Progress on Crowding Effect in Cell-like Structures
Abstract
1. Introduction
2. Impact of Crowding Media on the Physical and Chemical Properties of Biological Macromolecules
3. Crowding Effects Induced Actin Assembly Behavior in Confined Spaces
4. Aggregation Behavior of Tubulin in a Crowded Environment
5. Crowding Effect on Gene Expression
6. Summary and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Mittal, S.; Chowhan, R.K.; Singh, L.R. Macromolecular crowding: Macromolecules friend or foe. Acta Gen. Subj. 2015, 1850, 1822–1831. [Google Scholar] [CrossRef] [PubMed]
- Vibhute, M.A.; Schaap, M.H.; Maas, R.J.M.; Nelissen, F.H.T.; Spruijt, E.; Heus, H.A.; Hansen, M.M.K.; Huck, W.T.S. Transcription and Translation in Cytomimetic Protocells Perform Most Efficiently at Distinct Macromolecular Crowding Conditions. ACS Synth. Biol. 2020, 9, 2797–2807. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 2001, 26, 597–604. [Google Scholar] [CrossRef]
- Mourão, M.A.; Hakim, J.B.; Schnell, S. Connecting the Dots: The Effects of Macromolecular Crowding on Cell Physiology. Biophys. J. 2014, 107, 2761–2766. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, J.; Boersma, A.J.; Poolman, B. Microorganisms maintain crowding homeostasis. Nat. Rev. Microbiol. 2017, 15, 309–318. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Rivas, G.; Minton, A.P. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu. Rev. Biophys. 2008, 37, 375–397. [Google Scholar] [CrossRef] [PubMed]
- McGuffee, S.R.; Elcock, A.H. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010, 6, e1000694. [Google Scholar]
- Ge, X.; Luo, D.; Xu, J. Cell-free protein expression under macromolecular crowding conditions. PLoS ONE 2011, 6, e28707. [Google Scholar] [CrossRef]
- Zhao, H.; Ibrahimova, V.; Garanger, E.; Lecommandoux, S. Dynamic Spatial Formation and Distribution of Intrinsically Disordered Protein Droplets in Macromolecularly Crowded Protocells. Angew. Chem. Int. Ed. 2020, 132, 11121–11129. [Google Scholar] [CrossRef]
- Kulothungan, S.R.; Das, M.; Johnson, M.; Ganesh, C.; Varadarajan, R. Effect of Crowding Agents, Signal Peptide, and Chaperone SecB on the Folding and Aggregation of E. coli Maltose Binding Protein. Langmuir 2009, 25, 6637–6648. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. What Macromolecular Crowding Can Do to a Protein. Int. J. Mol. Sci. 2014, 15, 23090–23140. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. Excluded volume as a determinant of macromolecular structure and reactivity. Biopolymers 1981, 20, 2093–2120. [Google Scholar] [CrossRef]
- Minton, A.P. Quantitative assessment of the relative contributions of steric repulsion and chemical interactions to macromolecular crowding. Biopolymers 2013, 99, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Jeon, C.; Hyeon, C.; Jung, Y.; Ha, B.-Y. How are molecular crowding and the spatial organization of a biopolymer interrelated. Soft Matter 2016, 12, 9786–9796. [Google Scholar] [CrossRef]
