Nebivolol as a Potent TRPM8 Channel Blocker: A Drug-Screening Approach through Automated Patch Clamping and Ligand-Based Virtual Screening
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Preparation
2.2. Automated Patch Clamp Electrophysiology
2.3. Buffers and Compounds
2.4. Ligand-Based Virtual Screening
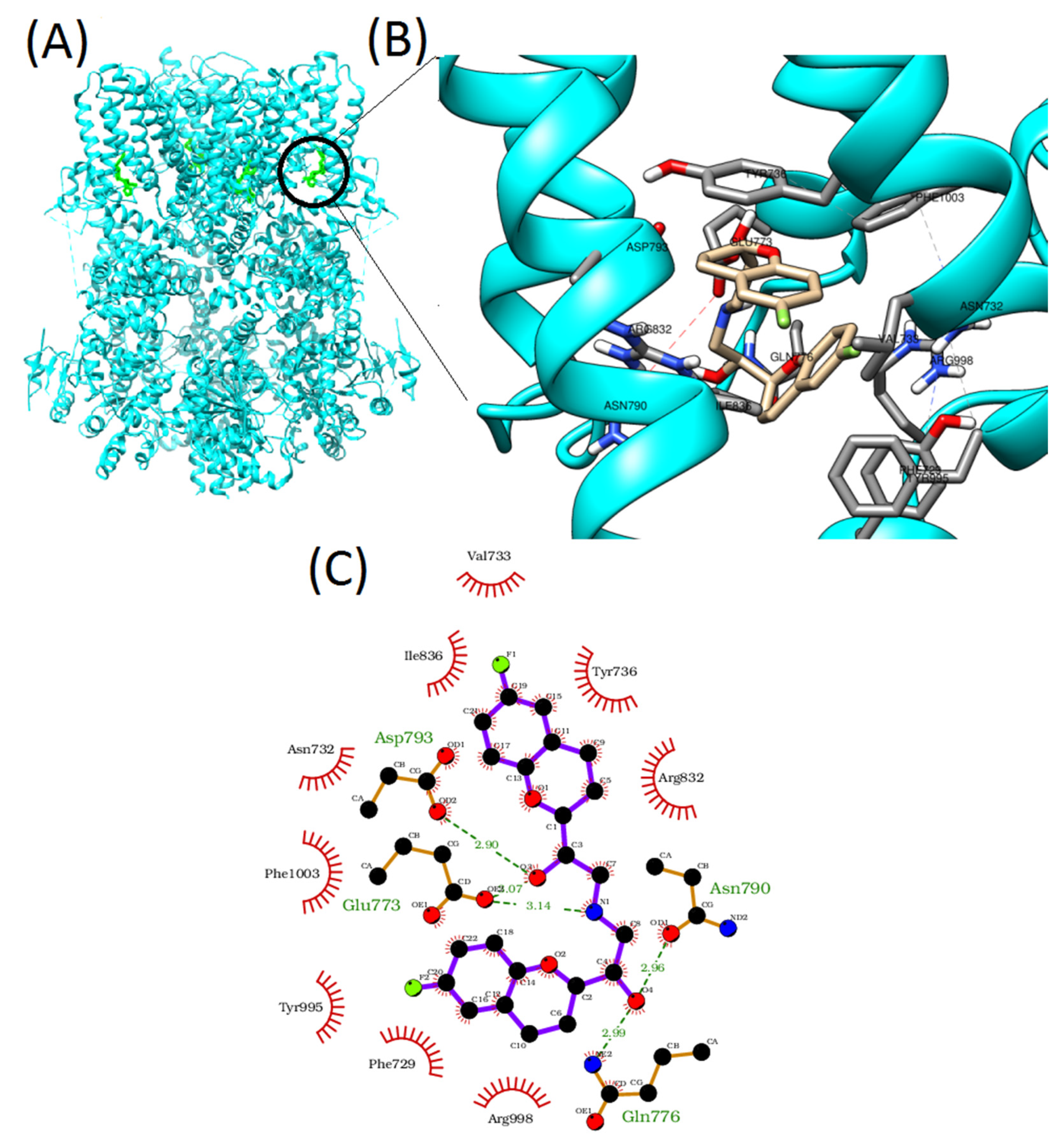
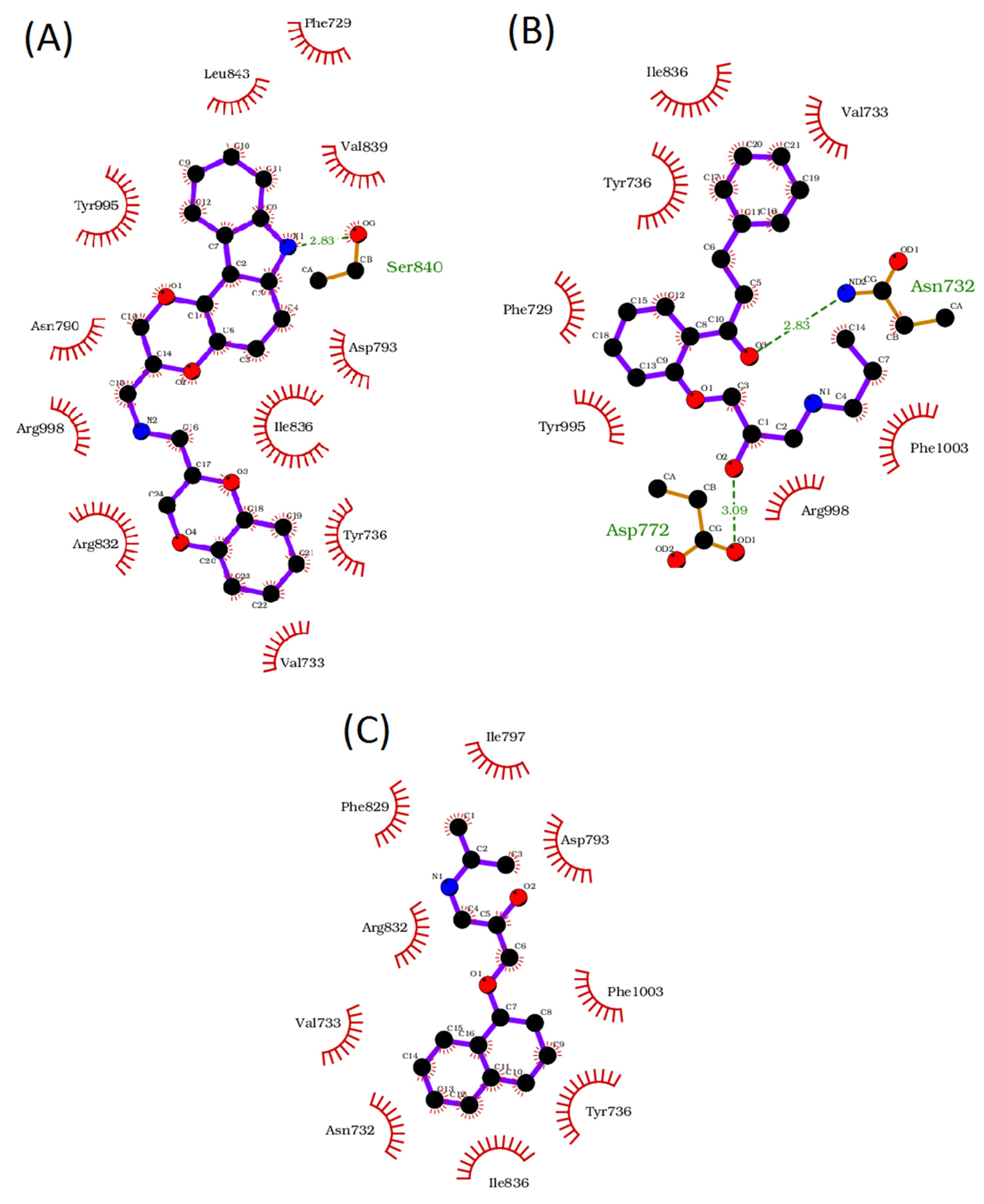
2.5. Molecular Docking Study
2.6. Data Analysis
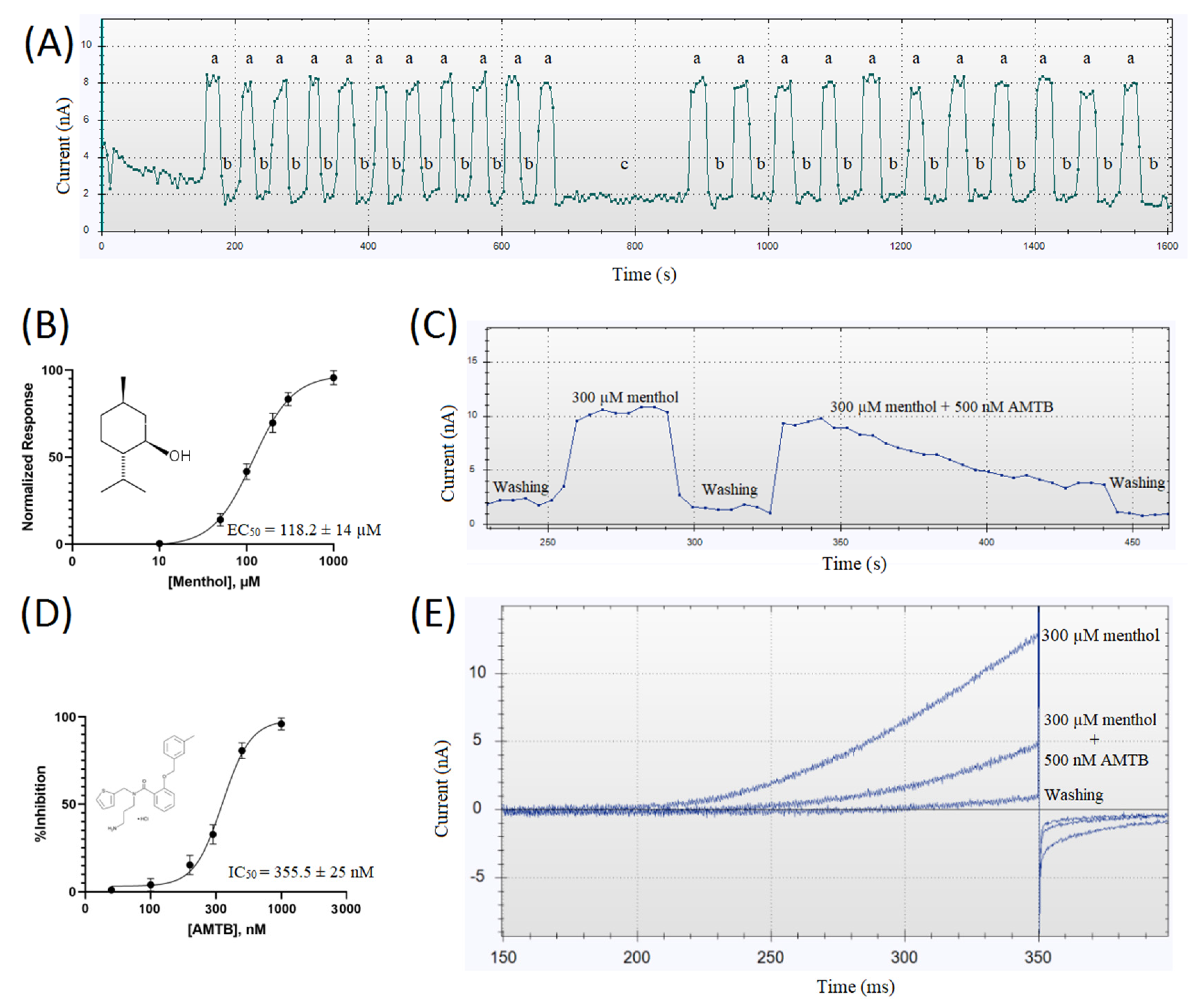
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaczorowski, G.J.; McManus, O.B.; Priest, B.T.; Garcia, M.L. Ion channels as drug targets: The next GPCRs. J. Gen. Physiol. 2008, 131, 399–405. [Google Scholar] [CrossRef]
- Venkatachalam, K.; Montell, C. TRP channels. Annu. Rev. Biochem. 2007, 76, 387–417. [Google Scholar] [CrossRef] [PubMed]
- McKemy, D.D.; Neuhausser, W.M.; Julius, D. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature 2002, 416, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, W.M.; Daniels, R.L.; Palkar, R.; McCoy, D.D.; McKemy, D.D. Pharmacological blockade of TRPM8 ion channels alters cold and cold pain responses in mice. PLoS ONE 2011, 6, e25894. [Google Scholar] [CrossRef] [PubMed]
- Tsavaler, L.; Shapero, M.H.; Morkowski, S.; Laus, R. Trp-p8, a novel prostate-specific gene, is up-regulated in prostate cancer and other malignancies and shares high homology with transient receptor potential calcium channel proteins. Cancer Res. 2001, 61, 3760–3769. [Google Scholar]
- Stein, R.J.; Santos, S.; Nagatomi, J.; Hayashi, Y.; Minnery, B.S.; Xavier, M.; Patel, A.S.; Nelson, J.B.; Futrell, W.J.; Yoshimura, N. Cool (TRPM8) and hot (TRPV1) receptors in the bladder and male genital tract. J. Urol. 2004, 172, 1175–1178. [Google Scholar] [CrossRef]
