The Contribution of Hydrophobic Interactions to Conformational Changes of Inward/Outward Transmembrane Transport Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. The FOD/FOD-M Models
3. Results
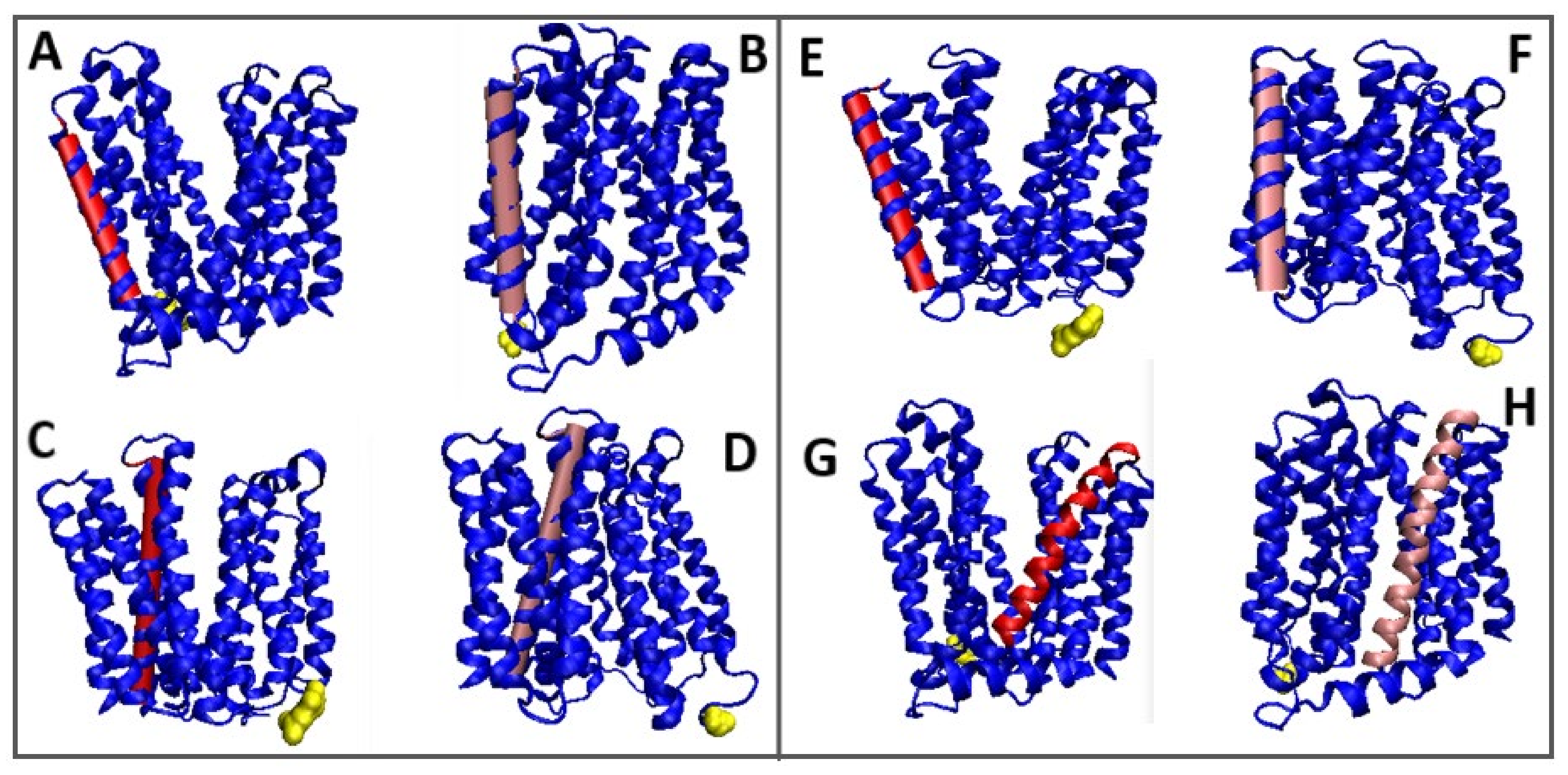
3.1. The Analysis of Individual Examples
- In the center of a protein there is a highly polar residue (for example lysine), which significantly reduces the level of hydrophobicity;
- In the center of a protein is a free space. In this situation, even residues with a high level of hydrophobicity (due to the reduced number of other residues with which interaction is possible) cause a reduction in the value of the observed hydrophobicity O (which is the sum of interactions).
3.1.1. Putative Bacterial Homologue of Ferroportin (BbFPN)
3.1.2. Representative of Solute Carrier 17 (SLC17)
3.1.3. Proton-Coupled Sugar Transporter Xy1E
3.1.4. Multidrug Transporter MdfA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vinothkumar, K.R.; Henderson, R. Structures of membrane proteins. Q. Rev. Biophys. 2010, 43, 65–158. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Mayor, S. Active organization of membrane constituents in living cells. Curr. Opin. Cell Biol. 2014, 29, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Bretscher, M.S. Membrane Structure: Some General Principles. Science 1973, 181, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Bretscher, M.S.; Raff, M.C. Mammalian plasma membranes. Nature 1975, 258, 43–49. [Google Scholar] [CrossRef]
- Von Heijne, G. Membrane proteins: From bench to bits. Biochem. Soc. Trans. 2011, 39, 747–750. [Google Scholar] [CrossRef]
- Bretscher, M.S. The Molecules of the Cell Membrane. Sci. Am. 1985, 253, 100–108. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid Rafts As a Membrane-Organizing Principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef]
- Levental, I.; Lingwood, D.; Grzybek, M.; Coskun, U.; Simons, K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 22050–22054. [Google Scholar] [CrossRef]
- Engelman, N.M.; Chen, Y.; Chin, C.-N.; Curran, A.R.; Dixon, A.M.; Dupuy, A.D.; Lee, A.S.; Lehnert, U.; Matthews, E.E.; Reshetnyak, Y.K.; et al. Membrane protein folding: Beyond the two stage model. FEBS Lett. 2003, 555, 122–125. [Google Scholar] [CrossRef]
- Curran, A.R.; Engelman, D.M. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 2003, 13, 412–417. [Google Scholar] [CrossRef]
- Martin, S.; Parton, R.G. Lipid droplets: A unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006, 7, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Hannesschlaeger, C.; Horner, A.; Pohl, P. Intrinsic Membrane Permeability to Small Molecules. Chem. Rev. 2019, 119, 5922–5953. [Google Scholar] [CrossRef] [PubMed]
- Kaldenhoff, R.; Kai, L.; Uehlein, N. Aquaporins and membrane diffusion of CO2 in living organisms. Biochim. Et Biophys. Acta 2014, 1840, 1592–1595. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.J. In Silico Prediction of Permeability Coefficients. Methods Mol. Biol. 2021, 2315, 243–261. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.H.; Morris, J.C. The Sodium-Iodide Symporter. Curr. Drug Targets Immune Endocr. Metab. Disord. 2004, 4, 167–174. [Google Scholar] [CrossRef]
- Abramson, J.; Wright, E.M. Structure and function of Na+-symporters with inverted repeats. Curr. Opin. Struct. Biol. 2009, 19, 425–432. [Google Scholar] [CrossRef]
- Dutta, D.; Fliegel, L. Structure and function of yeast and fungal Na+/H+antiporters. IUBMB Life 2018, 70, 23–31. [Google Scholar] [CrossRef]
- Alhadeff, R.; Warshel, A. Simulating the function of sodium/proton antiporters. Proc. Natl. Acad. Sci. USA 2015, 112, 12378–12383. [Google Scholar] [CrossRef]
- Brammer, A.E.; Stockbridge, R.; Miller, C. F−/Cl− selectivity in CLCF-type F−/H+ antiporters. J. Gen. Physiol. 2014, 144, 129–136. [Google Scholar] [CrossRef]
- Riedelsberger, J.; Miller, J.; Valdebenito-Maturana, B.; Piñeros, M.; González, W.; Dreyer, I. Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation. Int. J. Mol. Sci. 2021, 22, 1892. [Google Scholar] [CrossRef]
- Wolfersberger, M.G. Uniporters, symporters and antiporters. J. Exp. Biol. 1994, 196, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Gerencser, G.A.; Zhang, J. Existence and nature of the chloride pump. Biochim. Et Biophys. Acta 2003, 1618, 133–139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Skou, J.C. The sodium, potassium-pump. Scand. J. Clin. Lab. Invest. Suppl. 1986, 180, 11–23. [Google Scholar] [PubMed]
- Levitt, D.G. The mechanism of the sodium pump. Biochim. Et Biophys. Acta 1980, 604, 321–345. [Google Scholar] [CrossRef]
- Thomas, L.E.; Rocafull, M.A.; del Castillo, J.R. Is the Second Sodium Pump Electrogenic? BioMed Res. Int. 2013, 2013, 1–8. [Google Scholar] [CrossRef][Green Version]
- Kirk, K.L.; Halm, D.R.; Dawson, D.C. Active sodium transport by turtle colon via an electrogenic Na–K exchange pump. Nature 1980, 287, 237–239. [Google Scholar] [CrossRef]
- Functional and Phylogenetic Classification of Membrane Transport Proteins. Available online: https://www.tcdb.org/ (accessed on 22 July 2022).