- Hata, Y.; Sawada, T.; Serizawa, T. Macromolecular crowding for materials-directed controlled self-assembly. J. Mater. Chem. B 2018, 6, 6344–6359. [Google Scholar] [CrossRef] [PubMed]
- Roman, M.I.; Yang, G. Macromolecular Crowding Affects the Mechanical Unfolding Forces in Titin: The Size Effect. Biophys. J. 2011, 100, 214a. [Google Scholar] [CrossRef][Green Version]
- Mikaelsson, T.; Ådén, J.; Johansson, L.B.Å.; Wittung-Stafshede, P. Direct Observation of Protein Unfolded State Compaction in the Presence of Macromolecular Crowding. Biophys. J. 2013, 4, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Strulson, C.A.; Molden, R.C.; Keating, C.D.; Bevilacqua, P.C. RNA catalysis through compartmentalization. Nat. Chem. 2012, 4, 941–946. [Google Scholar] [CrossRef]
- De Pieri, A.; Rana, S.; Korntner, S.; Zeugolis, D.I. Seaweed polysaccharides as macromolecular crowding agents. Int. J. Biol. Macromol. 2020, 164, 434–446. [Google Scholar] [CrossRef]
- Candotti, M.; Orozco, M. The Differential Response of Proteins to Macromolecular Crowding. PLoS Comput. Biol. 2016, 12, e1005040. [Google Scholar] [CrossRef]
- Su, W.-C.; Gettel, D.L.; Chabanon, M.; Rangamani, P.; Parikh, A.N. Pulsatile Gating of Giant Vesicles Containing Macromolecular Crowding Agents Induced by Colligative Nonideality. J. Am. Chem. Soc. 2018, 140, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P.; Spulber, M.; Fischer, O.; Car, A.; Meier, W. Investigation of Horseradish Peroxidase Kinetics in an “Organelle-Like” Environment. Small 2017, 13, 1603943. [Google Scholar] [CrossRef] [PubMed]
- Shendi, D.; Marzi, J.; Linthicum, W.; Rickards, A.J.; Dolivo, D.M.; Keller, S.; Kauss, M.A.; Wen, Q.; McDevitt, T.C.; Dominko, T.; et al. Hyaluronic acid as a macromolecular crowding agent for production of cell-derived matrices. Acta Biomater. 2019, 100, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Garnica-Galvez, S.; Korntner, S.H.; Skoufos, I.; Tzora, A.; Diakakis, N.; Prassinos, N.; Zeugolis, D.I. Hyaluronic Acid as Macromolecular Crowder in Equine Adipose-Derived Stem Cell Cultures. Cells 2021, 10, 859. [Google Scholar] [CrossRef]
- Diniz, A.; Dias, J.S.; Jiménez-Barbero, J.; Marcelo, F.; Cabrita, E.J. Protein–Glycan Quinary Interactions in Crowding Environment Unveiled by NMR Spectroscopy. Chem.-Eur. J. 2017, 23, 13213–13220. [Google Scholar] [CrossRef]
- Bai, J.; Liu, M.; Pielak, G.J.; Li, C. Macromolecular and Small Molecular Crowding Have Similar Effects on α-Synuclein Structure. ChemPhysChem 2017, 18, 55–58. [Google Scholar] [CrossRef]
- Barro, L.; Burnouf, P.-A.; Chou, M.-L.; Nebie, O.; Wu, Y.-W.; Chen, M.-S.; Radosevic, M.; Knutson, F.; Burnouf, T. Human platelet lysates for human cell propagation. Platelets 2021, 32, 152–162. [Google Scholar] [CrossRef]