- Huang, F.; Ni, M.; Zhang, J.; Li, D.; Shen, F. TRPM8 downregulation by angiotensin II in vascular smooth muscle cells is involved in hypertension. Mol. Med. Rep. 2017, 15, 1900–1908. [Google Scholar] [CrossRef]
- Parra, A.; Madrid, R.; Echevarria, D.; Del Olmo, S.; Morenilla-Palao, C.; Acosta, M.C.; Gallar, J.; Dhaka, A.; Viana, F.; Belmonte, C. Ocular surface wetness is regulated by TRPM8-dependent cold thermoreceptors of the cornea. Nat. Med. 2010, 16, 1396–1399. [Google Scholar] [CrossRef]
- Acharya, T.K.; Tiwari, A.; Majhi, R.K.; Goswami, C. TRPM8 channel augments T-cell activation and proliferation. Cell Biol. Int. 2021, 45, 198–210. [Google Scholar] [CrossRef]
- Hodgkin, A.L.; Huxley, A.F.; Katz, B. Measurement of current-voltage relations in the membrane of the giant axon of Loligo. J. Physiol. 1952, 116, 424–448. [Google Scholar] [CrossRef]
- Proudfoot, C.J.; Garry, E.M.; Cottrell, D.F.; Rosie, R.; Anderson, H.; Robertson, D.C.; Fleetwood-Walker, S.M.; Mitchell, R. Analgesia mediated by the TRPM8 cold receptor in chronic neuropathic pain. Curr. Biol. 2006, 16, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Mukerji, G.; Yiangou, Y.; Corcoran, S.L.; Selmer, I.S.; Smith, G.D.; Benham, C.D.; Bountra, C.; Agarwal, S.K.; Anand, P. Cool and menthol receptor TRPM8 in human urinary bladder disorders and clinical correlations. BMC Urol. 2006, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Dussor, G.; Cao, Y. TRPM8 and migraine. Headache J. Head Face Pain 2016, 56, 1406–1417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Barritt, G.J. Evidence that TRPM8 is an androgen-dependent Ca2+ channel required for the survival of prostate cancer cells. Cancer Res. 2004, 64, 8365–8373. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Y.; Shuai, S.; Ding, D.; Li, R.; Luo, R. TRPM8 promotes aggressiveness of breast cancer cells by regulating EMT via activating AKT/GSK-3β pathway. Tumor Biol. 2014, 35, 8969–8977. [Google Scholar] [CrossRef]
