Lipid Specific Membrane Interaction of Aptamers and Cytotoxicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Liposome Binding Assays-FL Measurements on PS Bound Aptamers
2.2. Liposome Binding Assays-DDM to Measure Liposome Bound Aptamer Concentrations
2.3. Cell Culture and Imaging Experiments
2.4. Cell Culture and Cytotoxicity Experiments
3. Results
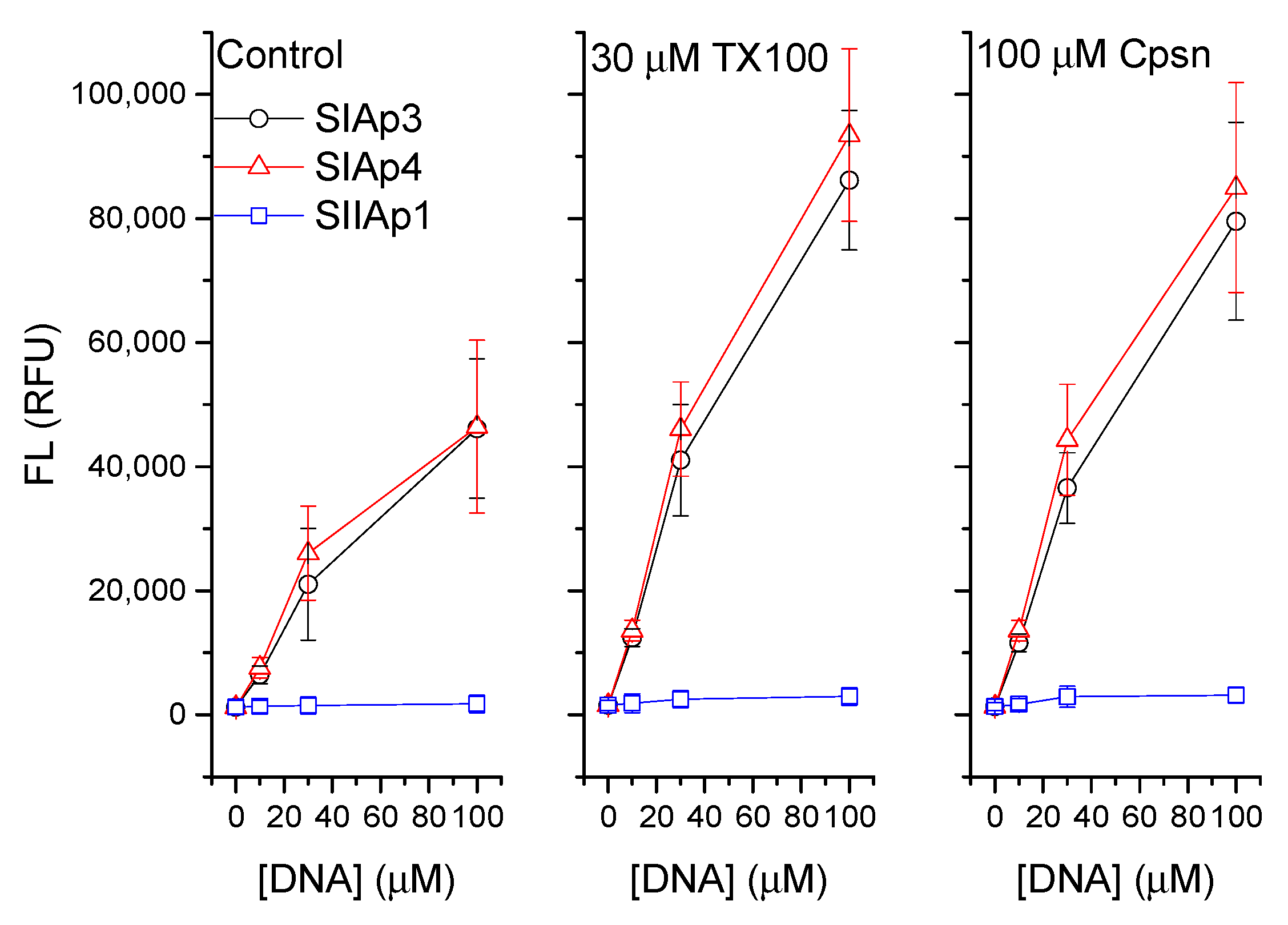
3.1. Amphiphiles Regulate the Liposome Adsorption of PS Aptamers
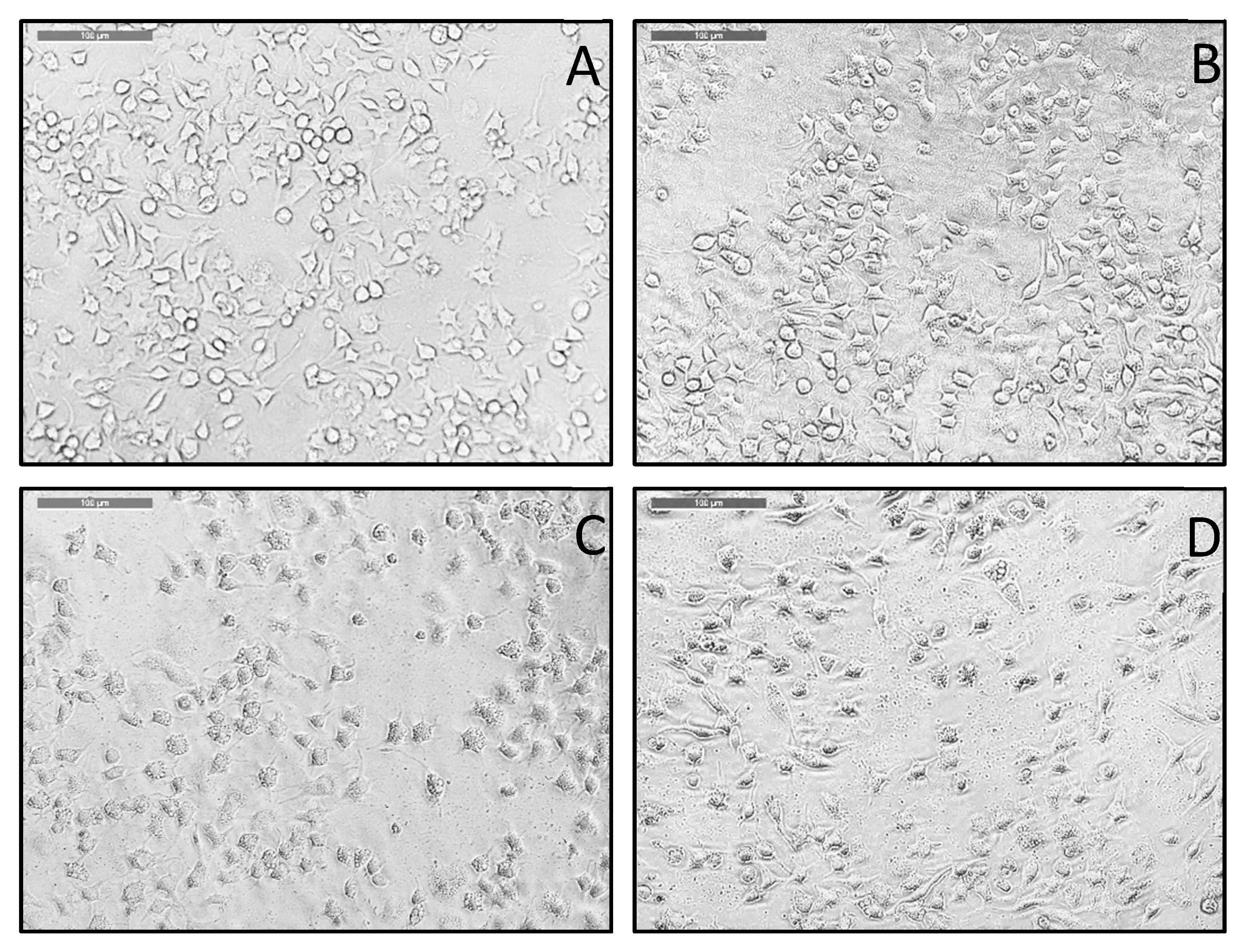
3.2. Imaging Experiments Tracking PS Aptamers on the Cell Surface
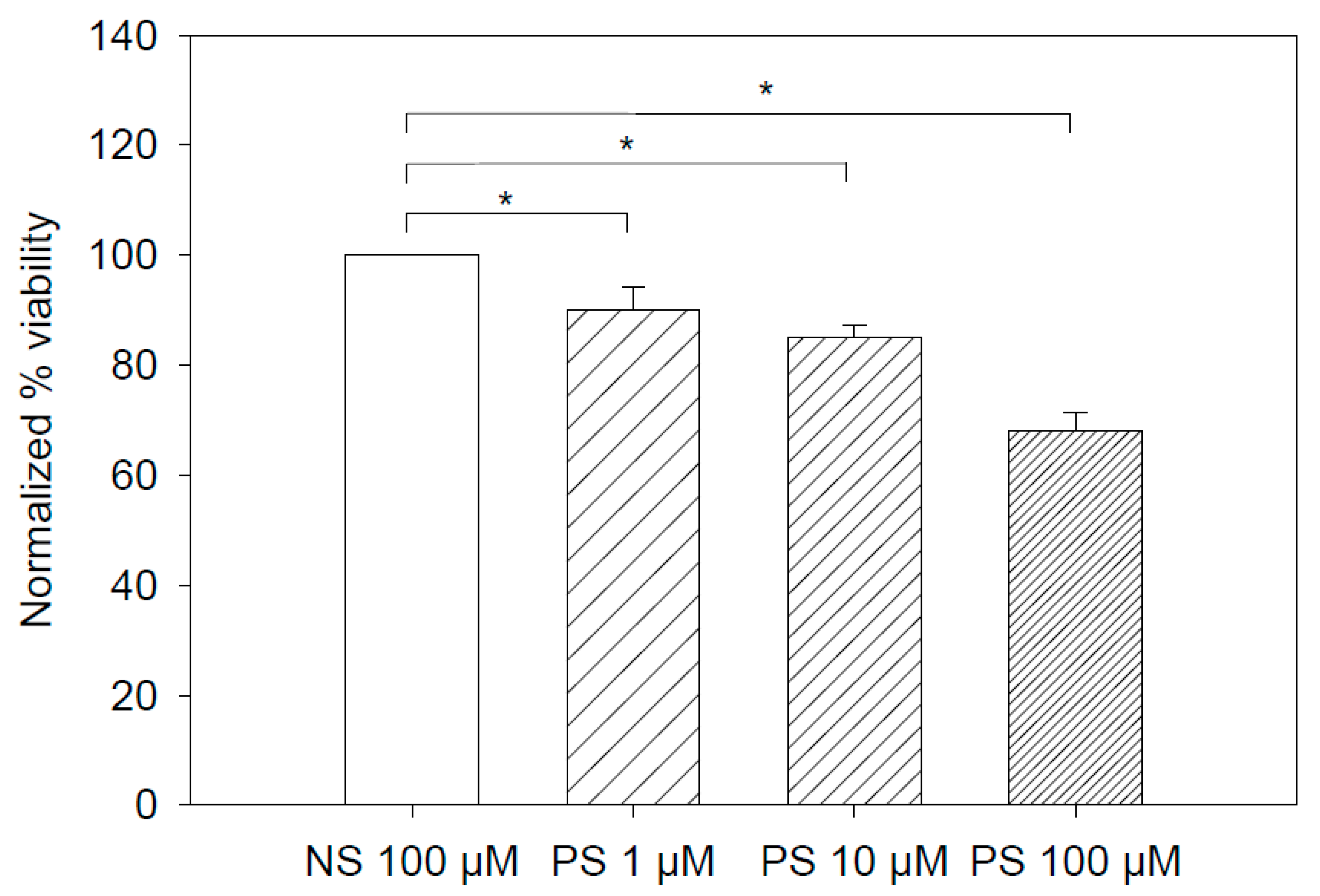
3.3. PS Aptamer-Induced Cytotoxicity Results
4. Discussion
4.1. Lipid Bilayer Physical Properties Regulate the Binding Mechanisms of PS Aptamers with Liposomes
4.2. Fluorescence Images Suggest That PS Aptamers Bind with Apoptosis-Induced Cancer Cell Surface Targets
4.3. PS Aptamers Are Modestly Cytotoxic
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ashrafuzzaman, M. Aptamers as both drugs and drug-carriers. BioMed Res. Int. 2014, 2014, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhang, K. The application of aptamers in cancer research: An up-to-date review. Future Oncol. 2013, 9, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Li, Y.; Tang, F. Nucleic acid aptamer: A novel potential diagnostic and therapeutic tool for leukemia. OncoTargets Ther. 2019, 12, 10597–10613. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Tseng, C.