- Wang, X.; Tian, L.; Ren, Y.; Zhao, Z.; Du, H.; Zhang, Z.; Drinkwater, B.W.; Mann, S.; Han, X. Chemical Information Exchange in Organized Protocells and Natural Cell Assemblies with Controllable Spatial Positions. Small 2020, 16, 1906394. [Google Scholar] [CrossRef]
- Wang, X.; Tian, L.; Du, H.; Li, M.; Mu, W.; Drinkwater, B.W.; Han, X.; Mann, S. Chemical communication in spatially organized protocell colonies and protocell/living cell micro-arrays. Chem. Sci. 2019, 10, 9446–9453. [Google Scholar] [CrossRef]
- Zong, W.; Ma, S.; Zhang, X.; Wang, X.; Li, Q.; Han, X. A Fissionable Artificial Eukaryote-like Cell Model. J. Am. Chem. Soc. 2017, 139, 9955–9960. [Google Scholar] [CrossRef]
- Zhao, Q.-H.; Cao, F.-H.; Luo, Z.-H.; Huck, W.T.S.; Deng, N.-N. Photoswitchable Molecular Communication between Programmable DNA-Based Artificial Membraneless Organelles. Angew. Chem. Int. Ed. 2022, 61, e202117500. [Google Scholar] [CrossRef] [PubMed]
- McClintic, W.T.; Taylor, G.J.; Simpson, M.L.; Collier, C.P. Macromolecular Crowding Affects Voltage-Dependent Alamethicin Pore Formation in Lipid Bilayer Membranes. J. Phys. Chem. B 2020, 124, 5095–5102. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, M.; Sakaue, T.; Yoshikawa, K. Chapter Seven-Characteristic Behavior of Crowding Macromolecules Confined in Cell-Sized Droplets. Int. Rev. Cell Mol. Biol. 2014, 307, 175–204. [Google Scholar] [PubMed]
- Zhu, C.; Li, Q.; Dong, M.; Han, X. Giant Unilamellar Vesicle Microarrays for Cell Function Study. Anal. Chem. 2018, 90, 14363–14367. [Google Scholar] [CrossRef] [PubMed]
- Elani, Y. Interfacing Living and Synthetic Cells as an Emerging Frontier in Synthetic Biology. Angew. Chem. Int. Ed. 2021, 60, 5602–5611. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Chiba, M.; Eguchi, H.; Ohki, T.; Ishiwata, S.I. Cell-sized spherical confinement induces the spontaneous formation of contractile actomyosin rings in vitro. Nat. Cell Biol. 2015, 17, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Groen, J.; Foschepoth, D.; te Brinke, E.; Boersma, A.J.; Imamura, H.; Rivas, G.; Heus, H.A.; Huck, W.T.S. Associative Interactions in Crowded Solutions of Biopolymers Counteract Depletion Effects. J. Am. Chem. Soc. 2015, 137, 13041–13048. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Takiguchi, K.; Hayashi, M. Repetitive stretching of giant liposomes utilizing the nematic alignment of confined actin. Commun. Phys. 2018, 1, 18. [Google Scholar] [CrossRef]
- Garenne, D.; Noireaux, V. Analysis of Cytoplasmic and Membrane Molecular Crowding in Genetically Programmed Synthetic Cells. Biomacromolecules 2020, 21, 2808–2817. [Google Scholar] [CrossRef]
- Snead, W.T.; Hayden, C.C.; Gadok, A.K.; Zhao, C.; Lafer, E.M.; Rangamani, P.; Stachowiak, J.C. Membrane fission by protein crowding. Proc. Natl. Acad. Sci. USA 2017, 114, E3258–E3267. [Google Scholar] [CrossRef]