- Morice, A.H.; Marshall, A.E.; Higgins, K.S.; Grattan, T.J. Effect of inhaled menthol on citric acid induced cough in normal subjects. Thorax 1994, 49, 1024–1026. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef]
- González-Muñiz, R.; Bonache, M.; Martín-Escura, C.; Gómez-Monterrey, I. Recent progress in TRPM8 modulation: An update. Int. J. Mol. Sci. 2019, 20, 2618. [Google Scholar] [CrossRef]
- Gaston, T.E.; Friedman, D. Pharmacology of cannabinoids in the treatment of epilepsy. Epilepsy Behav. 2017, 70, 313–318. [Google Scholar] [CrossRef]
- Moridani, M.; Harirforoosh, S. Drug development and discovery: Challenges and opportunities. Drug Discov. Today 2014, 19, 1679–1681. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.-U. Polypharmacology–foe or friend? J. Med. Chem. 2013, 56, 8955–8971. [Google Scholar] [CrossRef] [PubMed]
- Wermuth, C.G. Selective optimization of side activities: The SOSA approach. Drug Discov. Today 2006, 11, 160–164. [Google Scholar] [CrossRef]
- Zoete, V.; Daina, A.; Bovigny, C.; Michielin, O. SwissSimilarity: A web tool for low to ultra high throughput ligand-based virtual screening. J. Chem. Inf. Model. 2016, 56, 1399–1404. [Google Scholar] [CrossRef]
- Bragina, M.E.; Daina, A.; Perez, M.A.S.; Michielin, O.; Zoete, V. The SwissSimilarity 2021 Web Tool: Novel Chemical Libraries and Additional Methods for an Enhanced Ligand-Based Virtual Screening Experience. Int. J. Mol. Sci. 2022, 23, 811. [Google Scholar] [CrossRef]
- Golden, A.P.; Li, N.; Chen, Q.; Lee, T.; Nevill, T.; Cao, X.; Johnson, J.; Erdemli, G.; Ionescu-Zanetti, C.; Urban, L. IonFlux: A microfluidic patch clamp system evaluated with human ether-à-go-go related gene channel physiology and pharmacology. Assay Drug Dev. Technol. 2011, 9, 608–619. [Google Scholar] [CrossRef]