-Y.; Ashrafuzzaman, M.; Mane, J.Y.; Kapty, J.; Mercer, J.R.; Tuszynski, J.A. Entropic fragment-based approach to Aptamer Design. Chem. Biol. Drug Des. 2011, 78, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ashrafuzzaman, M.; Tseng, C.-Y.; Kapty, J.; Mercer, J.R.; Tuszynski, J.A. A computationally designed DNA aptamer template with specific binding to phosphatidylserine. Nucleic Acid Ther. 2013, 23, 418–426. [Google Scholar] [CrossRef]
- Ashrafuzzaman, M. Energy-Based Method for Drug Design. U.S. Patent No. US10916330B1, 9 February 2021. Available online: https://patents.google.com/patent/US10916330B1/en (accessed on 25 December 2021).
- Ashrafuzzaman, M.; Tuszynski, J. Membrane Biophysics; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-3-642-16105-6. [Google Scholar]
- Grunicke, H.H. The cell membrane as a target for cancer chemotherapy. Eur. J. Cancer Clin. Oncol. 1991, 27, 281–284. [Google Scholar] [CrossRef]
- Yuan, J.; Peng, R.; Su, D.; Zhang, X.; Zhao, H.; Zhuang, X.; Chen, M.; Zhang, X.; Yuan, L. Cell membranes targeted unimolecular prodrug for programmatic photodynamic-chemo therapy. Theranostics 2021, 11, 3502–3511. [Google Scholar] [CrossRef]
- Ashrafuzzaman, M.; Tseng, C.-Y.; Duszyk, M.; Tuszynski, J.A. Chemotherapy drugs form ion pores in membranes due to physical interactions with lipids. Chem. Biol. Drug Des. 2012, 80, 992–1002. [Google Scholar] [CrossRef]
- Ashrafuzzaman, M.; Tseng, C.-Y.; Tuszynski, J.A. Charge-based interactions of antimicrobial peptides and general drugs with lipid bilayers. J. Mol. Graph. Model. 2020, 95, 107502. [Google Scholar] [CrossRef]