- Hernández-Vega, A.; Braun, M.; Scharrel, L.; Jahnel, M.; Wegmann, S.; Hyman, B.T.; Alberti, S.; Diez, S.; Hyman, A.A. Local Nucleation of Microtubule Bundles through Tubulin Concentration into a Condensed Tau Phase. Cell Rep. 2017, 20, 2304–2312. [Google Scholar] [CrossRef] [PubMed]
- King, M.R.; Petry, S. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun. 2020, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, J.B.; Ferreira Gomes, B.; Widlund, P.O.; Mahamid, J.; Honigmann, A.; Hyman, A.A. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 2017, 169, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, N.; Sakuta, H.; Hayashi, M.; Tanaka, S.; Takiguchi, K.; Tsumoto, K.; Yoshikawa, K. Specific Spatial Localization of Actin and DNA in a Water/Water Microdroplet: Self-Emergence of a Cell-Like Structure. ChemBioChem 2018, 19, 1370–1374. [Google Scholar] [CrossRef] [PubMed]
- Stano, P.; D’Aguanno, E.; Bolz, J.; Fahr, A.; Luisi, P.L. A Remarkable Self-Organization Process as the Origin of Primitive Functional Cells. Angew. Chem. Int. Ed. 2013, 125, 13639–13642. [Google Scholar] [CrossRef]
- Hansen, M.M.K.; Meijer, L.H.H.; Spruijt, E.; Maas, R.J.M.; Rosquelles, M.V.; Groen, J.; Heus, H.A.; Huck, W.T.S. Macromolecular crowding creates heterogeneous environments of gene expression in picolitre droplets. Nat. Nanotechnol. 2016, 11, 191–197. [Google Scholar] [CrossRef]
- Tan, C.; Saurabh, S.; Bruchez, M.P.; Schwartz, R.; LeDuc, P. Molecular crowding shapes gene expression in synthetic cellular nanosystems. Nat. Nanotechnol. 2013, 8, 602–608. [Google Scholar] [CrossRef]
- Norred, S.E.; Caveney, P.M.; Chauhan, G.; Collier, L.K.; Collier, C.P.; Abel, S.M.; Simpson, M.L. Macromolecular Crowding Induces Spatial Correlations That Control Gene Expression Bursting Patterns. ACS Synth. Biol. 2018, 7, 1251–1258. [Google Scholar] [CrossRef]
- Stadmiller, S.S.; Pielak, G.J. Protein-complex stability in cells and in vitro under crowded conditions. Curr. Opin. Struct. Biol. 2021, 66, 183–192. [Google Scholar] [CrossRef]
- Schavemaker, P.E.; Boersma, A.J.; Poolman, B. How Important Is Protein Diffusion in Prokaryotes? Front. Mol. Biosci. 2018, 5, 93. [Google Scholar] [CrossRef]
- Popielec, A.; Ostrowska, N.; Wojciechowska, M.; Feig, M.; Trylska, J. Crowded environment affects the activity and inhibition of the NS3/4A protease. Biochimie 2020, 176, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Lemus, E.; Reyes, J.S.; Gamon, L.F.; Lopez-Alarcon, C.; Davies, M.J. Effect of macromolecular crowding on protein oxidation: Consequences on the rate, extent and oxidation pathways. Redox Biol. 2021, 48, 102202. [Google Scholar] [CrossRef] [PubMed]