- Tikhonov, D.B.; Zhorov, B.S. Mechanism of sodium channel block by local anesthetics, antiarrhythmics, and anticonvulsants. J. Gen. Physiol. 2017, 149, 465–481. [Google Scholar] [CrossRef]
- Taylor, J.C.; Brauer, S.; Espir, M.L. Long-term treatment of trigeminal neuralgia with carbamazepine. Postgrad. Med. J. 1981, 57, 16–18. [Google Scholar] [CrossRef]
- Coderre, T.J.; Kumar, N.; Lefebvre, C.D.; Yu, J.S.C. A comparison of the glutamate release inhibition and anti-allodynic effects of gabapentin, lamotrigine, and riluzole in a model of neuropathic pain. J. Neurochem. 2007, 100, 1289–1299. [Google Scholar] [CrossRef]
- Morgan, K.; Sadofsky, L.R.; Crow, C.; Morice, A.H. Human TRPM8 and TRPA1 pain channels, including a gene variant with increased sensitivity to agonists (TRPA1 R797T), exhibit differential regulation by SRC-tyrosine kinase inhibitor. Biosci. Rep. 2014, 34, e00131. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Diver, M.M.; Cheng, Y.; Julius, D. Structural insights into TRPM8 inhibition and desensitization. Science 2019, 365, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Beck, E.J.; Hutchinson, T.L.; Qin, N.; Flores, C.M.; Liu, Y. Development and validation of a secondary screening assay for TRPM8 antagonists using QPatch HT. Assay Drug Dev. Technol. 2010, 8, 63–72. [Google Scholar] [CrossRef]
- Andersson, D.A.; Chase, H.W.N.; Bevan, S. TRPM8 activation by menthol, icilin, and cold is differentially modulated by intracellular pH. J. Neurosci. 2004, 24, 5364–5369. [Google Scholar] [CrossRef]
- Sherkheli, M.A.; Gisselmann, G.; Vogt-Eisele, A.K.; Doerner, J.F.; Hatt, H. Menthol derivative WS-12 selectively activates transient receptor potential melastatin-8 (TRPM8) ion channels. Pak. J. Pharm. Sci. 2008, 21, 370–378. [Google Scholar]