- Ashrafuzzaman, M.; Khan, Z.; Alqarni, A.; Alanazi, M.; Alam, M.S. Cell surface binding and lipid interactions behind chemotherapy-drug-induced ion pore formation in membranes. Membranes 2021, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Gajate, C.; Mollinedo, F. Lipid rafts and FAS/CD95 signaling in cancer chemotherapy. Recent Pat. Anti-Cancer Drug Discov. 2011, 6, 274–283. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. Thematic review series: Glycerolipids. phosphatidylserine and phosphatidylethanolamine in mammalian cells: Two metabolically related aminophospholipids. J. Lipid Res. 2008, 49, 1377–1387. [Google Scholar] [CrossRef]
- Vance, J.E.; Tasseva, G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2013, 1831, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Adayev, T.; Estephan, R.; Meserole, S.; Mazza, B.; Yurkow, E.J.; Banerjee, P. Externalization of phosphatidylserine may not be an early signal of apoptosis in neuronal cells, but only the phosphatidylserine-displaying apoptotic cells are phagocytosed by Microglia. J. Neurochem. 2002, 71, 1854–1864. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Smrž, D.; Lebduška, P.; Dráberová, L.; Korb, J.; Dráber, P. Engagement of phospholipid scramblase 1 in activated cells. J. Biol. Chem. 2008, 283, 10904–10918. [Google Scholar] [CrossRef]
- Blankenberg, F.G. Imaging the molecular signatures of apoptosis and injury with radiolabeled annexin V. Proc. Am. Thorac. Soc. 2009, 6, 469–476. [Google Scholar] [CrossRef]