- Tabaka, M.; Kalwarczyk, T.; Szymanski, J.; Hou, S.; Holyst, R. The effect of macromolecular crowding on mobility of biomolecules, association kinetics, and gene expression in living cells. Front. Phys. 2014, 2, 54. [Google Scholar] [CrossRef]
- Miermont, A.; Waharte, F.; Hu, S.; McClean, M.N.; Bottani, S.; Léon, S.; Hersen, P. Severe osmotic compression triggers a slowdown of intracellular signaling, which can be explained by molecular crowding. Proc. Natl. Acad. Sci. USA 2013, 110, 5725–5730. [Google Scholar] [CrossRef] [PubMed]
- Golkaram, M.; Hellander, S.; Drawert, B.; Petzold, L.R. Macromolecular Crowding Regulates the Gene Expression Profile by Limiting Diffusion. PLoS Comput. Biol. 2016, 12, e1005122. [Google Scholar] [CrossRef] [PubMed]
- Junker, N.O.; Vaghefikia, F.; Albarghash, A.; Hofig, H.; Kempe, D.; Walter, J.; Otten, J.; Pohl, M.; Katranidis, A.; Wiegand, S.; et al. Impact of Molecular Crowding on Translational Mobility and Conformational Properties of Biological Macromolecules. J. Phys. Chem. B 2019, 123, 4477–4486. [Google Scholar] [CrossRef] [PubMed]
- Várkuti, B.H.; Yang, Z.; Kintses, B.; Erdélyi, P.; Bárdos-Nagy, I.; Kovács, A.L.; Hári, P.; Kellermayer, M.; Vellai, T.; Málnási-Csizmadia, A. A novel actin binding site of myosin required for effective muscle contraction. Nat. Struct. Mol. Biol. 2012, 19, 299–306. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Weirich Kimberly, L.; Dasbiswas, K.; Witten Thomas, A.; Vaikuntanathan, S.; Gardel Margaret, L. Self-organizing motors divide active liquid droplets. Proc. Natl. Acad. Sci. USA 2019, 116, 11125–11130. [Google Scholar] [CrossRef] [PubMed]
- Weirich Kimberly, L.; Banerjee, S.; Dasbiswas, K.; Witten Thomas, A.; Vaikuntanathan, S.; Gardel Margaret, L. Liquid behavior of cross-linked actin bundles. Proc. Natl. Acad. Sci. USA 2017, 114, 2131–2136. [Google Scholar] [CrossRef]
- Mishra, A.; Korlepara, D.B.; Kumar, M.; Jain, A.; Jonnalagadda, N.; Bejagam, K.K.; Balasubramanian, S.; George, S.J. Biomimetic temporal self-assembly via fuel-driven controlled supramolecular polymerization. Nat. Commun. 2018, 9, 1295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Grange, M.; Pospich, S.; Wagner, T.; Kho Ay, L.; Gautel, M.; Raunser, S. Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin. Science 2022, 375, eabn1934. [Google Scholar] [CrossRef] [PubMed]
- Baumann, H.; Surrey, T. Motor-mediated Cortical versus Astral Microtubule Organization in Lipid-monolayered Droplets. J. Biol. Chem. 2014, 289, 22524–22535. [Google Scholar] [CrossRef] [PubMed]
- Ganar, K.A.; Honaker, L.W.; Deshpande, S. Shaping synthetic cells through cytoskeleton-condensate-membrane interactions. Curr. Opin. Colloid Interface Sci. 2021, 54, 101459. [Google Scholar] [CrossRef]
- Lee, K.Y.; Park, S.-J.; Lee, K.A.; Kim, S.-H.; Kim, H.; Meroz, Y.; Mahadevan, L.; Jung, K.-H.; Ahn, T.K.; Parker, K.K.; et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 2018, 36, 530–535. [Google Scholar] [CrossRef]