- Lashinger, E.S.R.; Steiginga, M.S.; Hieble, J.P.; Leon, L.A.; Gardner, S.D.; Nagilla, R.; Davenport, E.A.; Hoffman, B.E.; Laping, N.J.; Su, X. AMTB, a TRPM8 channel blocker: Evidence in rats for activity in overactive bladder and painful bladder syndrome. Am. J. Physiol. Physiol. 2008, 295, F803–F810. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Van de Water, A.; Janssens, W.; Van Neuten, J.; Xhonneux, R.; De Cree, J.; Verhaegen, H.; Reneman, R.S.; Janssen, P.A. Pharmacological and hemodynamic profile of nebivolol, a chemically novel, potent, and selective beta 1-adrenergic antagonist. J. Cardiovasc. Pharmacol. 1988, 11, 552–563. [Google Scholar] [CrossRef]
- Dessy, C.; Saliez, J.; Ghisdal, P.; Daneau, G.; Lobysheva, I.I.; Frérart, F.; Belge, C.; Jnaoui, K.; Noirhomme, P.; Feron, O. Endothelial β3-adrenoreceptors mediate nitric oxide–dependent vasorelaxation of coronary microvessels in response to the third-generation β-blocker nebivolol. Circulation 2005, 112, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Talevi, A. Multi-target pharmacology: Possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Front. Pharmacol. 2015, 6, 205. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, C.; Wang, L.; Yang, Z.; Guo, D.; Fan, C. Repurposing old drugs as novel inhibitors of human MIF from structural and functional analysis. Bioorg. Med. Chem. Lett. 2022, 55, 128445. [Google Scholar] [CrossRef] [PubMed]
- Anand, N.M.; Liya, D.H.; Pradhan, A.K.; Tayal, N.; Bansal, A.; Donakonda, S.; Jainarayanan, A.K. A comprehensive SARS-CoV-2 genomic analysis identifies potential targets for drug repurposing. PLoS ONE 2021, 16, e0248553. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Han, Y.; Chen, X.; Aierken, A.; Wen, H.; Zheng, W.; Wang, H.; Lu, X.; Zhao, Z.; Ma, C. Molecular mechanisms underlying menthol binding and activation of TRPM8 ion channel. Nat. Commun. 2020, 11, 3790. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Van Bay, M.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock vina adopts more accurate binding poses but autodock4 forms better binding affinity. J. Chem. Inf. Model. 2019, 60, 204–211. [Google Scholar] [CrossRef]
