Glucoregulatory and Anti-Inflammatory Activities of Peptide Fractions Separated by Electrodialysis with Ultrafiltration Membranes from Salmon Protein Hydrolysate and Identification of Four Novel Glucoregulatory Peptides
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Electrodialysis Cell
2.1.1. Hydrolysate Preparation
2.1.2. Chemicals
2.1.3. Peptide Synthesis
2.1.4. Membranes
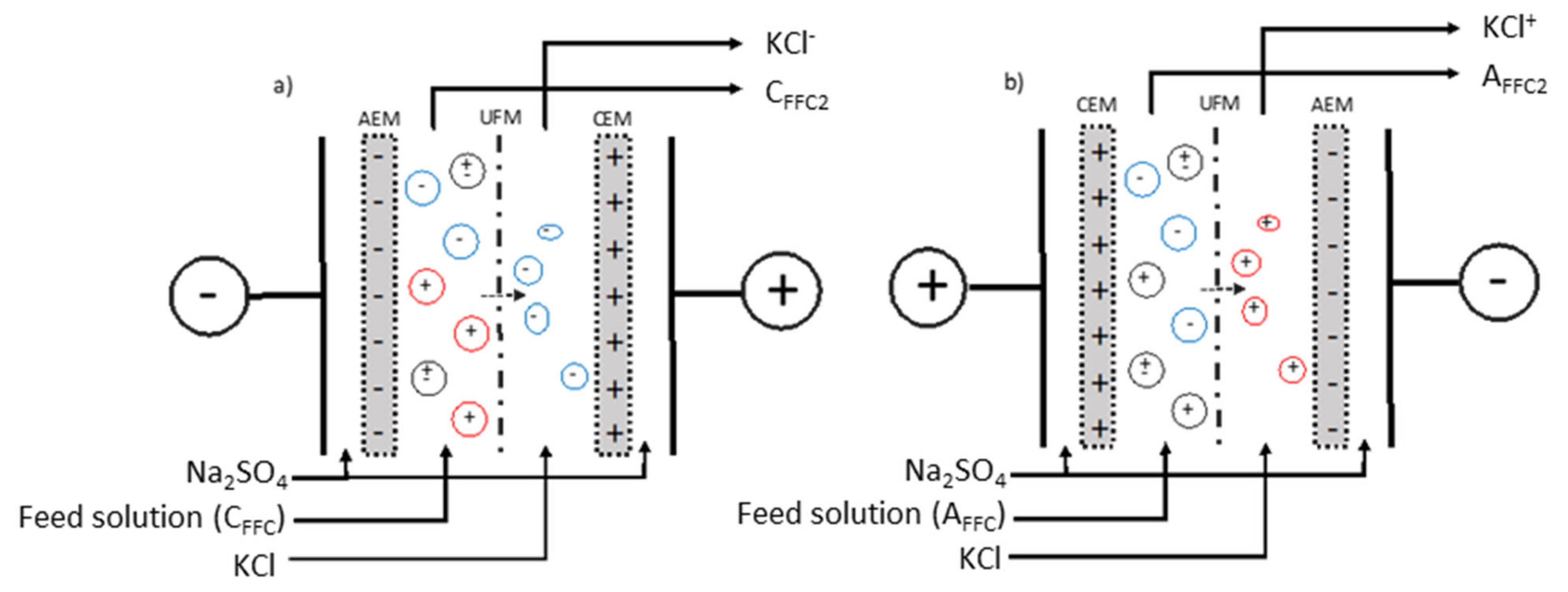
2.1.5. Electrodialysis Configurations
2.1.6. Electroseparation Protocol
2.2. In-Vitro Experiments and Analyses
2.2.1. Glucose Uptake Experiments
2.2.2. Hepatic Glucose Production Experiments
2.2.3. Anti-Inflammatory Experiments
2.2.4. Total Peptide Concentration in Dry Samples
2.2.5. RP-UPLC and Mass Spectrometry Analyses
2.2.6. Statistical Analyses
3. Results and Discussion
3.1. Effects of EDUF Fractions
3.1.1. Glucose Uptake
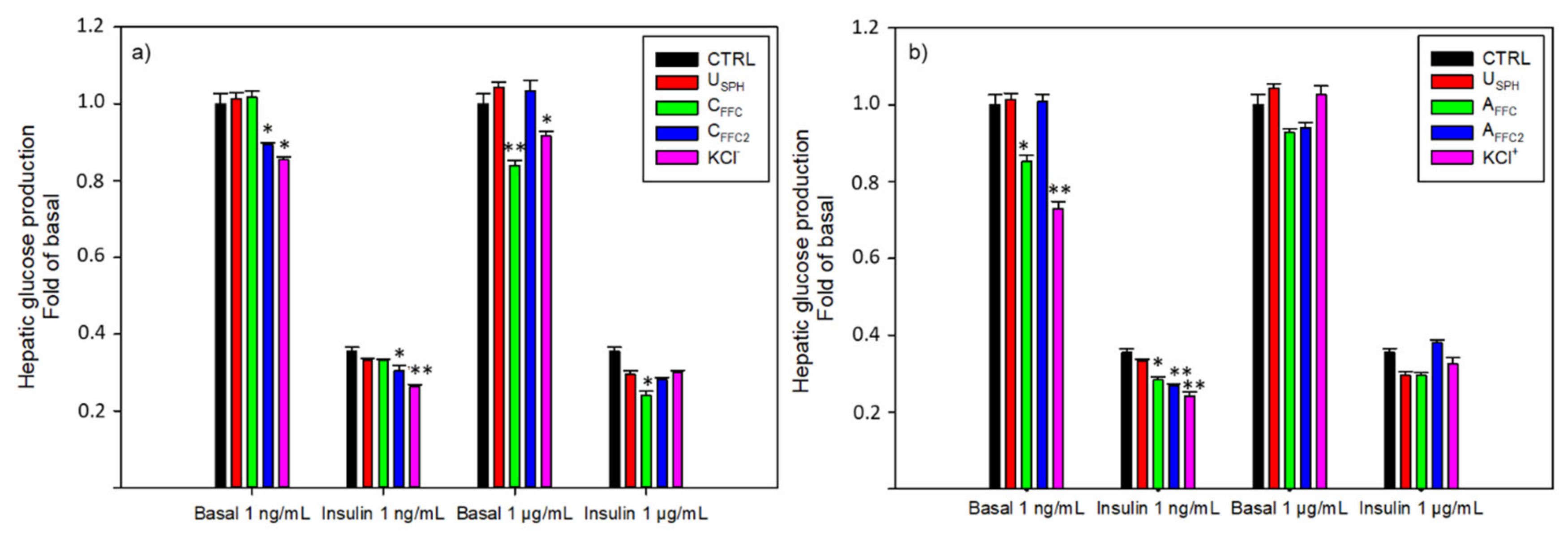
3.1.2. Hepatic Glucose Production
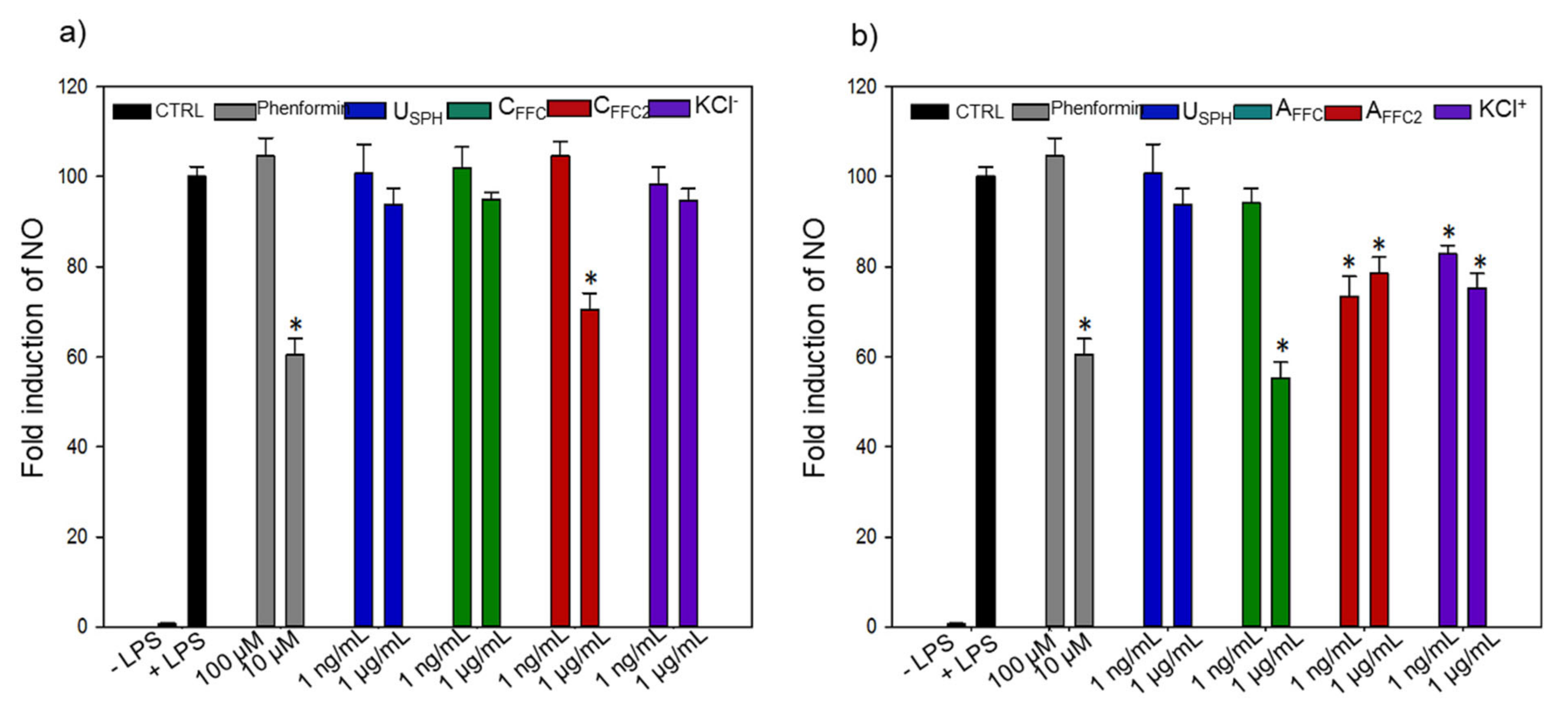
3.1.3. Inflammation
3.2. Peptide Sequence Identification and In-Vitro Experiments of Individual Synthesized Peptides
3.2.1. Peptide Sequence Identification
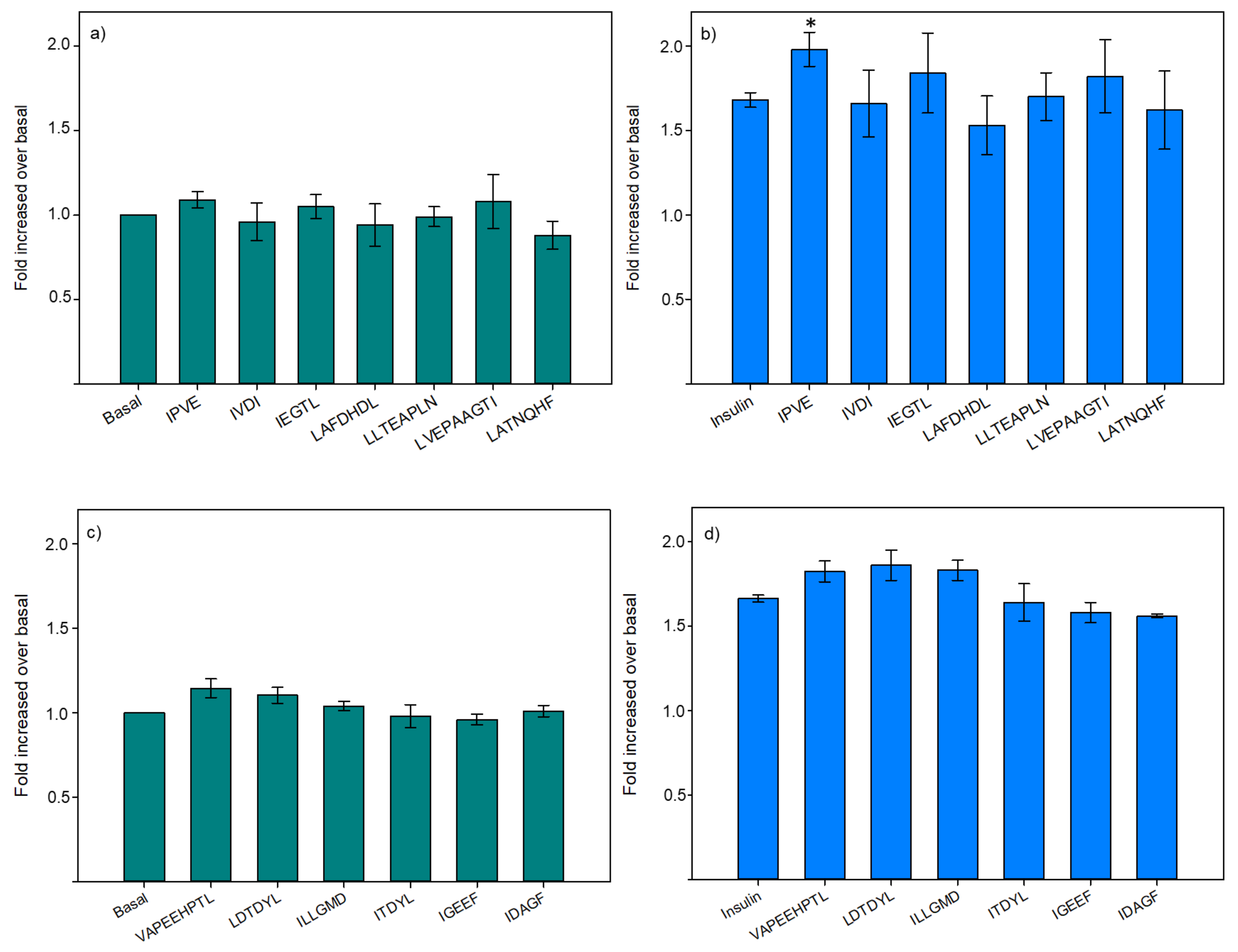
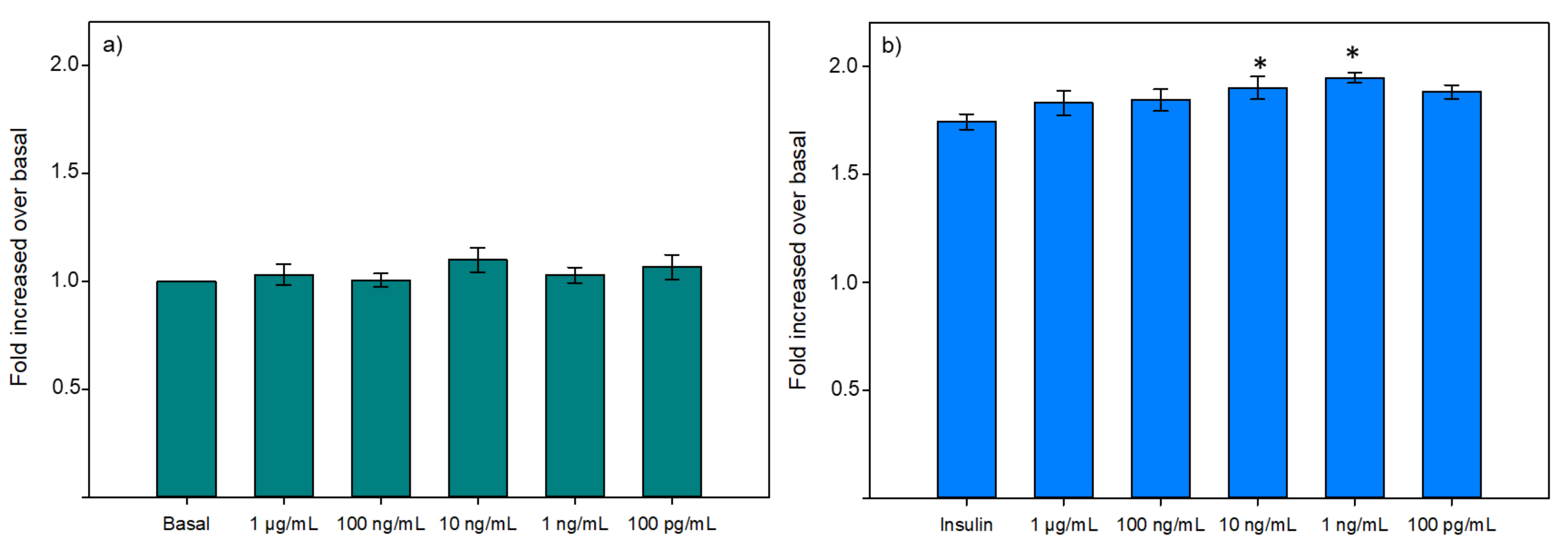
3.2.2. Glucose Uptake
3.2.3. Hepatic Glucose Production
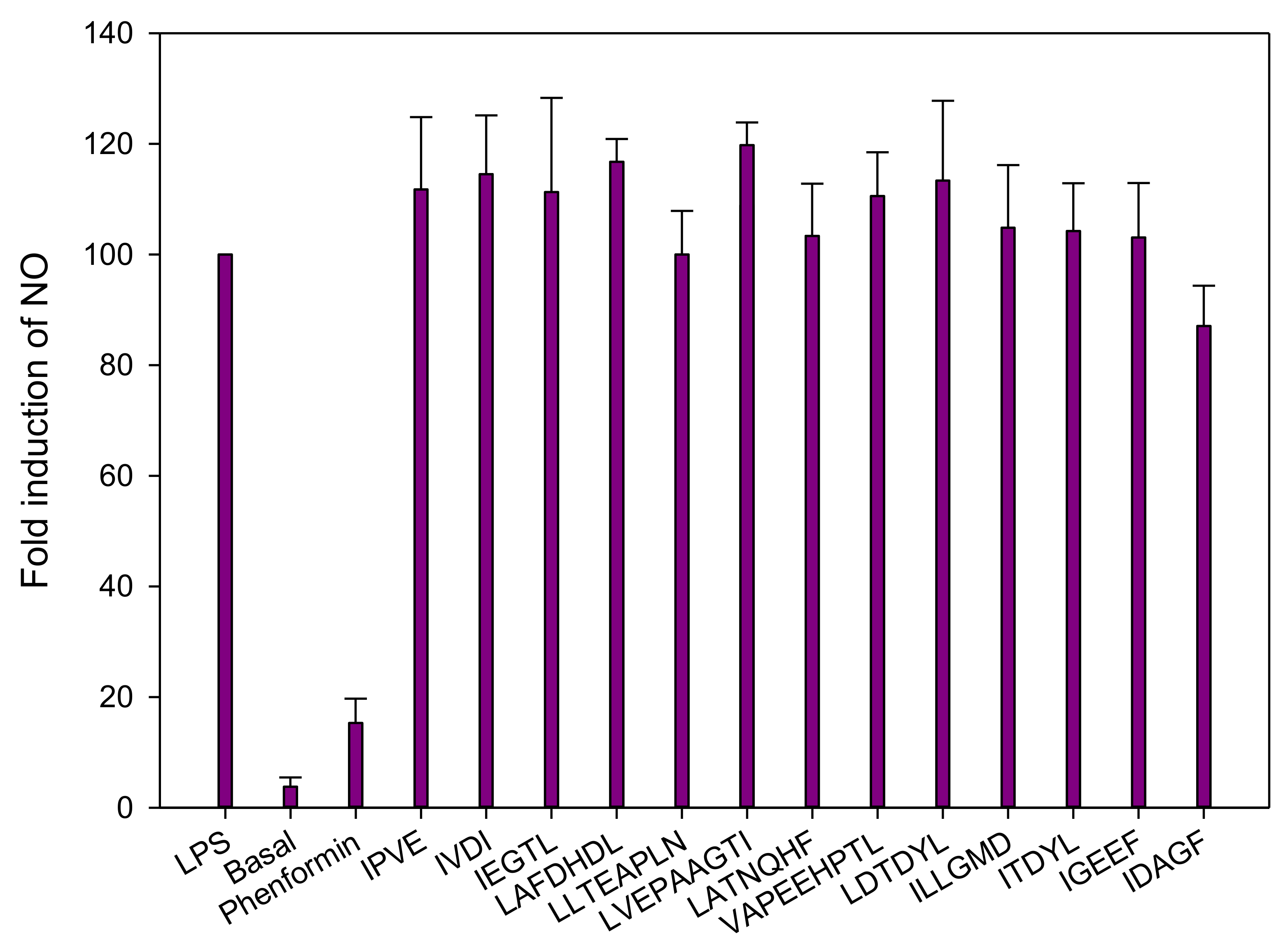
3.2.4. Inflammation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prato, S.; Bianchi, C.; Daniele, G. Abnormalities of Insulin Secretion and β-Cell Defects in Type 2 Diabetes. In Textbook of Diabetes, 5th ed.; John Wiley & Sons Ltd.: Chichester, UK, 2016; pp. 161–173, Chapter 12, Part 3. [Google Scholar]
- Marteau, T.M.; Hollands, G.J.; Fletcher, P.C. Changing human behavior to prevent disease: The importance of targeting automatic processes. Science 2012, 337, 1492–1495. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Action Plan for the Prevention and Control of Noncommunicable Diseases 2013–2020; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- International Diabetes Federation. Atlas du Diabète de la FID; International Diabetes Federation: Brussels, Belgium, 2019. [Google Scholar]