- Sayer, M.H., Jr.; Reddy, V.S.; Tamang, D.G.; Västermark, Å. The Transporter Classification Database. Nucleic Acids Res. 2014, 42, D251–D258. [Google Scholar] [CrossRef]
- Saier, M.H.; Yen, M.R.; Noto, K.; Tamang, D.G.; Elkan, C. The Transporter Classification Database: Recent advances. Nucleic Acids Res. 2009, 37, D274–D278. [Google Scholar] [CrossRef]
- Saier, M.H. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006, 34, D181–D186. [Google Scholar] [CrossRef]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.B.; Meyer, E.F., Jr.; Brice, M.D.; Rodgers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M. The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 1977, 112, 535–542. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Berezovsky, I.N.; Kirzhner, V.M.; Kirzhner, A.; Trifonov, E.N. Protein folding: Looping from hydrophobic nuclei. Proteins Struct. Funct. Bioinform. 2001, 45, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Klevanik, A.V. Hydrophobicity and prediction of the secondary structure of membrane proteins and peptides. Membr. Cell Biol. 2001, 14, 673–697. [Google Scholar] [PubMed]
- Reuben, M.A.; Lasater, L.S.; Sachs, G. Characterization of a beta subunit of the gastric H+/K(+)-transporting ATPase. Proc. Natl. Acad. Sci. USA 1990, 87, 6767–6771. [Google Scholar] [CrossRef] [PubMed]
- Kister, A.E.; Phillips, J.C. A stringent test for hydrophobicity scales: Two proteins with 88% sequence identity but different structure and function. Proc. Natl. Acad. Sci. USA 2008, 105, 9233–9237. [Google Scholar] [CrossRef] [PubMed]
- Tcheremenskaia, O.; Giuliani, A.; Tomasi, M. PROFALIGN Algorithm Identifies the Regions Containing Folding Determinants by Scoring Pairs of Hydrophobic Profiles of Remotely Related Proteins. J. Comput. Biol. 2008, 15, 445–455. [Google Scholar] [CrossRef]
- Juretić, D.; Zucić, D.; Lučić, B.; Trinajstić, N. Preference functions for prediction of membrane-buried helices in integral membrane proteins. Comput. Chem. 1998, 22, 279–294. [Google Scholar] [CrossRef]