- Lundbæk, J.A. Lipid bilayer–mediated regulation of ion channel function by amphiphilic drugs. J. Gen. Physiol. 2008, 131, 421–429. [Google Scholar] [CrossRef]
- Morenilla-Palao, C.; Pertusa, M.; Meseguer, V.; Cabedo, H.; Viana, F. Lipid raft segregation modulates TRPM8 channel activity. J. Biol. Chem. 2009, 284, 9215–9224. [Google Scholar] [CrossRef]
- Sarria, I.; Ling, J.; Zhu, M.X.; Gu, J.G. TRPM8 acute desensitization is mediated by calmodulin and requires PIP2: Distinction from tachyphylaxis. J. Neurophysiol. 2011, 106, 3056–3066. [Google Scholar] [CrossRef] [PubMed]
- Benedikt, J.; Teisinger, J.; Vyklicky, L.; Vlachova, V. Ethanol inhibits cold-menthol receptor TRPM8 by modulating its interaction with membrane phosphatidylinositol 4, 5-bisphosphate. J. Neurochem. 2007, 100, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, T.; Ponce-Balbuena, D.; López-Izquierdo, A.; Aréchiga-Figueroa, I.A.; de Boer, T.P.; van der Heyden, M.A.G.; Sánchez-Chapula, J.A. Carvedilol inhibits Kir2. 3 channels by interference with PIP2-channel interaction. Eur. J. Pharmacol. 2011, 668, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Amorós, I.; Dolz-Gaitón, P.; Gómez, R.; Matamoros, M.; Barana, A.; de la Fuente, M.G.; Núñez, M.; Pérez-Hernández, M.; Moraleda, I.; Gálvez, E. Propafenone blocks human cardiac Kir2. x channels by decreasing the negative electrostatic charge in the cytoplasmic pore. Biochem. Pharmacol. 2013, 86, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Rusinova, R.; Koeppe, R.E.; Andersen, O.S. A general mechanism for drug promiscuity: Studies with amiodarone and other antiarrhythmics. J. Gen. Physiol. 2015, 146, 463–475. [Google Scholar] [CrossRef]

