bilayers: A molecular dynamics study of chlorhexidine. J. Mol. Graph. Model. 2014, 48, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Decherchi, S.; Cavalli, A. Thermodynamics and kinetics of drug-target binding by molecular simulation. Chem. Rev. 2020, 120, 12788–12833. [Google Scholar] [CrossRef] [PubMed]
- Kasparyan, G.; Poojari, C.; Róg, T.; Hub, J.S. Cooperative effects of an antifungal moiety and DMSO on pore formation over lipid membranes revealed by free energy calculations. J. Phys. Chem. B 2020, 124, 8811–8821. [Google Scholar] [CrossRef] [PubMed]
- Lajunen, T.; Kontturi, L.S.; Viitala, L.; Manna, M.; Cramariuc, O.; Rog, T.; Bunker, A.; Laaksonen, T.; Viitala, T.; Murtomaki, L.; et al. Indocyanine green-loaded liposomes for light-triggered drug release. Mol. Pharm. 2016, 13, 2095–2107. [Google Scholar] [CrossRef] [PubMed]
- Van Oosten, B.; Marquardt, D.; Harroun, T.A. Testing high concentrations of membrane active antibiotic chlorhexidine via computational titration and calorimetry. J. Phys. Chem. B 2017, 121, 4657–4668. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Lorent, J.; Levental, K.; Ganesan, L.; Rivera-Longsworth, G.; Sezgin, E.; Doktorova, M.; Lyman, E.; Levental, I. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020, 16, 644–652. [Google Scholar] [CrossRef]
- Reglinski, K.; Steinfort-Effelsberg, L.; Sezgin, E.; Klose, C.; Platta, H.W.; Girzalsky, W.; Eggeling, C.; Erdmann, R. Fluidity and lipid composition of membranes of peroxisomes, mitochondria and the ER from oleic acid-induced Saccharomyces cerevisiae. Front. Cell Dev. Biol. 2020, 8, 1205. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Davila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Klauda, J.B.; Monje, V.; Kim, T.; Im, W. Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B 2012, 116, 9424–9431. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.V.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N·log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Hub, J.S.; De Groot, B.L.; Van Der Spoel, D. g_wham—A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 2010, 6, 3713–3720. [Google Scholar] [CrossRef]
- Hub, J.S.; Awasthi, N. Probing a continuous polar defect: A reaction coordinate for pore formation in lipid membranes. J. Chem. Theory Comput. 2017, 13, 2352–2366. [Google Scholar] [CrossRef] [PubMed]
- Ting, C.L.; Awasthi, N.; Müller, M.; Hub, J.S. Metastable prepores in tension-free lipid bilayers. Phys. Rev. Lett. 2018, 120, 128103. [Google Scholar] [CrossRef] [PubMed]
- Enkavi, G.; Javanainen, M.; Kulig, W.; Róg, T.; Vattulainen, I. Multiscale simulations of biological membranes: The challenge to understand biological phenomena in a living substance. Chem. Rev. 2019, 119, 5607–5774. [Google Scholar] [CrossRef] [PubMed]
- Javanainen, M.; Martinez-Seara, H.; Vattulainen, I. Nanoscale membrane domain formation driven by cholesterol. Sci. Rep. 2017, 7, 1143. [Google Scholar] [CrossRef] [PubMed]
- Bennett, W.D.; Sapay, N.; Tieleman, D.P. Atomistic simulations of pore formation and closure in lipid bilayers. Biophys. J. 2014, 106, 210–219. [Google Scholar] [CrossRef]
- Bennett, W.D.; Tieleman, D.P. The importance of membrane defects-lessons from simulations. Acc. Chem. Res. 2014, 47, 2244–2251. [Google Scholar] [CrossRef]
- Casares, D.; Escriba, P.V.; Rossello, C.A. Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef]
- Komljenović, I.; Marquardt, D.; Harroun, T.A.; Sternin, E. Location of chlorhexidine in DMPC model membranes: A neutron diffraction study. Chem. Phys. Lipids 2010, 163, 480–487. [Google Scholar] [CrossRef]
- Barrett-Bee, K.; Newboult, L.; Edwards, S. The membrane destabilising action of the antibacterial agent chlorhexidine. FEMS Microbiol. Lett. 1994, 119, 249–253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheung, H.-Y.; Wong, M.M.-K.; Cheung, S.-H.; Liang, L.Y.; Lam, Y.-W.; Chiu, S.-K. Differential actions of chlorhexidine on the cell wall of Bacillus subtilis and Escherichia coli. PLoS ONE 2012, 7, e36659. [Google Scholar] [CrossRef] [PubMed]