- Gribskov, M.; Burgess, R.R.; Devereux, J. PEPPLOT, a protein secondary structure analysis program for the UWGCG sequence analysis software package. Nucleic Acids Res. 1986, 14, 327–334. [Google Scholar] [CrossRef][Green Version]
- Orgel, J.P. Sequence context and modified hydrophobic moment plots help identify ‘horizontal’ surface helices in transmembrane protein structure prediction. J. Struct. Biol. 2004, 148, 51–65. [Google Scholar] [CrossRef] [PubMed]
- De Marothy, M.T.; Elofsson, A. Marginally hydrophobic transmembraneα-helices shaping membrane protein folding. Protein Sci. 2015, 24, 1057–1074. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Rath, A.; Johnson, R.M.; Deber, C.M. Distinctions between Hydrophobic Helices in Globular Proteins and Transmembrane Segments as Factors in Protein Sorting. J. Biol. Chem. 2009, 284, 5395–5402. [Google Scholar] [CrossRef] [PubMed]
- White, S.H.; Wimley, W.C. Membrane protein folding and stability: Physical Principles. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 319–365. [Google Scholar] [CrossRef] [PubMed]
- Hristova, K.; Wimley, W.C.; Mishra, V.K.; Anantharamiah, G.M.; Segrest, J.P.; White, S.H. An amphipathic α-helix at a membrane interface: A structural study using a novel X-ray diffraction method. J. Mol. Biol. 1999, 290, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Pogozheva, I.D.; Mosberg, H.I.; Lomize, A.L. Life at the border: Adaptation of proteins to anisotropic membrane environment. Protein Sci. 2014, 23, 1165–1196. [Google Scholar] [CrossRef]
- Whitelegge, J.P. Integral Membrane Proteins and Bilayer Proteomics. Anal. Chem. 2013, 85, 2558–2568. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Fabian, P.; Konieczny, L.; Banach, M. Model of Environmental Membrane Field for Transmembrane Proteins. Int. J. Mol. Sci. 2021, 22, 3619. [Google Scholar] [CrossRef]
- Banach, M.; Konieczny, L.; Roterman, I. Protein-protein interaction encoded as an exposure of hydrophobic residues on the surface. In From Globular Proteins to Amyloids; Elsevier: Amsterdam, The Netherlands, 2019; pp. 79–89. [Google Scholar] [CrossRef]