- Weinberg, R.A. The Biology of Cancer; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Zhang, G.; Gurtu, V.; Kain, S.R.; Yan, G. Early detection of apoptosis using a fluorescent conjugate of annexin V. BioTechniques 1997, 23, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; Reutelingsperger, C.P.; McGahon, A.J.; Rader, J.A.; van Schie, R.C.; LaFace, D.M.; Green, D.R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: Inhibition by overexpression of bcl-2 and Abl. J. Exp. Med. 1995, 182, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Boersma, H.H.; Kietselaer, B.L.; Stolk, L.M.; Bennaghmouch, A.; Hofstra, L.; Narula, J.; Heidendal, G.A.; Reutelingsperger, C.P. Past, present, and future of annexin A5: From protein discovery to clinical applications. J. Nucl. Med. 2005, 46, 2035–2050. [Google Scholar]
- Lundbæk, J.A.; Birn, P.; Tape, S.E.; Toombes, G.E.; Søgaard, R.; Koeppe, R.E.; Gruner, S.M.; Hansen, A.J.; Andersen, O.S. Capsaicin regulates voltage-dependent sodium channels by altering lipid bilayer elasticity. Mol. Pharmacol. 2005, 68, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Greisen, P.; Lum, K.; Ashrafuzzaman, M.; Greathouse, D.V.; Andersen, O.S.; Lundbaek, J.A. Linear Rate-equilibrium relations arising from Ion Channel-bilayer energetic coupling. Proc. Natl. Acad. Sci. USA 2011, 108, 12717–12722. [Google Scholar] [CrossRef]