- Tsai, F.-C.; Koenderink, G.H. Shape control of lipid bilayer membranes by confined actin bundles. Soft Matter 2015, 11, 8834–8847. [Google Scholar] [CrossRef]
- Park, S.; Barnes, R.; Lin, Y.; Jeon, B.-j.; Najafi, S.; Delaney, K.T.; Fredrickson, G.H.; Shea, J.-E.; Hwang, D.S.; Han, S. Dehydration entropy drives liquid-liquid phase separation by molecular crowding. Commun. Chem. 2020, 3, 83. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Wei, S.-P.; Qian, Z.-G.; Hu, C.-F.; Pan, F.; Chen, M.-T.; Lee, S.Y.; Xia, X.-X. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol. 2020, 16, 1143–1148. [Google Scholar] [CrossRef]
- Kohata, K.; Miyoshi, D. RNA phase separation–mediated direction of molecular trafficking under conditions of molecular crowding. Biophys. Rev. 2020, 12, 669–676. [Google Scholar] [CrossRef]
- Lin, Y.; Fichou, Y.; Longhini, A.P.; Llanes, L.C.; Yin, P.; Bazan, G.C.; Kosik, K.S.; Han, S. Liquid-Liquid Phase Separation of Tau Driven by Hydrophobic Interaction Facilitates Fibrillization of Tau. J. Mol. Biol. 2021, 433, 166731. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, S.; Eftekharzadeh, B.; Tepper, K.; Zoltowska, K.M.; Bennett, R.E.; Dujardin, S.; Laskowski, P.R.; MacKenzie, D.; Kamath, T.; Commins, C.; et al. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 2018, 37, e98049. [Google Scholar] [CrossRef] [PubMed]
- Alfaro-Aco, R.; Thawani, A.; Petry, S. Structural analysis of the role of TPX2 in branching microtubule nucleation. J. Cell Biol. 2017, 216, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.C. Centrosome structure and biogenesis: Variations on a theme? Cell Dev. Biol. 2021, 110, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, H.; Ueda, T. Purified cell-free systems as standard parts for synthetic biology. Curr. Opin. Chem. Biol. 2014, 22, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Timm, A.C.; Shankles, P.G.; Foster, C.M.; Doktycz, M.J.; Retterer, S.T. Microreactors: Toward Microfluidic Reactors for Cell-Free Protein Synthesis at the Point-of-Care. Small 2016, 12, 690. [Google Scholar] [CrossRef]
- Shang, L.; Zhao, Y. Droplet-Templated Synthetic Cells. Matter 2021, 4, 95–115. [Google Scholar] [CrossRef]
- Wang, C.; Geng, Y.; Sun, Q.; Xu, J.; Lu, Y. A Sustainable and Efficient Artificial Microgel System: Toward Creating a Configurable Synthetic Cell. Small 2020, 16, 2002313. [Google Scholar] [CrossRef]
- Skóra, T.; Vaghefikia, F.; Fitter, J.; Kondrat, S. Macromolecular Crowding: How Shape and Interactions Affect Diffusion. J. Phys. Chem. B 2020, 124, 7537–7543. [Google Scholar] [CrossRef]
- Vazquez, A.; Oltvai, Z.N. Macromolecular crowding explains overflow metabolism in cells. Sci. Rep. 2016, 6, 31007. [Google Scholar] [CrossRef]