- Banach, M.; Konieczny, L.; Roterman, I. Ligand binding cavity encoded as a local hydrophobicity deficiency. In From Globular Proteins to Amyloids; Roterman-Konieczna, I., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 91–95. [Google Scholar]
- Taniguchi, R.; Kato, H.E.; Font, J.; Deshpande, C.N.; Wada, M.; Ito, K.; Ishitani, R.; Jormakka, M.; Nureki, O. Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Leano, J.B.; Batarni, S.; Eriksen, J.; Juge, N.; Pak, J.E.; Kimura-Someya, T.; Robles-Colmenares, Y.; Moriyama, Y.; Stroud, R.M.; Edwards, R.H. Structures suggest a mechanism for energy coupling by a family of organic anion transporters. PLoS Biol. 2019, 17, e3000260. [Google Scholar] [CrossRef]
- Wisedchaisri, G.; Park, M.-S.; Iadanza, M.G.; Zheng, H.; Gonen, T. Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat. Commun. 2014, 5, 4521. [Google Scholar] [CrossRef]
- Sun, L.; Zeng, X.; Yan, C.; Sun, X.; Gong, X.; Rao, Y.; Yan, N. Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 2012, 490, 361–366. [Google Scholar] [CrossRef]
- Heng, J.; Zhao, Y.; Liu, M.; Liu, Y.; Fan, J.; Wang, X.; Zhao, Y.; Zhang, X.C. Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res. 2015, 25, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Nagarathinam, K.; Nakada-Nakura, Y.; Parthier, C.; Terada, T.; Juge, N.; Jaenecke, F.; Liu, K.; Hotta, Y.; Miyaji, T.; Omote, H.; et al. Outward open conformation of a Major Facilitator Superfamily multidrug/H+ antiporter provides insights into switching mechanism. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kauzmann, W. Structural factors in protein denaturation. J. Cell. Comp. Physiol. 1956, 47, 113–131. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, L.; Roterman, I. Description of Fuzzy Oil Drop Model in: From Globular Proteins to Amyloids; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–12. [Google Scholar]