- Ashrafuzzaman, M. Amphiphiles capsaicin and Triton X-100 regulate the chemotherapy drug colchicine’s membrane adsorption and ion pore formation potency. Saudi J. Biol. Sci. 2021, 28, 3100–3109. [Google Scholar] [CrossRef] [PubMed]
- Lundbaek, J.A.; Andersen, O.S. Amphiphile-regulation of ion channel function by changes in a transferable lipid bilayer spring constant. Biophys. J. 2010, 98, 482a. [Google Scholar] [CrossRef][Green Version]
- Lundbaek, J.A.; Koeppe, R.E.; Andersen, O.S. Amphiphile regulation of ion channel function by changes in the bilayer spring constant. Proc. Natl. Acad. Sci. USA 2010, 107, 15427–15430. [Google Scholar] [CrossRef]
- Smrž, D.; Dráberová, L.; Dráber, P. Non-apoptotic phosphatidylserine externalization induced by engagement of glycosylphosphatidylinositol-anchored proteins. J. Biol. Chem. 2007, 282, 10487–10497. [Google Scholar] [CrossRef]
- Martin, S.; Pombo, I.; Poncet, P.; David, B.; Arock, M.; Blank, U. Immunologic stimulation of mast cells leads to the reversible exposure of phosphatidylserine in the absence of apoptosis. Int. Arch. Allergy Immunol. 2000, 123, 249–258. [Google Scholar] [CrossRef]
- Bevers, E.M.; Comfurius, P.; Dekkers, D.W.C.; Zwaal, R.F.A. Lipid translocation across the plasma membrane of mammalian cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 1999, 1439, 317–330. [Google Scholar] [CrossRef]
- Ashrafuzzaman, M.; Tseng, C.-Y. Method for Direct Detection of Lipid Binding Agents in Membrane. U.S. Patent No. US9529006B1, 27 December 2016. Available online: https://patents.google.com/patent/US9529006B1/en (accessed on 25 December 2021).