- Shim, A.R.; Nap, R.J.; Huang, K.; Almassalha, L.M.; Matusda, H.; Backman, V.; Szleifer, I. Dynamic Crowding Regulates Transcription. Biophys. J. 2020, 118, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Benny, P.; Raghunath, M. Making microenvironments: A look into incorporating macromolecular crowding into in vitro experiments, to generate biomimetic microenvironments which are capable of directing cell function for tissue engineering applications. J. Tissue Eng. 2017, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Assuncao, M.; Wong, C.W.; Richardson, J.J.; Tsang, R.; Beyer, S.; Raghunath, M.; Blocki, A. Macromolecular dextran sulfate facilitates extracellular matrix deposition by electrostatic interaction independent from a macromolecular crowding effect. Mater. Sci. Eng. C 2020, 106, 110280. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.-I.; Ko, K.-W.; Cha, S.-G.; Park, S.-Y.; Woo, J.; Han, D.K. Highly effective induction of cell-derived extracellular matrix by macromolecular crowding for osteogenic differentiation of mesenchymal stem cells. J. Ind. Eng. Chem. 2022, 107, 391–400. [Google Scholar] [CrossRef]
- Bascetin, R.; Laurent-Issartel, C.; Blanc-Fournier, C.; Vendrely, C.; Kellouche, S.; Carreiras, F.; Gallet, O.; Leroy-Dudal, J. A biomimetic model of 3D fluid extracellular macromolecular crowding microenvironment fine-tunes ovarian cancer cells dissemination phenotype. Biomaterials 2021, 269, 120610. [Google Scholar] [CrossRef] [PubMed]
- Lowe, M.; Kalacheva, M.; Boersma, A.J.; Kedrov, A. The more the merrier: Effects of macromolecular crowding on the structure and dynamics of biological membranes. FEBS J. 2020, 287, 5039–5067. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jiang, N.; Lai, Y.T.; Chang, Y.Y.; Yang, X.; Sun, H.; Li, H. Green Fluorescent Probe for Imaging His6-Tagged Proteins Inside Living Cells. ACS Sens. 2019, 4, 1190–1196. [Google Scholar] [CrossRef]
- Horton, M.R.; Höfling, F.; Rädler, J.O.; Franosch, T. Development of anomalous diffusion among crowding proteins. Soft Matter 2010, 6, 2648–2656. [Google Scholar] [CrossRef]
- Garenne, D.; Libchaber, A.; Noireaux, V. Membrane molecular crowding enhances MreB polymerization to shape synthetic cells from spheres to rods. Proc. Natl. Acad. Sci. USA 2020, 117, 1902–1909. [Google Scholar] [CrossRef]
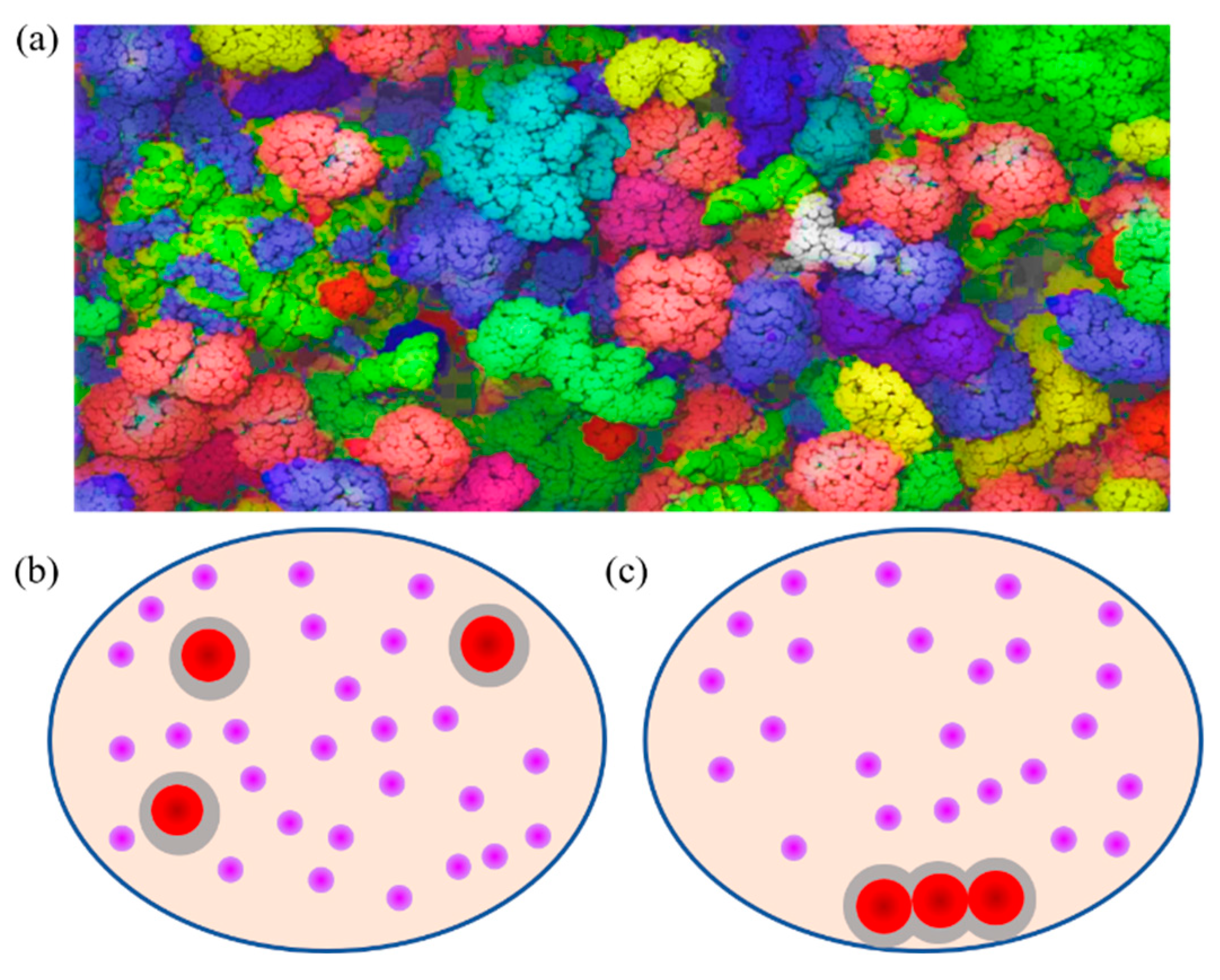
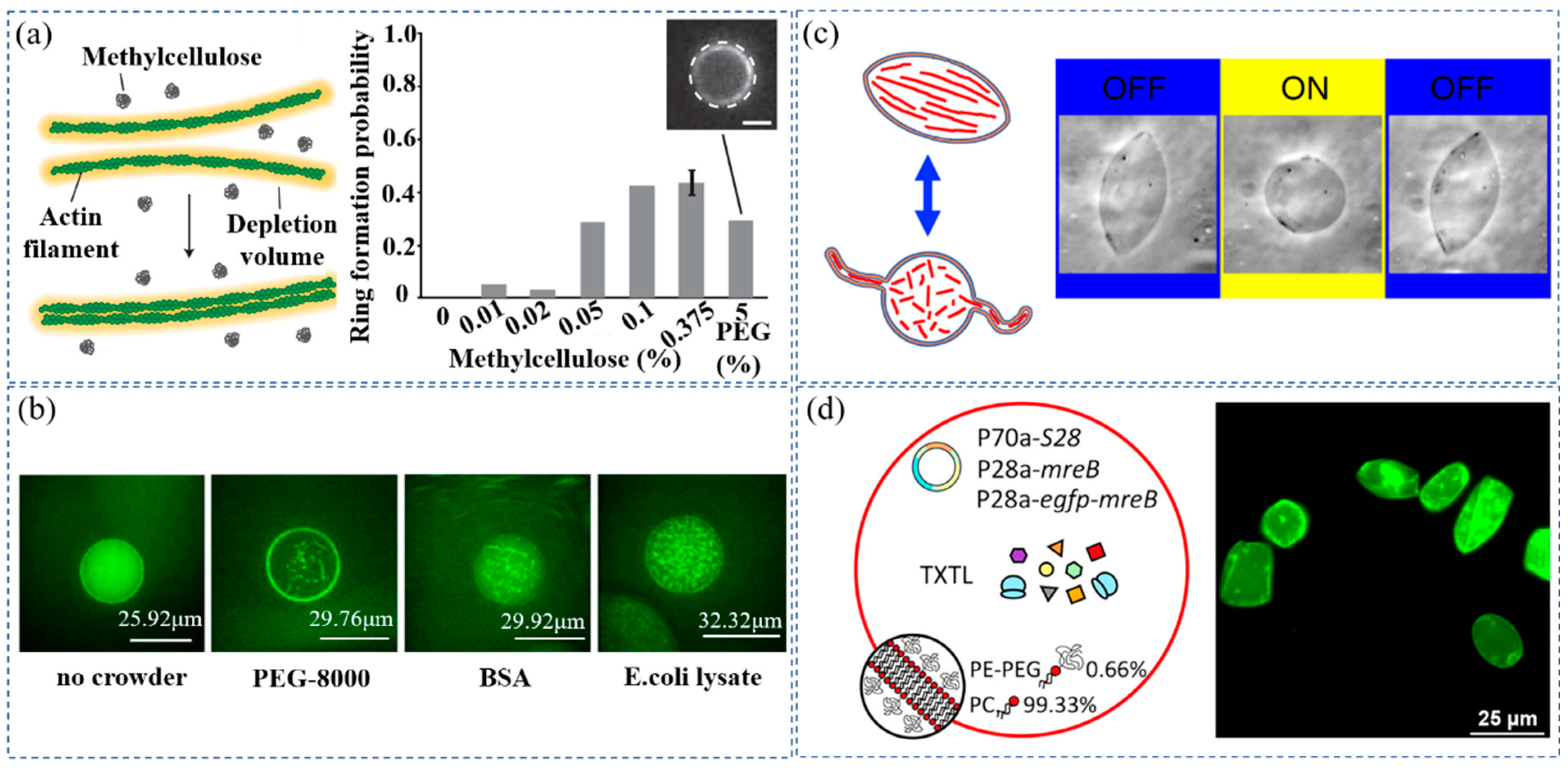
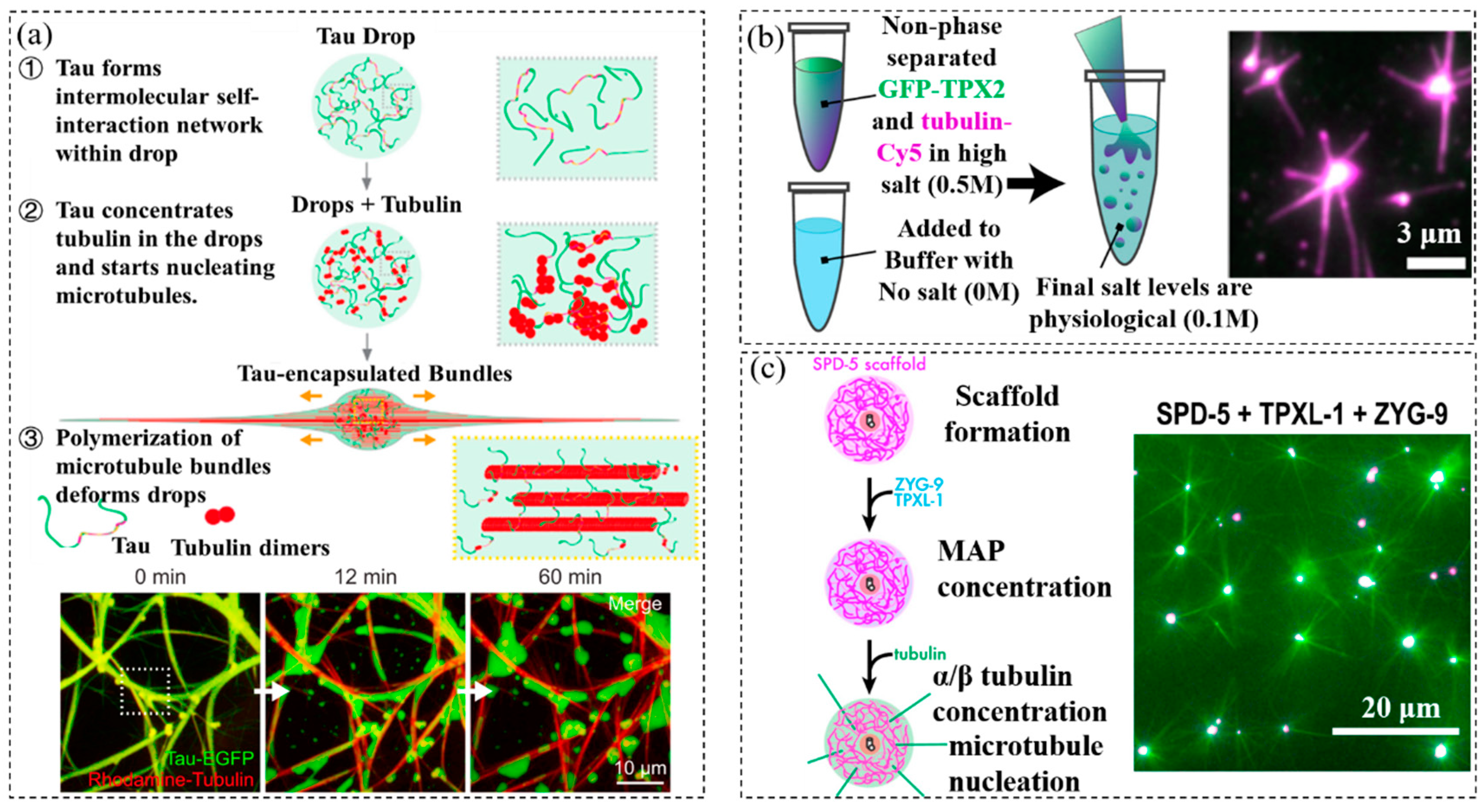
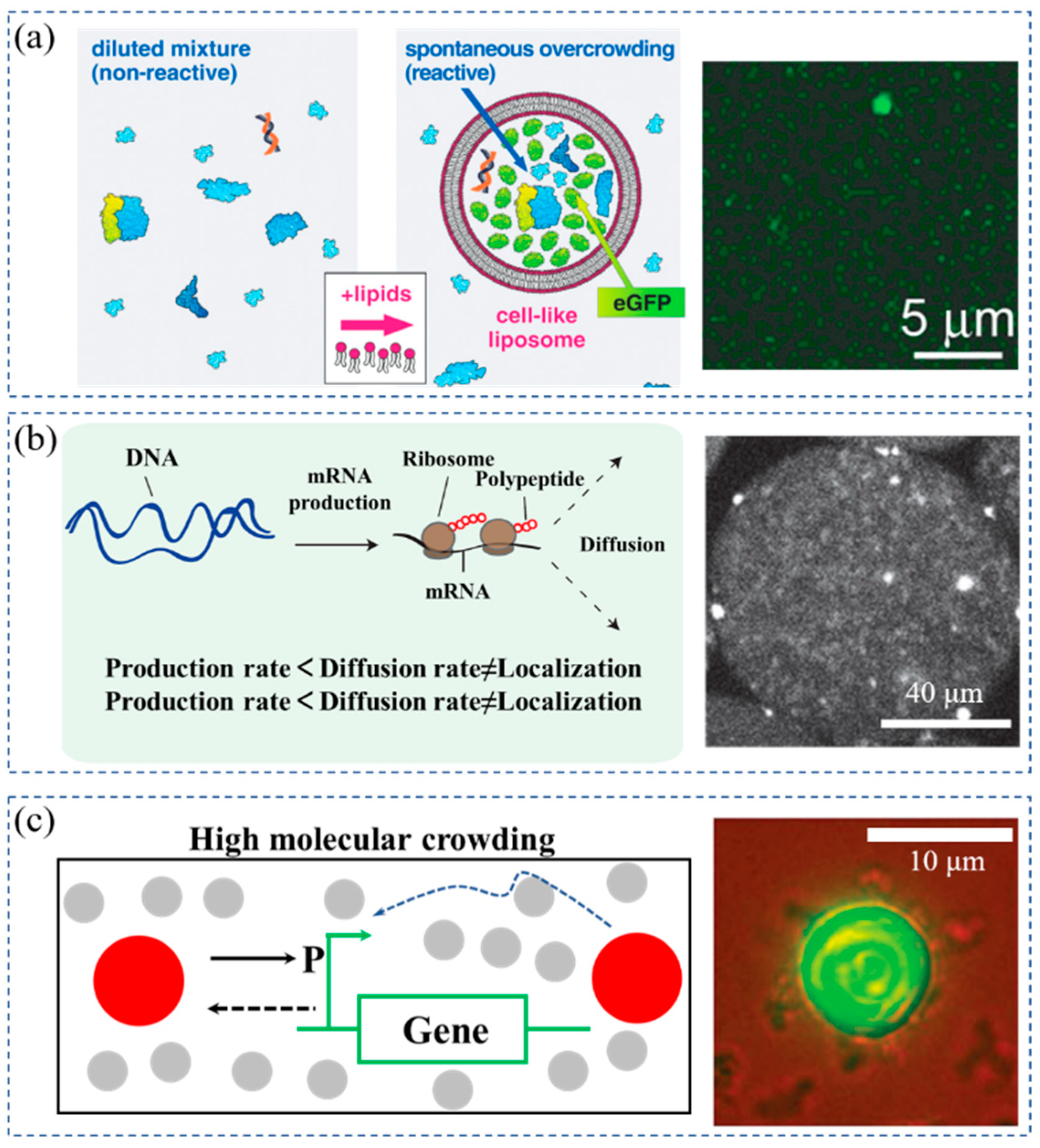
| Crowded System | Physiological Function of Crowding Effect | Confined Space | Reference |
|---|---|---|---|
| Methylcellulose, actin | Ring-shaped actin bundles assembled at the inner peripheries | Water-in-oil droplets | [36] |
| FtsZ, PEG-8000, BSA, E. coli lysates | FtsZ bundle formation in microdroplets | Water-in-oil droplets | [37] |
| Actin | Actin filaments aggregated into thick actin bundles within Giant unillamellar vesicles (GUVs) to deform GUVs into spindle shapes | Giant unillamellar vesicles | [38] |