- Levitt, M. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976, 104, 59–107. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Gądek, K.; Gubała, T.; Nowakowski, P.; Fabian, P.; Konieczny, L. Dependence of Protein Structure on Environment: FOD Model Applied to Membrane Proteins. Membranes 2022, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Roterman, I.; Stapor, K.; Fabian, P.; Konieczny, L. The Functional Significance of Hydrophobic Residue Distribution in Bacterial Beta-Barrel Transmembrane Proteins. Membranes 2021, 11, 580. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Gądek, K.; Gubała, T.; Nowakowski, P.; Fabian, P.; Konieczny, L. On the Dependence of Prion and Amyloid Structure on the Folding Environment. Int. J. Mol. Sci. 2021, 22, 13494. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Fabian, P.; Konieczny, L. Connexins and Pannexins—Similarities and Differences According to the FOD-M Model. Biomedicines 2022, 10, 1504. [Google Scholar] [CrossRef]
- Yu, J.; Wessling-Resnick, M. Structural and Functional Analysis of SFT, a Stimulator of Fe Transport. J. Biol. Chem. 1998, 273, 21380–21385. [Google Scholar] [CrossRef]
- Cappellini, M.D.; Musallam, K.M.; Taher, A.T. Iron deficiency anaemia revisited. J. Intern. Med. 2020, 287, 153–170. [Google Scholar] [CrossRef]
- Zobnina, V.; Roterman, I. Application of the fuzzy-oil-drop model to membrane protein simulation. Proteins Struct. Funct. Bioinform. 2009, 77, 378–394. [Google Scholar] [CrossRef] [PubMed]
- Serpell, L.C. Alzheimer’s amyloid fibrils: Structure and assembly. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2000, 1502, 16–30. [Google Scholar] [CrossRef]
- Roterman, I.; Sieradzan, A.; Stapor, K.; Fabian, P.; Wesołowski, P.; Konieczny, L. On the need to introduce environmental characteristics in ab initio protein structure prediction using a coarse-grained UNRES force field. J. Mol. Graph. Model. 2022, 114, 108166. [Google Scholar] [CrossRef] [PubMed]