| Drug Name | Average Percentage Inhibition at 10 µM | |
|---|---|---|
| Propafenone | 26 | (n = 7) |
| Flecainide | 0 | (n = 6) |
| Phenytoin | 0 | (n = 6) |
| Oxcarbazepine | 0 | (n = 6) |
| Lamotrigine | 0 | (n = 6) |
| Riluzole | 0 | (n = 6) |
| Carbamazepine | 0 | (n = 6) |
| Gabapentin | 0 | (n = 6) |
| Rufinamide | 0 | (n = 6) |
| Ranolazine | 0 | (n = 6) |
| Tetracaine | 0 | (n = 6) |
| Mexiletine | 0 | (n = 6) |
| Drug Name | Average Percentage Inhibition at 10 μM | |
|---|---|---|
| Nebivolol | 100 | (n = 8) |
| Carvedilol | 57 | (n = 7) |
| Propranolol | 12 | (n = 6) |
| Metoprolol | 0 | (n = 6) |
| Atenolol | 0 | (n = 6) |
| Nadolol | 0 | (n = 6) |
| Acebutolol | 0 | (n = 6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahanfar, F.; Sadofsky, L.; Morice, A.; D’Amico, M. Nebivolol as a Potent TRPM8 Channel Blocker: A Drug-Screening Approach through Automated Patch Clamping and Ligand-Based Virtual Screening. Membranes 2022, 12, 954. https://doi.org/10.3390/membranes12100954
Jahanfar F, Sadofsky L, Morice A, D’Amico M. Nebivolol as a Potent TRPM8 Channel Blocker: A Drug-Screening Approach through Automated Patch Clamping and Ligand-Based Virtual Screening. Membranes. 2022; 12(10):954. https://doi.org/10.3390/membranes12100954
Chicago/Turabian StyleJahanfar, Farhad, Laura Sadofsky, Alyn Morice, and Massimo D’Amico. 2022. "Nebivolol as a Potent TRPM8 Channel Blocker: A Drug-Screening Approach through Automated Patch Clamping and Ligand-Based Virtual Screening" Membranes 12, no. 10: 954. https://doi.org/10.3390/membranes12100954
APA StyleJahanfar, F., Sadofsky, L., Morice, A., & D’Amico, M. (2022). Nebivolol as a Potent TRPM8 Channel Blocker: A Drug-Screening Approach through Automated Patch Clamping and Ligand-Based Virtual Screening. Membranes, 12(10), 954. https://doi.org/10.3390/membranes12100954