- Davì, G.; Santilli, F.; Patrono, C. Nutraceuticals in diabetes and metabolic syndrome. Cardiovasc. Ther. 2010, 28, 216–226. [Google Scholar] [CrossRef]
- Daviglus, M.L.; Stamler, J.; Orencia, A.J.; Dyer, A.R.; Liu, K.; Greenland, P.; Walsh, M.K.; Morris, D.; Shekelle, R.B. Fish consumption and the 30-year risk of fatal myocardial infarction. N. Engl. J. Med. 1997, 336, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Nkondjock, A.; Receveur, O. Fish-seafood consumption, obesity, and risk of type 2 diabetes: An ecological study. Diabetes Metab. 2003, 29, 635–642. [Google Scholar] [CrossRef]
- Friedberg, C.E.; Janssen, M.J.; Heine, R.J.; Grobbee, D.E. Fish oil and glycemic control in diabetes: A meta-analysis. Diabetes Care 1998, 21, 494–500. [Google Scholar] [CrossRef]
- Feskens, E.J.; Bowles, C.H.; Kromhout, D. Inverse association between fish intake and risk of glucose intolerance in normoglycemic elderly men and women. Diabetes Care 1991, 14, 935–941. [Google Scholar] [CrossRef]
- Tremblay, F.; Lavigne, C.; Jacques, H.; Marette, A. Dietary cod protein restores insulin-induced activation of phosphatidylinositol 3-kinase/Akt and GLUT4 translocation to the T-tubules in skeletal muscle of high-fat-fed obese rats. Diabetes 2003, 52, 29–37. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tremblay, F.; Jacques, H.; Marette, A. Modulation of insulin action by dietary proteins and amino acids: Role of the mammalian target of rapamycin nutrient sensing pathway. Curr. Opin. Clin. Nutr. Metab. Care 2005, 8, 457–462. [Google Scholar] [CrossRef]
- Lavigne, C.; Tremblay, F.; Asselin, G.; Jacques, H.; Marette, A. Prevention of skeletal muscle insulin resistance by dietary cod protein in high fat-fed rats. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E62–E71. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, F.; Lavigne, C.; Jacques, H.; Marette, A. Role of dietary proteins and amino acids in the pathogenesis of insulin resistance. Annu. Rev. Nutr. 2007, 27, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, V.; Marois, J.; Weisnagel, S.J.; Jacques, H. Dietary cod protein improves insulin sensitivity in insulin-resistant men and women: A randomized controlled trial. Diabetes Care 2007, 30, 2816–2821. [Google Scholar] [CrossRef]
- Ouellet, V.; Weisnagel, S.J.; Marois, J.; Bergeron, J.; Julien, P.; Gougeon, R.; Tchernof, A.; Holub, B.J.; Jacques, H. Dietary cod protein reduces plasma C-reactive protein in insulin-resistant men and women. J. Nutr. 2008, 138, 2386–2391. [Google Scholar] [CrossRef]
- Pilon, G.; Ruzzin, J.; Rioux, L.-E.; Lavigne, C.; White, P.J.; Frøyland, L.; Jacques, H.; Bryl, P.; Beaulieu, L.; Marette, A. Differential effects of various fish proteins in altering body weight, adiposity, inflammatory status, and insulin sensitivity in high-fat–fed rats. Metabolism 2011, 60, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Chevrier, G.; Mitchell, P.L.; Rioux, L.-E.; Hasan, F.; Jin, T.; Roblet, C.R.; Doyen, A.; Pilon, G.; St-Pierre, P.; Lavigne, C.; et al. Low-molecular-weight peptides from salmon protein prevent obesity-linked glucose intolerance, inflammation, and dyslipidemia in LDLR−/−/ApoB100/100 mice. J. Nutr. 2015, 145, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Danquah, M.K.; Agyei, D. Pharmaceutical applications of bioactive peptides. OA Biotechnol. 2012, 1, 1–7. [Google Scholar] [CrossRef]
- Li-Chan, E.C.Y. Bioactive peptides and protein hydrolysates: Research trends and challenges for application as nutraceuticals and functional food ingredients. Curr. Opin. Food Sci. 2015, 1, 28–37. [Google Scholar] [CrossRef]
- del Mar Contreras, M.; Lpez-Expsito, I.; Hernndez-Ledesma, B.; Ramos, M.; Recio, I. Application of mass spectrometry to the characterization and quantification of food-derived bioactive peptides. J. AOAC Int. 2008, 91, 981–994. [Google Scholar] [CrossRef]