- Chelli, R.; Pagliai, M.; Procacci, P.; Cardini, G.; Schettino, V. Polarization response of water and methanol investigated by a polarizable force field and density functional theory calculations: Implications for charge transfer. J. Chem. Phys. 2005, 122, 074504. [Google Scholar] [CrossRef] [PubMed]
- Ben-Amotz, D. Electric buzz in a glass of pure water. Science 2022, 376, 800–801. [Google Scholar] [CrossRef] [PubMed]
- Pullanchery, S.; Kulik, S.; Rehl, B.; Hassanali, A.; Roke, S. Charge transfer across C–H⋅⋅⋅O hydrogen bonds stabilizes oil droplets in water. Science 2021, 374, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Wise, P.K.; Ben-Amotz, D. Interfacial Adsorption of Neutral and Ionic Solutes in a Water Droplet. J. Phys. Chem. B 2017, 122, 3447–3453. [Google Scholar] [CrossRef]
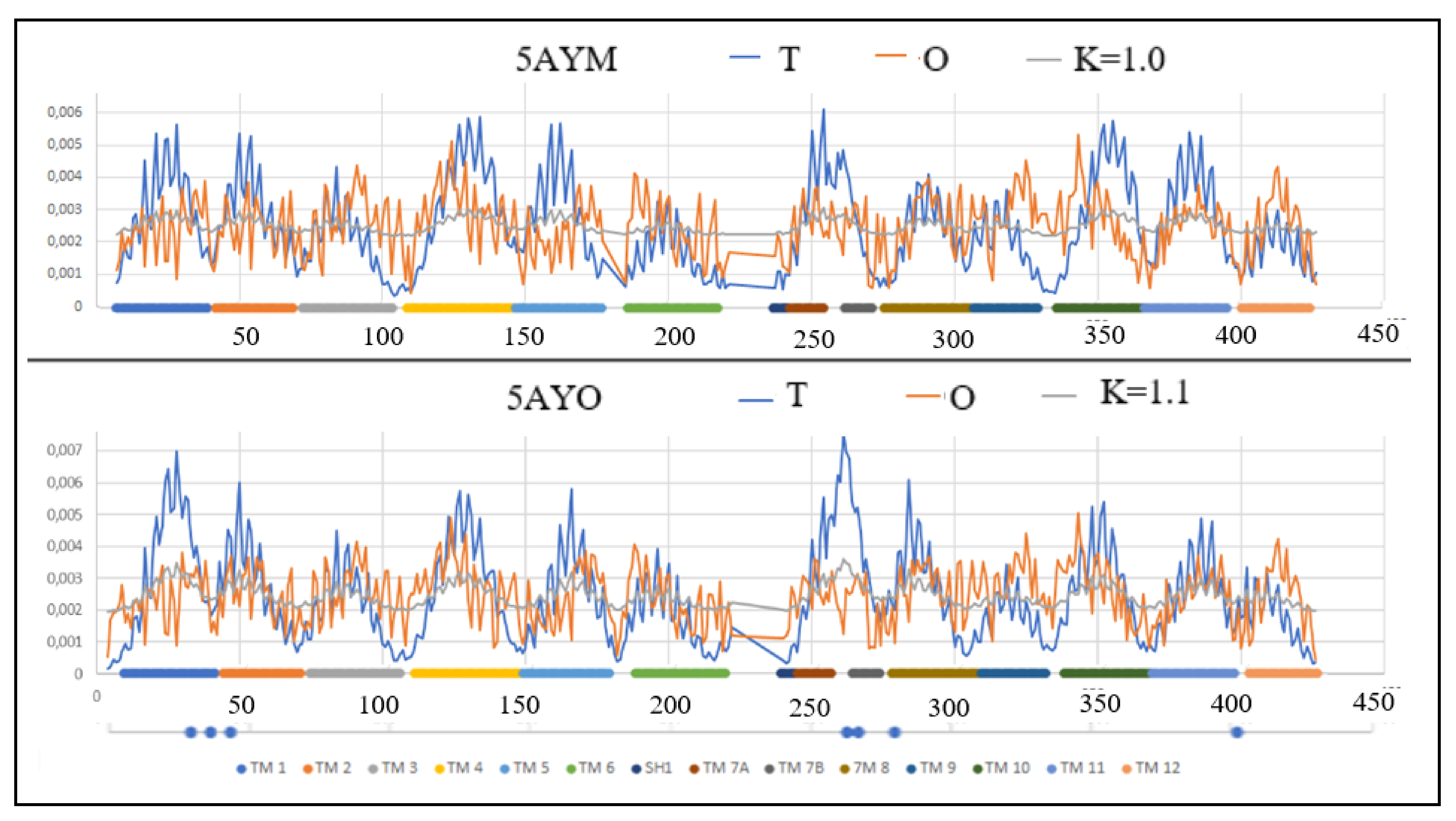
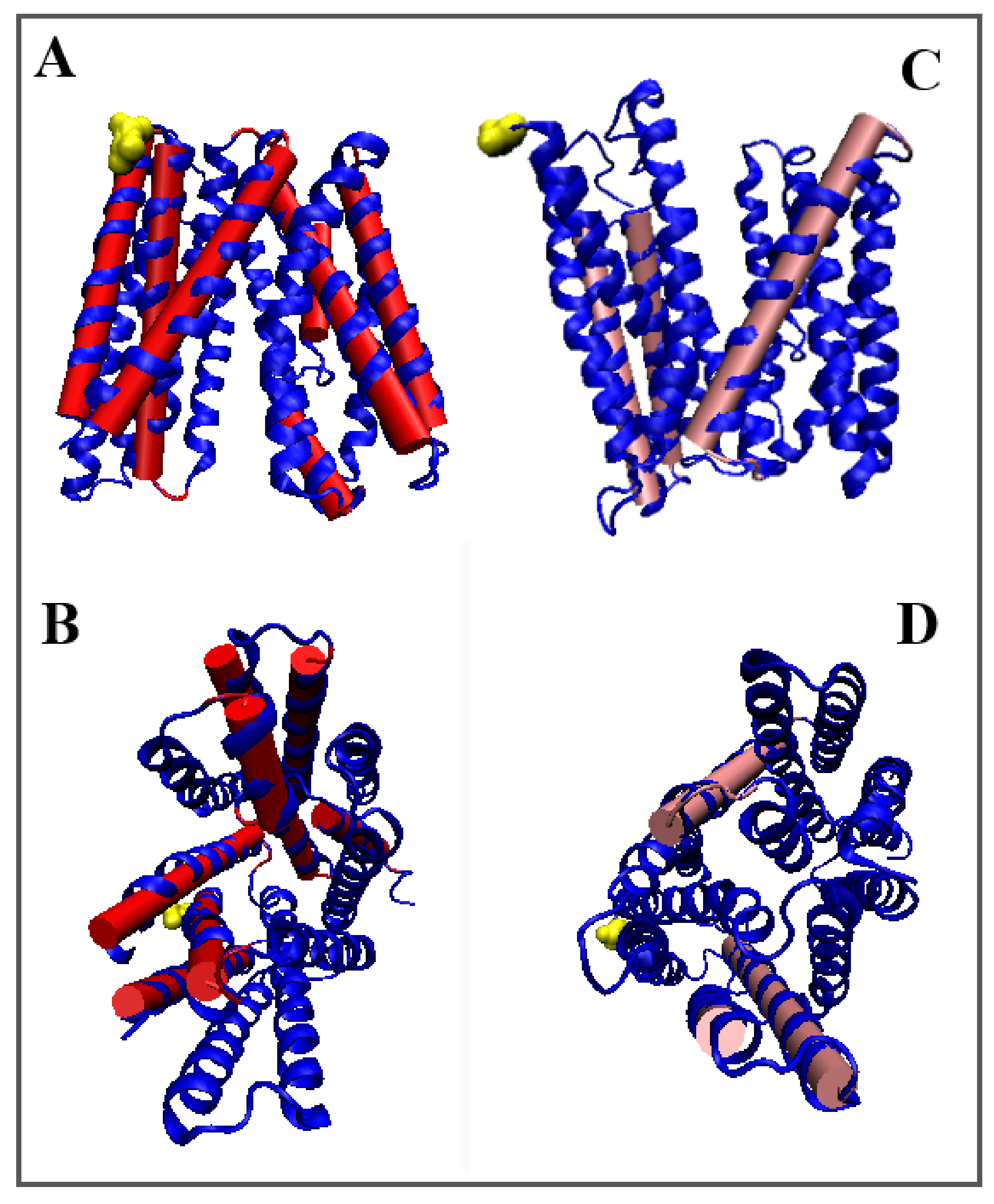
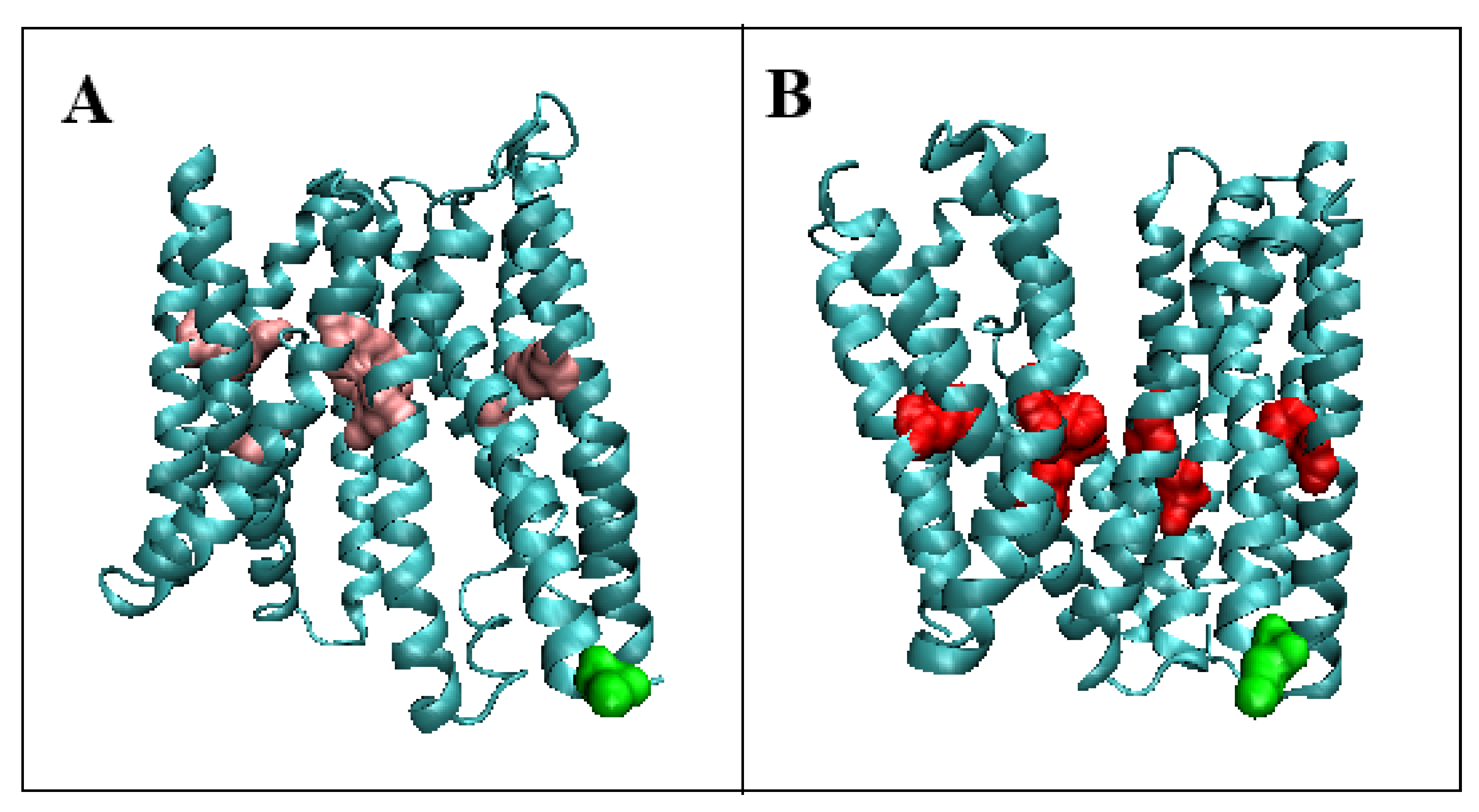
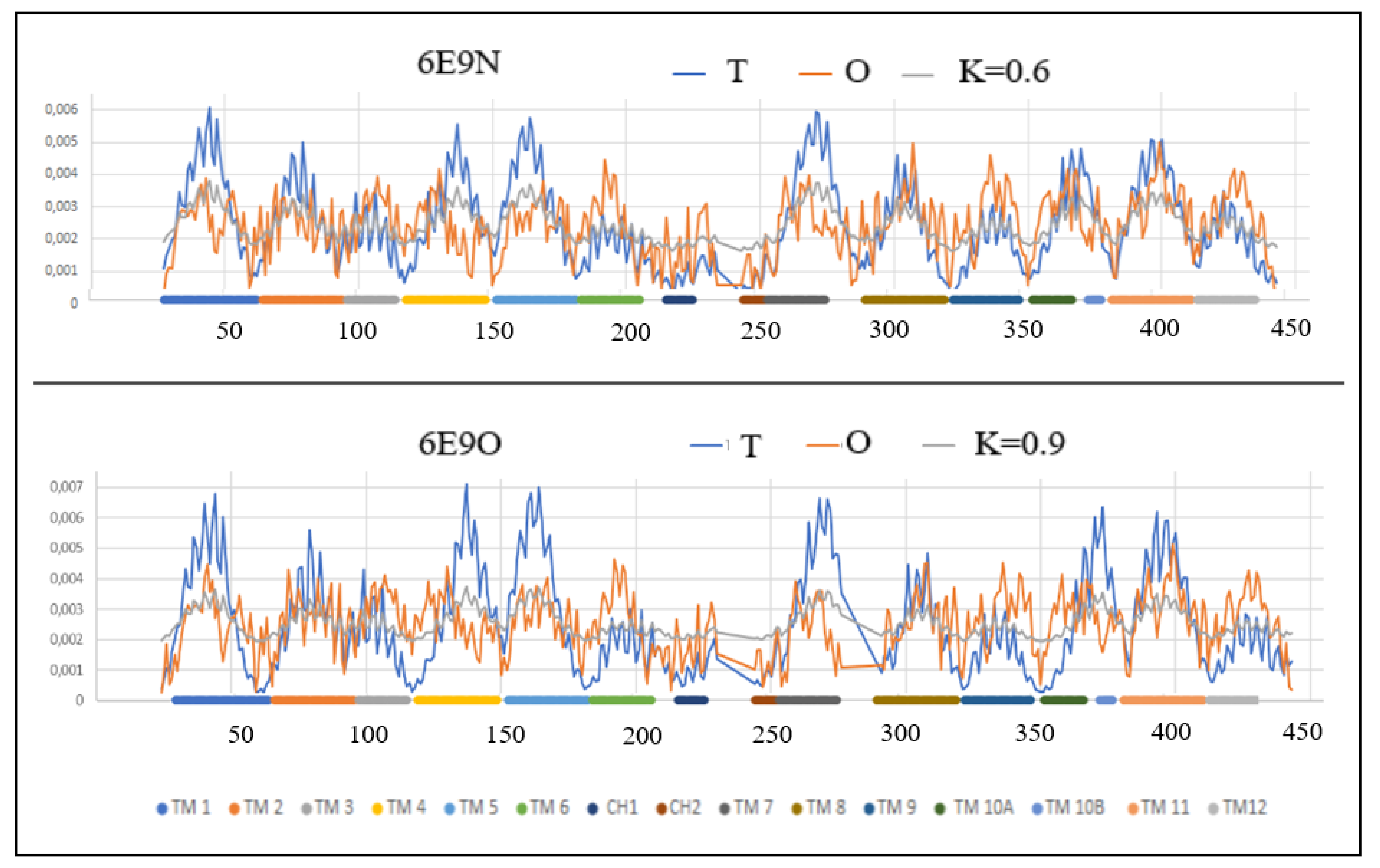
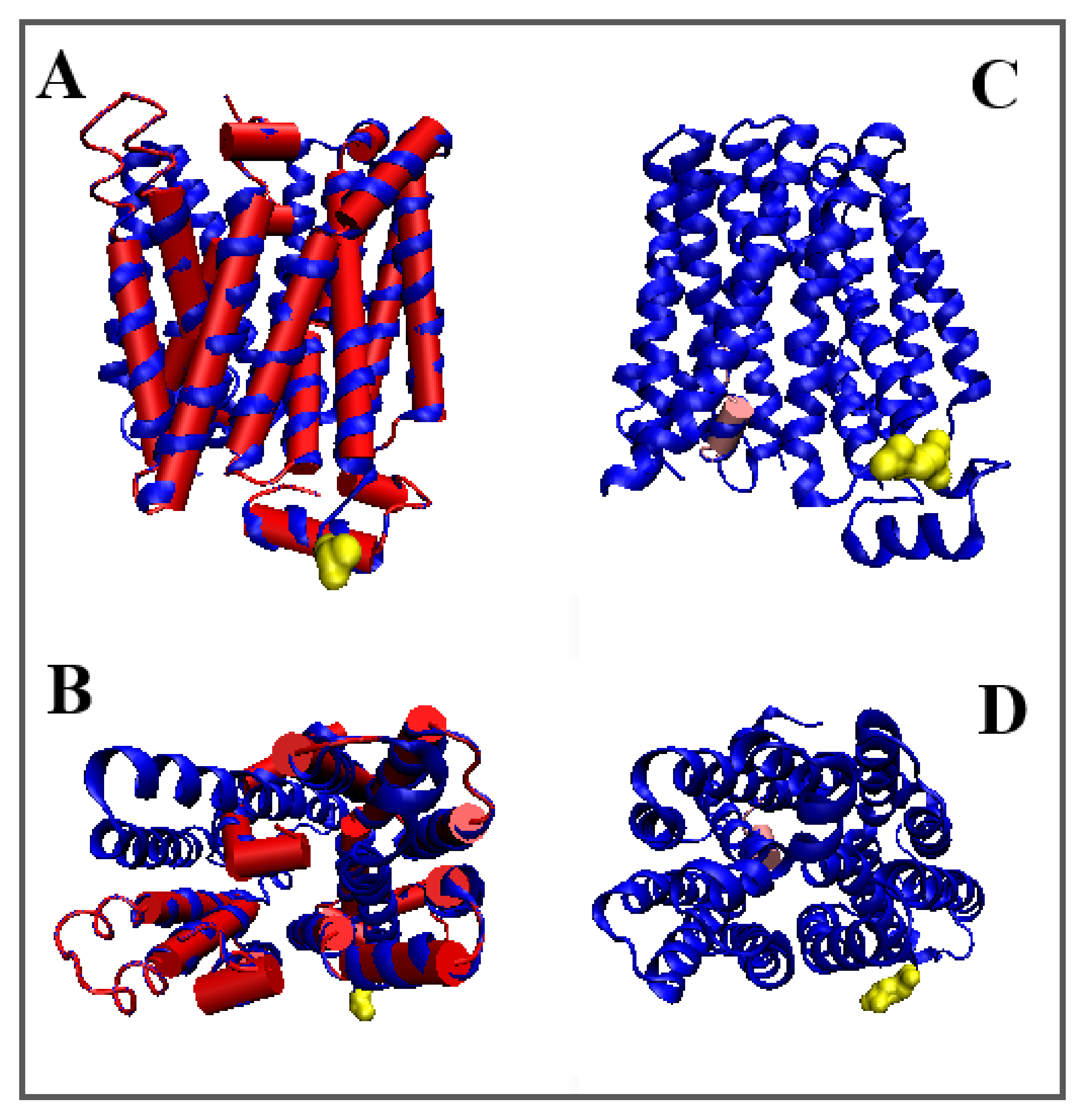
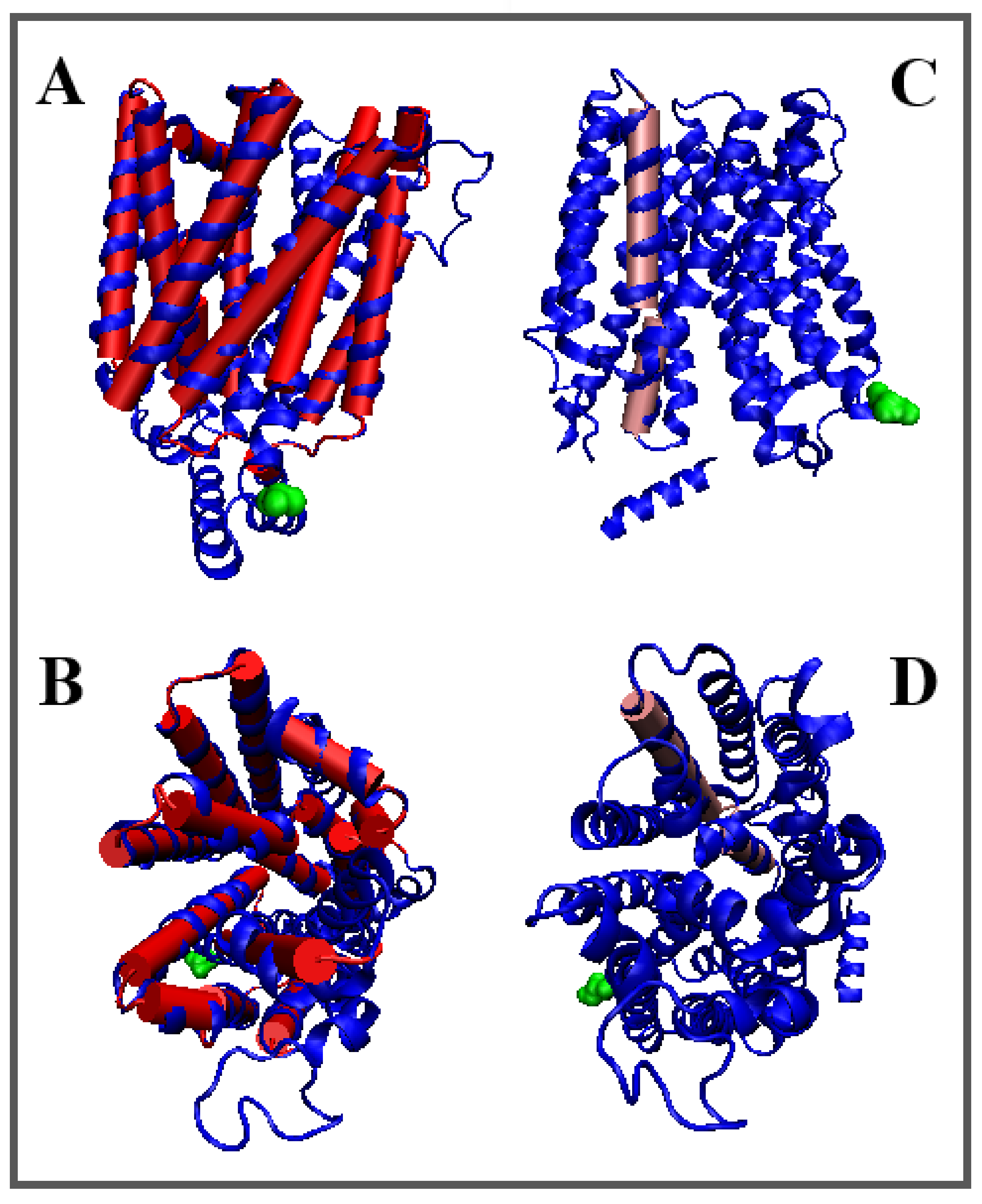
| Inward | Outward | |||||
|---|---|---|---|---|---|---|
| PDB ID [REF] | Chain Length | Transported Molecules | Transport | Source Organism | Chain Length | PDB ID [REF] |
| 5AYO [50] | (4–426) | Proton/Fe2+ | Symport | Bdellovibrio bacterovorus | (4–426) | 5AYM [50] |
| 6E9N [51] | (27–234) (243–443) | Proton/D-galactonate | Symport | E. coli | (24–230) (243–289) (295–442) | 6E9O [51] |
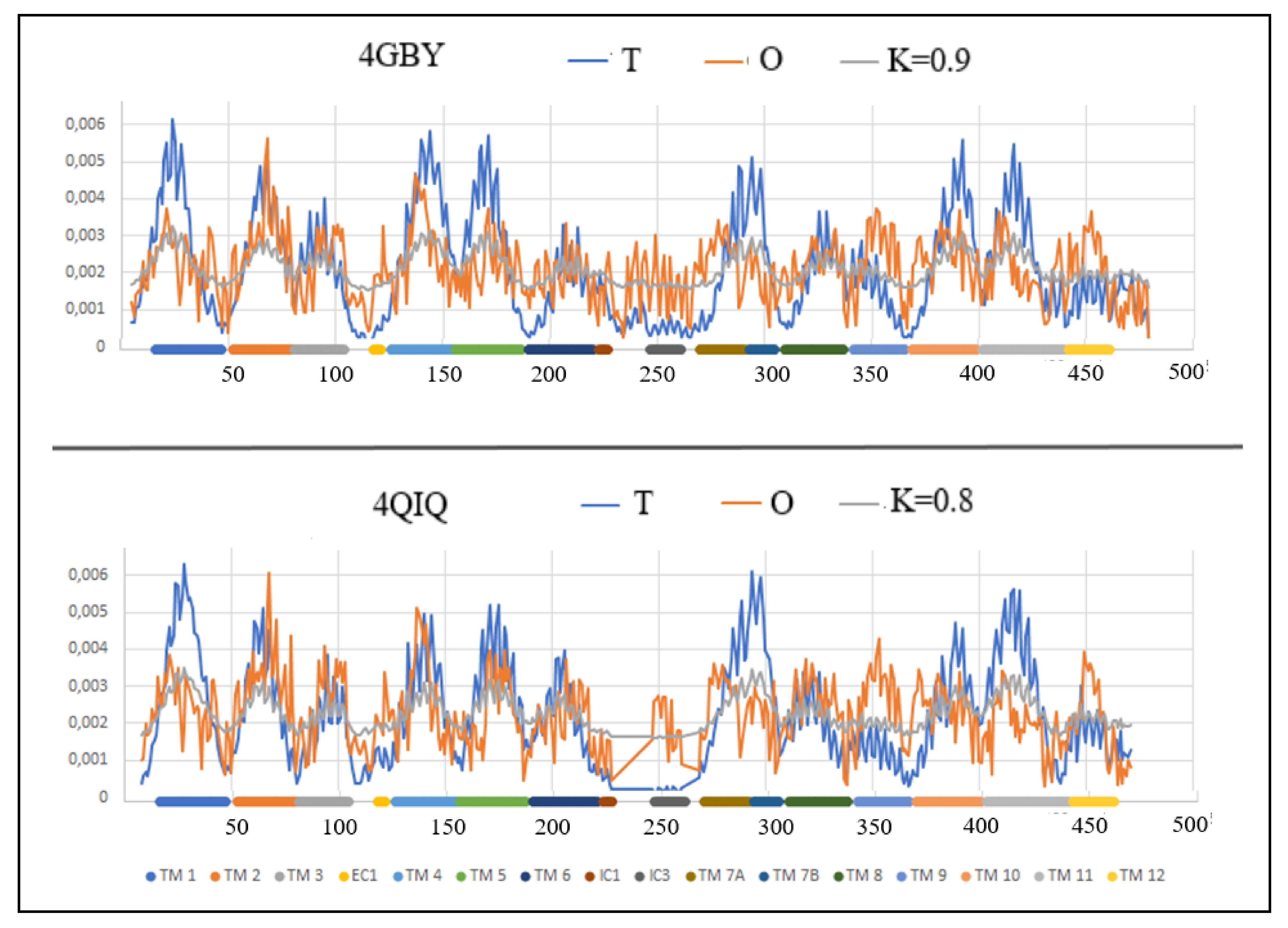
| 4QIQ [52] | (8–228) (247–471) | Proton/sugar | Symport | E. coli | (5–479) | 4GBY [53] |
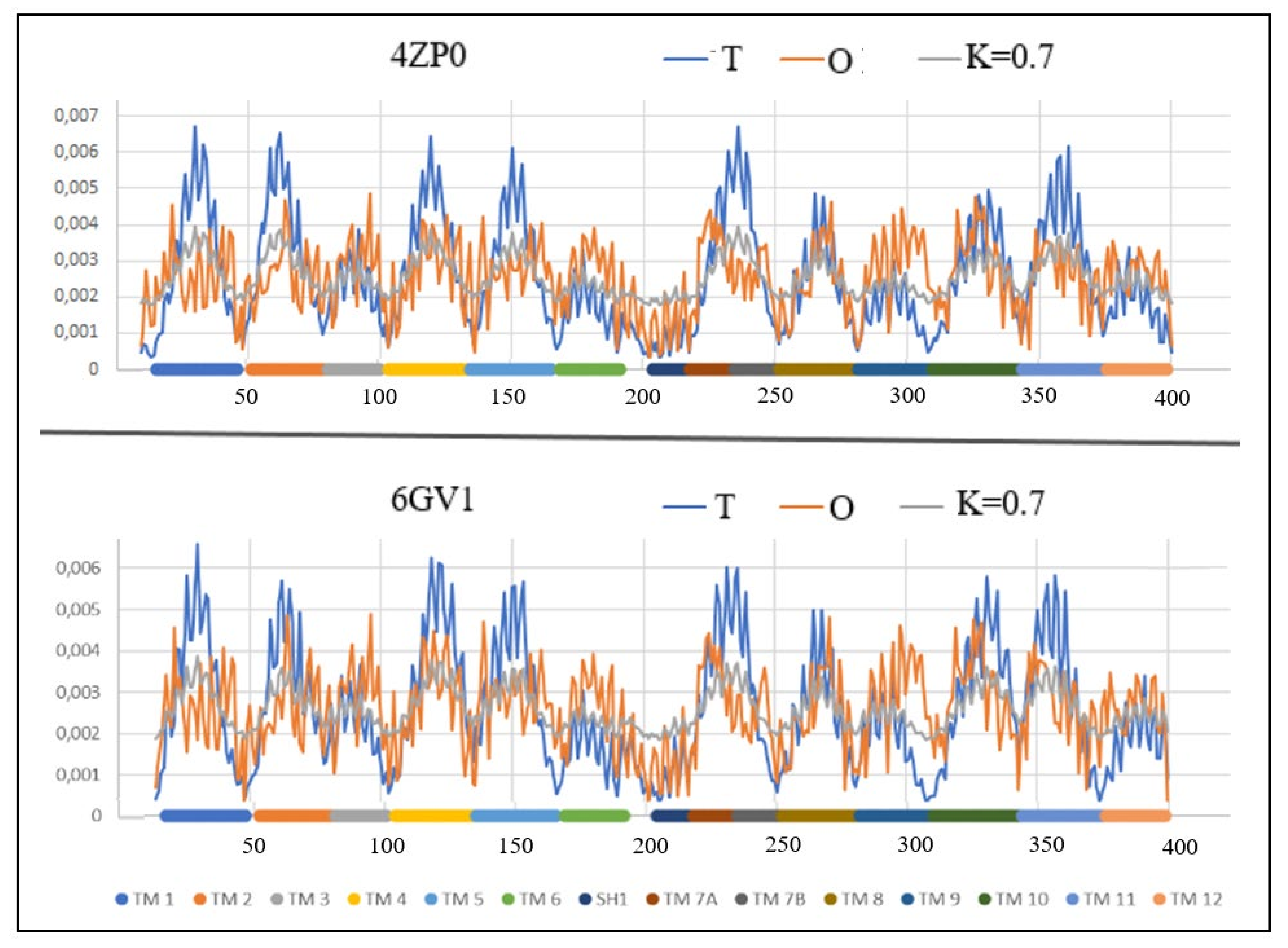
| 4ZP0 [54] | (9–400) | Proton/antibiotic | Antiport | E. coli | (14–400) | 6GVI [55] |
| INWARD | OUTWARD | ||||
|---|---|---|---|---|---|
| PDB ID | RD | K | K | RD | PDB |
| 6E9N | 0.596 | 0.6 | 0.9 | 0.711 | 6E9O |
| 4QIQ | 0.693 | 0.8 | 0.9 | 0.697 | 4GBY |
| 5AYO | 0.725 | 1.1 | 1.0 | 0.711 | 5AYM |
| 4ZP0 | 0.647 | 0.7 | 0.7 | 0.642 | 6GV1 |
| HELIX | OUTWARD PDP ID—5AYM | INWARD PDB ID—5AYO | |||
|---|---|---|---|---|---|
| TM | FRAGMENT | RD | K | RD | K |