- Eslami, H.; Das, S.; Zhou, T.; Müller-Plathe, F. How alcoholic disinfectants affect coronavirus model membranes: Membrane fluidity, permeability, and disintegration. J. Phys. Chem. B 2020, 124, 10374–10385. [Google Scholar] [CrossRef] [PubMed]
- Gurtovenko, A.A.; Anwar, J. Interaction of ethanol with biological membranes: The formation of non-bilayer structures within the membrane interior and their significance. J. Phys. Chem. B 2009, 113, 1983–1992. [Google Scholar] [CrossRef]
- Kranenburg, M.; Vlaar, M.; Smit, B. Simulating induced interdigitation in membranes. Biophys. J. 2004, 87, 1596–1605. [Google Scholar] [CrossRef]
- Oh, J.Y.; Yu, J.M.; Ko, J.H. Analysis of ethanol effects on corneal epithelium. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3852–3856. [Google Scholar] [CrossRef]
- Sonmez, M.; Ince, H.Y.; Yalcin, O.; Ajdžanović, V.; Spasojević, I.; Meiselman, H.J.; Baskurt, O.K. The effect of alcohols on red blood cell mechanical properties and membrane fluidity depends on their molecular size. PLoS ONE 2013, 8, e76579. [Google Scholar] [CrossRef]
- Al-Tannir, M.A.; Goodman, H.S. A review of chlorhexidine and its use in special populations. Spec. Care Dent. 1994, 14, 116–122. [Google Scholar] [CrossRef]

| Outer Leaflet Plasma Membrane Composition Used in Simulations | ||
|---|---|---|
| Lipid | Structure | Mol % |
| PLPC | PC 16:0–18:2 | 15 |
| PAPC | PC 16:0–20:4 | 8 |
| POPC | PC 16:0–18:1 | 5 |
| PSM | PC 18:1–16:0 | 12 |
| LSM | PC 18:1–24:0 | 8 |
| NSM | PC 18:1–24:1 | 8 |
| SAPS | PS 18:0–20:4 | 2 |
| SAPI | PI 18:0–20:4 | 2 |
| CHOL | Cholesterol | 40 |
| Viral Membrane Composition Used in Simulations | ||
|---|---|---|
| Lipid | Structure | Mol % |
| POPC | PC 16:0–18:1 | 15 |
| DOPC | PC 18:1–18:1 | 35 |
| POPE | PE16:0–18:1 | 10 |
| DOPE | PE 18:1–18:1 | 20 |
| POPS | PS 16:0–18:1 | 2 |
| DOPS | PS 18:1–18:1 | 3 |
| SAPI | PI 18:0–20:4 | 10 |
| CHOL | Cholesterol | 5 |
| Simulation Type | Membrane | No. of CHX | No. of Lipids | No. of Waters | System Size (nm × nm × nm) | No. of Repeats | Total Time (µs) |
|---|---|---|---|---|---|---|---|
| Conventional | PM | - | 320 | 12,482 | 8.31 × 8.31 × 10.07 | 1 | 1 |
| VM | - | 320 | 16,401 | 9.88 × 9.88 × 9.09 | 1 | 1 | |
| PM | 8 | 320 | 18,215 | 8.35 × 8.35 × 12.50 | 3 | 3 | |
| VM | 8 | 320 | 22,991 | 10.13 × 10.13 × 10.63 | 3 | 3 | |
| PM | 1 | 160 | 7278 | 6.27 × 5.43 × 11.13 | 1 | 1 | |
| VM | 1 | 160 | 9598 | 7.51 × 6.50 × 9.95 | 1 | 1 | |
| Umbrella Sampling-Binding energy | PM | 1 | 160 | 7278 | 6.27 × 5.43 × 11.13 | 1 | 6.6 |
| PM | 1 | 160 | 9598 | 7.51 × 6.50 × 9.95 | 1 | 6.6 | |
| Umbrella Sampling-Pore formation | PM | - | 320 | 11,861 | 8.40 × 8.40 × 9.59 | 1 | 6.75 |
| PM | 8 | 320 | 11,877 | 8.39 × 8.39 × 9.66 | 1 | 6.75 | |
| PM | 24 | 320 | 11,845 | 8.54 × 8.54 × 9.50 | 1 | 6.75 | |
| VM | - | 320 | 13,342 | 9.97 × 9.97 × 7.98 | 1 | 6.75 | |
| VM | 8 | 320 | 13,342 | 10.03 × 10.03 × 7.94 | 1 | 6.75 | |
| VM | 24 | 320 | 13,342 | 10.17 × 10.17 × 7.86 | 1 | 6.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathod, A.K.; Poojari, C.S.; Manna, M. Is Lipid Specificity Key to the Potential Antiviral Activity of Mouthwash Reagent Chlorhexidine against SARS-CoV-2? Membranes 2022, 12, 616. https://doi.org/10.3390/membranes12060616
Rathod AK, Poojari CS, Manna M. Is Lipid Specificity Key to the Potential Antiviral Activity of Mouthwash Reagent Chlorhexidine against SARS-CoV-2? Membranes. 2022; 12(6):616. https://doi.org/10.3390/membranes12060616
Chicago/Turabian StyleRathod, Arun K., Chetan S. Poojari, and Moutusi Manna. 2022. "Is Lipid Specificity Key to the Potential Antiviral Activity of Mouthwash Reagent Chlorhexidine against SARS-CoV-2?" Membranes 12, no. 6: 616. https://doi.org/10.3390/membranes12060616
APA StyleRathod, A. K., Poojari, C. S., & Manna, M. (2022). Is Lipid Specificity Key to the Potential Antiviral Activity of Mouthwash Reagent Chlorhexidine against SARS-CoV-2? Membranes, 12(6), 616. https://doi.org/10.3390/membranes12060616