- Henaux, L.; Thibodeau, J.; Pilon, G.; Gill, T.; Marette, A.; Bazinet, L. How Charge and Triple Size-Selective Membrane Separation of Peptides from Salmon Protein Hydrolysate Orientate their Biological Response on Glucose Uptake. Int. J. Mol. Sci. 2019, 20, 1939. [Google Scholar] [CrossRef]
- Roden, M.; Petersen, K.; Shulman, G. Insulin resistance in type 2 diabetes. In Textbook of Diabetes; John Wiley & Sons, Ltd. Publishers: Hoboken, NJ, USA, 2017; pp. 174–186. [Google Scholar]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef]
- Hammami, R.; Bédard, F.; Gomaa, A.; Subirade, M.; Biron, E.; Fliss, I. Lasso-inspired peptides with distinct antibacterial mechanisms. Amino Acids 2015, 47, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Bollhagen, R.; Schmidberger, M.; Barlos, K.; Grelle, E. A new reagent for the cleavage of fully protected peptides synthesised on 2-chlorotrityl chloride resin. J. Chem. Soc. Chem. Commun. 1994, 22, 2559–2560. [Google Scholar] [CrossRef]
- Suwal, S.; Roblet, C.; Amiot, J.; Bazinet, L. Presence of free amino acids in protein hydrolysate during electroseparation of peptides: Impact on system efficiency and membrane physicochemical properties. Sep. Purif. Technol. 2015, 147, 227–236. [Google Scholar] [CrossRef]
- Roblet, C.; Doyen, A.; Amiot, J.; Pilon, G.; Marette, A.; Bazinet, L. Enhancement of glucose uptake in muscular cell by soybean charged peptides isolated by electrodialysis with ultrafiltration membranes (EDUF): Activation of the AMPK pathway. Food Chem. 2014, 147, 124–130. [Google Scholar] [CrossRef]
- Tremblay, F.; Marette, A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway—A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J. Biol. Chem. 2001, 276, 38052–38060. [Google Scholar] [CrossRef] [PubMed]
- Durand, R.; Fraboulet, E.; Marette, A.; Bazinet, L. Simultaneous double cationic and anionic molecule separation from herring milt hydrolysate and impact on resulting fraction bioactivities. Sep. Purif. Technol. 2019, 210, 431–441. [Google Scholar] [CrossRef]
- Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z. The metabolic syndrome. Lancet 2005, 365, 1415–1428. [Google Scholar] [CrossRef]
- Martínez-Maqueda, D.; Miralles, B.; Recio, I.; Hernández-Ledesma, B. Antihypertensive peptides from food proteins: A review. Food Funct. 2012, 3, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Roblet, C.; Akhtar, M.J.; Mikhaylin, S.; Pilon, G.; Gill, T.; Marette, A.; Bazinet, L. Enhancement of glucose uptake in muscular cell by peptide fractions separated by electrodialysis with filtration membrane from salmon frame protein hydrolysate. J. Funct. Foods 2016, 22, 337–346. [Google Scholar] [CrossRef]
- Durand, R.; Pellerin, G.; Thibodeau, J.; Fraboulet, E.; Marette, A.; Bazinet, L. Screening for metabolic syndrome application of a herring by-product hydrolysate after its separation by electrodialysis with ultrafiltration membrane and identification of novel anti-inflammatory peptides. Sep. Purif. Technol. 2020, 235, 116205. [Google Scholar] [CrossRef]
- Kadel, S.; Daigle, G.; Thibodeau, J.; Perreault, V.; Pellerin, G.; Lainé, C.; Bazinet, L. How physicochemical properties of filtration membranes impact peptide migration and selectivity during electrodialysis with filtration membranes: Development of predictive statistical models and understanding of mechanisms involved. J. Membr. Sci. 2021, 619, 118175. [Google Scholar] [CrossRef]
- Suwal, S.; Roblet, C.; Doyen, A.; Amiot, J.; Beaulieu, L.; Legault, J.; Bazinet, L. Electrodialytic separation of peptides from snow crab by-product hydrolysate: Effect of cell configuration on peptide selectivity and local electric field. Sep. Purif. Technol. 2014, 127, 29–38. [Google Scholar] [CrossRef]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/protein/?term=salmonidae (accessed on 11 July 2018).
- Agilent Technologies. Agilent G2721AA/G2733AA Spectrum Mill MS Proteomics Workbench: User Guide; Agilent Technologies: Santa Clara, CA, USA, 2012. [Google Scholar]