| 1 | 3–39 | 0.772 | 1.5 | 0.858 | 1.1 |
| 2 | 42–69 | 0.624 | 0.5 | 0.630 | 0.5 |
| 3 | 72–104 | 0.681 | 0.3 | 0.610 | 0.5 |
| 4 | 109–146 | 0.700 | 0.5 | 0.835 | 0.7 |
| 4” | 138–146 | 0.844 | 0.4 | ||
| 5 | 147–177 | 0.768 | 1.5 | 0.462 | 0.8 |
| 6 | 186–218 | 0.593 | 0.5 | 0.580 | 0.4 |
| SH1 | 237–242 | 0.557 | 0.1 | ||
| 7A | 243–255 | 0.803 | 0.8 | 0.805 | 0.6 |
| 7B | 262–272 | 0.414 | 0.2 | 0.351 | 0.0 |
| 8 | 276–306 | 0.413 | 0.2 | 0.647 | 0.6 |
| 9 | 307–330 | 0.759 | 0.8 | 0.616 | 0.5 |
| 10 | 336–366 | 0.758 | 0.9 | 0.648 | 0.6 |
| 11 | 367–396 | 0.548 | 0.3 | 0.561 | 0.3 |
| 12 | 401–424 | 0.243 | 0.0 | 0.590 | 0.4 |
| HELIX | OUTWARD PDB ID—6E9O | INWARD PDB ID—6E9N | |||
|---|---|---|---|---|---|
| TM | FRAGMENT | RD | K | RD | K |
| 1 | 28–62 | 0.683 | 0.5 | 0.554 | 0.3 |
| 2 | 64–94 | 0.457 | 0.3 | 0.534 | 0.3 |
| 3 | 96–114 | 0.800 | 0.8 | 0.670 | 0.5 |
| 4 | 118–148 | 0.891 | 1.9 | 0.751 | 1.4 |
| 5 | 152–182 | 0.738 | 0.5 | 0.634 | 0.4 |
| 6 | 184–206 | 0.578 | 0.4 | 0.465 | 0.2 |
| CH1 | 216–226 | 0.286 | 0.1 | 0.270 | 0.0 |
| CH2 | 245–253 | 0.478 | 0.3 | 0.501 | 0.3 |
| 7 | 254–276 | 0.503 | 0.2 | 0.501 | 0.3 |
| 8 | 291–321 | 0.537 | 0.4 | 0.568 | 0.3 |
| 9 | 324–349 | 0.714 | 0.7 | 0.445 | 0.2 |
| 10A | 354–369 | 0.932 | 1.0 | 0.661 | 0.4 |
| 10B | 375–380 | 0.822 | 0.3 | 0.681 | 0.6 |
| 11 | 384–414 | 0.600 | 0.3 | 0.502 | 0.2 |
| 12 | 416–438 | 0.556 | 0.2 | 0.557 | 0.2 |
| HELIX | OUTWARD PDB ID—4GBY | INWARD PDB ID—4QIQ | |||
|---|---|---|---|---|---|
| TM | FRAGMENT | RD | K | RD | K |
| 1 | 16–47 | 0.684 | 0.6 | 0.672 | 0.5 |
| 2 | 52–80 | 0.356 | 0.2 | 0.505 | 0.3 |
| 3 | 81–104 | 0.610 | 0.5 | 0.308 | 0.0 |
| EC1 | 118–121 | 0.385 | 0.1 | 0.473 | 0.1 |
| 4 | 126–155 | 0.419 | 0.1 | 0.457 | 0.2 |
| 5 | 156–187 | 0.776 | 0.6 | 0.475 | 0.2 |
| 6 | 190–220 | 0.783 | 0.6 | 0.642 | 0.5 |
| IC1 | 223–228 | 0.621 | 0.4 | 0.382 | 0.1 |
| IC3 | 247–261 | 0.349 | 0.1 | 0.716 | 1.1 |
| 7A | 270–291 | 0.899 | 1.6 | 0.734 | 0.9 |
| 7B | 293–305 | 0.664 | 0.4 | 0.535 | 0.2 |