| PC, PE-PEG, cell-free protein-synthesis system of MreB | The polymerization of the protein MreB at the inner membrane into a sturdy cytoskeleton capable of transforming spherical GUVs into elongated shapes | Giant unillamellar vesicles | [39] |
| Endocytosis protein (Epsin1), green fluorescent protein | Epsin1 or GFP were able to drive fission efficiently when bound to the membrane at high coverage | Surface of phospholipid vesicle membrane | [40] |
| Dextran, polyethylene glycol, ficoll, Tau protein, tubulin | Tubulin partitioned into Tau drops, efficiently increasing tubulin concentration and driving the nucleation of microtubules | Phase-separated protein droplets | [41] |
| TPX2 protein, tubulin | Phase separation of TPX2 and tubulin could underlie the tenfold improvement in the branching MT nucleation efficiency | Phase-separated protein droplets | [42] |
| SPD-5 protein, ficoll, dextran, lysozyme | Tubulin was concentrated 4-fold over background to promote tubulin nucleation | Phase-separated protein droplets | [43] |
| Polyethylene glycol, dextran, actin, long DNA | Actin bundles distributed across the phase interface, deforming the interface and pushing DNA to their ends | Interfacial layer of liquid–liquid phase separation | [44] |
| Polyethylene glycol, dextran, ELP protein | ELP protein droplets were distributed near the interface between the two phases | Interfacial layer of liquid–liquid phase separation | [9] |
| Cell-free protein-synthesis system | Green fluorescent protein was expressed | Phospholipid vesicles | [45] |
| Ficoll, cell-free protein synthesis-system of cyan and yellow fluorescent protein | An order-of-magnitude decrease in the diffusion coefficients of RNA and proteins | Water-in-oil droplets | [46] |
| Dextran, cell-free protein-synthesis system of GFP | An increase in the robustness of gene expression | Giant unillamellar vesicles | [47] |
| Ficoll, cell-free protein-synthesis system of GFP | A 10-fold increase in protein noise | Giant unillamellar vesicles | [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Zhang, X.; Dong, M.; Han, X. Progress on Crowding Effect in Cell-like Structures. Membranes 2022, 12, 593. https://doi.org/10.3390/membranes12060593
Li C, Zhang X, Dong M, Han X. Progress on Crowding Effect in Cell-like Structures. Membranes. 2022; 12(6):593. https://doi.org/10.3390/membranes12060593
Chicago/Turabian StyleLi, Chao, Xiangxiang Zhang, Mingdong Dong, and Xiaojun Han. 2022. "Progress on Crowding Effect in Cell-like Structures" Membranes 12, no. 6: 593. https://doi.org/10.3390/membranes12060593
APA StyleLi, C., Zhang, X., Dong, M., & Han, X. (2022). Progress on Crowding Effect in Cell-like Structures. Membranes, 12(6), 593. https://doi.org/10.3390/membranes12060593