- Pangestuti, R.; Kim, S.-K. Bioactive peptide of marine origin for the prevention and treatment of non-communicable diseases. Mar. Drugs 2017, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Kleinert, M.; Liao, Y.-H.; Nelson, J.L.; Bernard, J.R.; Wang, W.; Ivy, J.L. An amino acid mixture enhances insulin-stimulated glucose uptake in isolated rat epitrochlearis muscle. J. Appl. Physiol. 2011, 111, 163–169. [Google Scholar] [CrossRef]
- Mîinea, C.P.; Sano, H.; Kane, S.; Sano, E.; Fukuda, M.; Peränen, J.; Lane, W.S.; Lienhard, G.E. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem. J. 2005, 391, 87–93. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A. Three peptides from soy glycinin modulate glucose metabolism in human hepatic HepG2 cells. Int. J. Mol. Sci. 2015, 16, 27362–27370. [Google Scholar] [CrossRef] [PubMed]
- Turban, S.; Stretton, C.; Drouin, O.; Green, C.J.; Watson, M.L.; Gray, A.; Ross, F.; Lantier, L.; Viollet, B.; Hardie, D.G. Defining the contribution of AMP-activated protein kinase (AMPK) and protein kinase C (PKC) in regulation of glucose uptake by metformin in skeletal muscle cells. J. Biol. Chem. 2012, 287, 20088–20099. [Google Scholar] [CrossRef]
- Brunmair, B.; Staniek, K.; Gras, F.; Scharf, N.; Althaym, A.; Clara, R.; Roden, M.; Gnaiger, E.; Nohl, H.; Waldhäusl, W. Thiazolidinediones, like metformin, inhibit respiratory complex I: A common mechanism contributing to their antidiabetic actions? Diabetes 2004, 53, 1052–1059. [Google Scholar] [CrossRef]
- Peyrollier, K.; Hajduch, E.; Blair, A.S.; Hyde, R.; Hundal, H.S. L-leucine availability regulates phosphatidylinositol 3-kinase, p70 S6 kinase and glycogen synthase kinase-3 activity in L6 muscle cells: Evidence for the involvement of the mammalian target of rapamycin (mTOR) pathway in the L-leucine-induced up-regulation of system A amino acid transport. Biochem. J. 2000, 350, 361–368. [Google Scholar] [PubMed]