- Chen, X.-M.; Liu, J.; Wang, T.; Shang, J. Colchicine-induced apoptosis in human normal liver L-02 cells by mitochondrial mediated pathways. Toxicol. In Vitro 2012, 26, 649–655. [Google Scholar] [CrossRef]
- Huang, Z.H.E.N.; Xu, Y.E.; Peng, W.E.I. Colchicine induces apoptosis in HT-29 human colon cancer cells via the Akt and c-jun N-terminal kinase signaling pathways. Mol. Med. Rep. 2015, 12, 5939–5944. [Google Scholar] [CrossRef] [PubMed]
- Cory, A.H.; Owen, T.C.; Barltrop, J.A.; Cory, J.G. Use of an aqueous soluble tetrazolium/Formazan assay for cell growth assays in culture. Cancer Commun. 1991, 3, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Bratton, D.L.; Fadok, V.A.; Richter, D.A.; Kailey, J.M.; Guthrie, L.A.; Henson, P.M. Appearance of phosphatidylserine on apoptotic cells requires calcium-mediated nonspecific flip-flop and is enhanced by loss of the aminophospholipid translocase. J. Biol. Chem. 1997, 272, 26159–26165. [Google Scholar] [CrossRef]
- Elliott, J.I.; Sardini, A.; Cooper, J.C.; Alexander, D.R.; Davanture, S.; Chimini, G.; Higgins, C.F. Phosphatidylserine exposure in B lymphocytes: A role for lipid packing. Blood 2006, 108, 1611–1617. [Google Scholar] [CrossRef]
- Mariño, G.; Kroemer, G. Mechanisms of apoptotic phosphatidylserine exposure. Cell Res. 2013, 23, 1247–1248. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashrafuzzaman, M.; AlMansour, H.A.M.; AlOtaibi, M.A.S.; Khan, Z.; Shaik, G.M. Lipid Specific Membrane Interaction of Aptamers and Cytotoxicity. Membranes 2022, 12, 37. https://doi.org/10.3390/membranes12010037
Ashrafuzzaman M, AlMansour HAM, AlOtaibi MAS, Khan Z, Shaik GM. Lipid Specific Membrane Interaction of Aptamers and Cytotoxicity. Membranes. 2022; 12(1):37. https://doi.org/10.3390/membranes12010037
Chicago/Turabian StyleAshrafuzzaman, Md., Hanouf A. M. AlMansour, Maha A. S. AlOtaibi, Zahid Khan, and Gouse M. Shaik. 2022. "Lipid Specific Membrane Interaction of Aptamers and Cytotoxicity" Membranes 12, no. 1: 37. https://doi.org/10.3390/membranes12010037
APA StyleAshrafuzzaman, M., AlMansour, H. A. M., AlOtaibi, M. A. S., Khan, Z., & Shaik, G. M. (2022). Lipid Specific Membrane Interaction of Aptamers and Cytotoxicity. Membranes, 12(1), 37. https://doi.org/10.3390/membranes12010037