| 8 | 310–337 | 0.648 | 0.4 | 0.560 | 0.3 |
| 9 | 342–366 | 0.657 | 0.6 | 0.523 | 0.3 |
| 10 | 369–399 | 0.885 | 0.9 | 0.880 | 1.2 |
| 11 | 402–440 | 0.602 | 0.5 | 0.625 | 0.4 |
| 12 | 442–461 | 0.425 | 0.2 | 0.455 | 0.2 |
| HELIX | OUTWARD PDB ID—6GV1 | INWARD PDB ID—4ZP0 | |||
|---|---|---|---|---|---|
| TM | FRAGMENT | RD | K | RD | K |
| 1 | 16–47 | 0.769 | 1.2 | 0.763 | 1.2 |
| 2 | 52–80 | 0.676 | 0.5 | 0.693 | 0.5 |
| 3 | 81–101 | 0.506 | 0.2 | 0.362 | 0.1 |
| 4 | 104–134 | 0.514 | 0.3 | 0.577 | 0.3 |
| 5 | 135–166 | 0.723 | 0.6 | 0.685 | 0.6 |
| 6 | 169–192 | 0.308 | 0.1 | 0.331 | 0.2 |
| SH1 | 204–217 | 0.415 | 0.2 | 0.283 | 0.1 |
| 7A | 218–234 | 0.597 | 0.3 | 0.660 | 0.5 |
| 7B | 234–251 | 0.781 | 0.8 | 0.878 | 0.6 |
| 8 | 251–281 | 0.427 | 0.2 | 0.276 | 0.1 |
| 9 | 281–309 | 0.760 | 0.7 | 0.628 | 0.4 |
| 10 | 309–343 | 0.641 | 0.4 | 0.509 | 0.2 |
| 11 | 343–375 | 0.629 | 0.4 | 0.557 | 0.8 |
| 12 | 375–399 | 0.660 | 0.6 | 0.630 | 0.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roterman, I.; Stapor, K.; Konieczny, L. The Contribution of Hydrophobic Interactions to Conformational Changes of Inward/Outward Transmembrane Transport Proteins. Membranes 2022, 12, 1212. https://doi.org/10.3390/membranes12121212
Roterman I, Stapor K, Konieczny L. The Contribution of Hydrophobic Interactions to Conformational Changes of Inward/Outward Transmembrane Transport Proteins. Membranes. 2022; 12(12):1212. https://doi.org/10.3390/membranes12121212
Chicago/Turabian StyleRoterman, Irena, Katarzyna Stapor, and Leszek Konieczny. 2022. "The Contribution of Hydrophobic Interactions to Conformational Changes of Inward/Outward Transmembrane Transport Proteins" Membranes 12, no. 12: 1212. https://doi.org/10.3390/membranes12121212
APA StyleRoterman, I., Stapor, K., & Konieczny, L. (2022). The Contribution of Hydrophobic Interactions to Conformational Changes of Inward/Outward Transmembrane Transport Proteins. Membranes, 12(12), 1212. https://doi.org/10.3390/membranes12121212