- Morifuji, M.; Koga, J.; Kawanaka, K.; Higuchi, M. Branched-chain amino acid-containing dipeptides, identified from whey protein hydrolysates, stimulate glucose uptake rate in L6 myotubes and isolated skeletal muscles. J. Nutr. Sci. Vitaminol. 2009, 55, 81–86. [Google Scholar] [CrossRef]
- Schmitz-Peiffer, C.; Biden, T.J. Protein kinase C function in muscle, liver, and β-cells and its therapeutic implications for type 2 diabetes. Diabetes 2008, 57, 1774–1783. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Chandrasekara, A. Millet grain phenolics and their role in disease risk reduction and health promotion: A review. J. Funct. Foods 2013, 5, 570–581. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Darewicz, M.; Dziuba, B.; Minkiewicz, P.; Dziuba, J. The preventive potential of milk and colostrum proteins and protein fragments. Food Rev. Int. 2011, 27, 357–388. [Google Scholar] [CrossRef]
- Pauletti, G.M.; Gangwar, S.; Knipp, G.T.; Nerurkar, M.M.; Okumu, F.W.; Tamura, K.; Siahaan, T.J.; Borchardt, R.T. Structural requirements for intestinal absorption of peptide drugs. J. Control. Release 1996, 41, 3–17. [Google Scholar] [CrossRef]
- Witt, K.A.; Davis, T.P. CNS drug delivery: Opioid peptides and the blood-brain barrier. AAPS J. 2006, 8, E76–E88. [Google Scholar] [CrossRef] [PubMed]
- Salamat-Miller, N.; Johnston, T.P. Current strategies used to enhance the paracellular transport of therapeutic polypeptides across the intestinal epithelium. Int. J. Pharm. 2005, 294, 201–216. [Google Scholar] [CrossRef]


| Sequences Identified | Molecular Weight (Da) | Retention Time (min) | Net Charge (at pH 6) | pI | Scores * | % SPI ** |
|---|---|---|---|---|---|---|
| IPVE | 456.2581 | 10.174 | - | 4.60 | 7.97 | 73.1 |
| IVDI | 458.2738 | 22.342 | - | 3.80 | 6.6 | 78 |
| IEGTL | 531.2890 | 17.270 | - | 4.00 | 9.93 | 77.3 |
| LAFDHDL | 829.3972 | 24.588 | - | 4.19 | 10.19 | 79.3 |
| LATNQHF | 829.3972 | 24.588 | + | 6.74 | 11.26 | 80,8 |
| LVEPAAGTI | 869.4857 | 18.700 | - | 3.99 | 7.74 | 77.9 |
| LLTEAPLN | 869.4857 | 18.700 | - | 3.99 | 8.93 | 79.2 |
| VAPEEHPTL | 991.4970 | 15.796 | - | 4.50 | 17.53 | 93.7 |
| LDTDYL | 738.3424 | 40.044 | - | 3.56 | 8.26 | 81.3 |
| ILLGMD | 660.3504 | 29.454 | - | 4.30 | 10.63 | 81.3 |
| ITDYL | 623.3160 | 25.851 | - | 3.80 | 11.59 | 89.1 |
| IGEEF | 593.2770 | 20.050 | - | 3.79 | 9.05 | 77.8 |
| IDAGF | 521.2483 | 27.307 | - | 3.79 | 7.83 | 81.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henaux, L.; Pereira, K.D.; Thibodeau, J.; Pilon, G.; Gill, T.; Marette, A.; Bazinet, L. Glucoregulatory and Anti-Inflammatory Activities of Peptide Fractions Separated by Electrodialysis with Ultrafiltration Membranes from Salmon Protein Hydrolysate and Identification of Four Novel Glucoregulatory Peptides. Membranes 2021, 11, 528. https://doi.org/10.3390/membranes11070528
Henaux L, Pereira KD, Thibodeau J, Pilon G, Gill T, Marette A, Bazinet L. Glucoregulatory and Anti-Inflammatory Activities of Peptide Fractions Separated by Electrodialysis with Ultrafiltration Membranes from Salmon Protein Hydrolysate and Identification of Four Novel Glucoregulatory Peptides. Membranes. 2021; 11(7):528. https://doi.org/10.3390/membranes11070528
Chicago/Turabian StyleHenaux, Loïc, Karina Danielle Pereira, Jacinthe Thibodeau, Geneviève Pilon, Tom Gill, André Marette, and Laurent Bazinet. 2021. "Glucoregulatory and Anti-Inflammatory Activities of Peptide Fractions Separated by Electrodialysis with Ultrafiltration Membranes from Salmon Protein Hydrolysate and Identification of Four Novel Glucoregulatory Peptides" Membranes 11, no. 7: 528. https://doi.org/10.3390/membranes11070528
APA StyleHenaux, L., Pereira, K. D., Thibodeau, J., Pilon, G., Gill, T., Marette, A., & Bazinet, L. (2021). Glucoregulatory and Anti-Inflammatory Activities of Peptide Fractions Separated by Electrodialysis with Ultrafiltration Membranes from Salmon Protein Hydrolysate and Identification of Four Novel Glucoregulatory Peptides. Membranes, 11(7), 528. https://doi.org/10.3390/membranes11070528








